Abstract
Evolution by natural selection is the conceptual foundation for nearly every branch of biology and increasingly also for biomedicine and medical research. In cancer biology, evolution explains how populations of cells in tumors change over time. It is a fundamental question whether this evolutionary process is driven primarily by natural selection and adaptation or by other evolutionary processes such as founder effects and drift. In cancer biology, as in organismal evolutionary biology, there is controversy about this question and also about the use of adaptation through natural selection as a guiding framework for research. In this review, we discuss the differences and similarities between evolution among somatic cells versus evolution among organisms. We review what is known about the parameters and rate of evolution in neoplasms, as well as evidence for adaptation. We conclude that adaptation is a useful framework that accurately explains the defining characteristics of cancer. Further, convergent evolution through natural selection provides the only satisfying explanation both for how a group of diverse pathologies have enough in common to usefully share the descriptive label of “cancer” and for why this convergent condition becomes life-threatening.
Cancers exhibit hallmark characteristics, but molecular variations indicate that perhaps, like snowflakes, no two cancers are alike. This apparent contradiction is resolved by understanding the underlying evolutionary processes.
Although evolutionary adaptation by somatic cells is different from adaptation by organisms, it is an important part of cancer biology and needs to be understood. One centrally important application lies in reconciling two major and contrasting patterns in cancer biology, which we refer to as trait “hallmarks” and molecular “snowflakes.” The first of these two patterns to be described was the striking consistency among different types of cancer at the level of cell traits. Although they originate from different tissues and cell types, virtually all cancers consist of cells with the same essential “hallmark” traits (Hanahan and Weinberg 2000, 2011). This quickly became one of the few organizing frameworks to impose order on the bewildering diversity of cancer. The superficially contrasting pattern that has since emerged from molecular analysis is that virtually any molecular category of cancers can, on closer inspection, be broken down into subcategories by looking for, and invariably finding, molecular variations. Indeed, molecular differences can usually be found between any two cases, leading to the impression that perhaps, like snowflakes, no two cancer cases are exactly alike (Kurzrock and Giles 2015).
The apparent contradiction between trait “hallmarks” and molecular “snowflakes” is resolved by understanding the underlying evolutionary and ecological process of cancer, as distinct from its static manifestations (Merlo et al. 2006; Pepper et al. 2014). A cancer is a dynamic population of abnormal somatic cells evolving through natural selection. In classical evolutionary biology, it is well understood that even when two such populations arise from different genetic backgrounds, and thus have few similarities at the molecular level, a shared ecological niche and its shared selective pressures can drive the process of evolutionary adaptation by natural selection toward the same traits, causing the two populations to converge onto those traits despite their different starting points and persistent molecular differences (Fig. 1).
Figure 1.
An example of convergent evolution in two species of cave fish descended from different ancestral populations. Amblyopsis rosae (top), and Astyanax mexicanus (bottom). Cave fish live in freshwater caves and have adapted to these specialized niches. Although these are different species, they have independently evolved similar phenotypes, such as loss of pigmentation and eyesight. (Images are under public domain, creative commons license.)
In evolutionary biology, there has long been controversy over the power and limitations of the “adaptationist program” as a way to generate hypotheses and guide research (Gould and Lewontin 1979; Mayr 1983). The adaptationist approach emphasizes that natural selection leads populations to evolve functional adaptations. This is contrasted with a null model of neutral evolution and an approach that emphasizes the importance of constraints and side effects in influencing the evolutionary trajectory (Gould and Lewontin 1979). A similar debate is now present in cancer biology, with some investigators arguing against “excessive adaptationism,” partly on the grounds that a few decades is probably not long enough for cancer cells to evolve all their observed complex traits (Arnal et al. 2015). This debate can be summarized by two questions: First, are cancers characterized by natural selection or by neutral evolution? And, second, does somatic evolution produce complex and novel adaptations in cancer cells or does it simply remove or activate cell functions that are already encoded in the constitutive human genome?
In fact, these are false dichotomies. The apparent alternatives are both true. There is good evidence for both neutral evolution and natural selection in neoplasms, and the relative importance of the two probably changes during progression. Neoplastic cells produce complex and novel adaptations, often by removing or activating cell functions that are already in the human genome, but also sometimes through genetic novelties.
IS A HUMAN LIFETIME LONG ENOUGH FOR SOMATIC CELLS TO EVOLVE COMPLEX ADAPTATIONS?
One of the arguments against applying an adaptationist framework to cancer cells is that the scope of resulting adaptation may be limited because the human lifetime is too short for complex cell adaptations to evolve de novo (Arnal et al. 2015). In this section, we examine the validity of this claim by reviewing the literature on the pace of evolutionary change in neoplasms. The rate of evolutionary change depends critically on parameters such as mutation rate, population size, and population turnover rate (inverse of cell generation time). Considering these factors suggests that somatic cell evolution can be much more rapid than evolution among multicellular organisms. Many cancer cells have the capacity to divide daily, evolve over a period of decades, and comprise populations numbering in the billions to trillions (Table 1). Based solely on population size and generation time, there are more reproductive events among the cells within one host individual than there have been among individuals in the entire history of the human species. Each such reproductive event is an opportunity for mutation and selective reproduction and, thus, for adaptive evolution.
Table 1.
Estimates and ranges for the parameters of somatic evolution in neoplasms
| Parameter | Estimates | References |
|---|---|---|
| Point mutation rate | 10−9–10−10 bp/cell division | Knudson 1971; Wang et al. 2002; Araten et al. 2005; Jones et al. 2008; Tomasetti et al. 2013 |
| Amplification and deletion rate | 10−4–10−5 gene/cell division | Tlsty et al. 1989 |
| Rate of loss of heterozygosity | 10−4–10−6 gene/cell division | de Nooij-van Dalen et al. 1998; Chan et al. 2001 |
| CpG methylation rate | 10−4 CpG/cell division | Chan et al. 2001; Sottoriva et al. 2013 |
| Total cell population size | 1 cm3 of solid tumor: 108 cells Multiple myeloma: 1012 Breast cancer: 107–1011 Lung cancer: 108–1012 |
Del Monte 2009; Sullivan and Salmon 1972; Friberg and Mattson 1997 |
| Stem cell population sizea | Multiple myeloma: 107–1010 Colorectal cancer: frequency 10−2–10−3 |
Hamburger and Salmon 1977a,b; Ricci-Vitiani et al. 2007; Merlos-Suarez et al. 2011; Sottoriva et al. 2013 |
| Stem cell generation time | Colorectal cancer in vitro: ∼10 days Colonic stem cells in vivo: ∼7 days |
Potten et al. 2003; Ricci-Vitiani et al. 2007 |
| Clonal expansion rate | 1.6 cm2 per year in Barrett’s esophagus | Martinez et al. 2016 |
| Selective coefficients | Driver gene mean: 0.004 Probability of replacing a competitor cell: 62%–78% |
Bozic et al. 2010; Vermeulen et al. 2013 |
| Number of cell generations | Colorectal cancer: 103–104 | Yachida et al. 2010 |
| Mutational burden | Point mutations: 0.3–111/Mb 108 coding mutations across a tumor |
Berger et al. 2012; Kandoth et al. 2013 |
| Copy number alterations: Median 40 per genome Covering 33% of the genome |
Ling et al. 2015; Zack et al. 2013 | |
| Translocations/genome: Colorectal cancer: 0–10 Prostate cancer: 43–213 Breast cancer: 1–231 |
Cancer Genome Atlas 2012; Berger et al. 2011; Stephens et al. 2009 | |
| Time from initiation to diagnosis | Colorectal cancer: 15–65 years Pancreatic cancer: 50–60 years |
Meza et al. 2008; Yachida et al. 2010 |
These parameters should be relevant to the clinical outcomes of all types of cancer, and so be universal biomarkers, applicable to all cancers. Most of these estimates are imprecise and future studies are needed to quantify and refine the estimates of the parameters of somatic evolution.
aCalculated from experimental observations of the frequency of putative cancer stem cells.
Regarding a third crucial parameter of evolution, mutation rate, current estimates suggest that every base pair in the cancer genome is probably mutated in some cell, in every cell generation within a neoplasm (Table 1). This too suggests that somatic evolution can be rapid and that its molecular bases can be diverse, as the same gene may mutate independently multiple times within the same neoplasm. This appears to be true empirically (Anderson et al. 2011; Gerlinger et al. 2012, 2014; Kovac et al. 2015; Yates et al. 2015). In fact, the exact same mutation is seen to occur multiple times in the same neoplasm, causing multiple independent origins, or “homoplasy” (e.g., Shpak et al. 2015). This mutational redundancy also explains why there are likely to be multiple mechanisms of acquired resistance available for a response to the selection imposed by therapy (Murugaesu et al. 2015).
In neoplasms, the number of cell generations per unit time may be high and only loosely related to the size of the tumor, because of high levels of apoptosis and rapid cell turnover (Lowe and Lin 2000; Liu et al. 2001). To estimate number of cell generations, the total number of cell divisions necessary to generate a tumor's size must be integrated with the rate of cell death.
Another factor accelerating cellular evolution in neoplasms is that it can be driven not only by genetic, but also by epigenetic modifications. Epigenetic changes are subject to somatic natural selection because they are heritable across mitosis and can affect the fitness of the cell by changing its phenotype. The rate of epigenetic modifications is sometimes much higher than the rate of genetic mutations (Table 1) and so may be an important driver of evolution in neoplasms.
Taken together, all these considerations suggest that a human lifetime is sufficient for cells to evolve substantial adaptation. Large population sizes, short generation time, and high mutation rates all contribute to a rapid pace of evolution within neoplasms. In the framework of the adaptationism debate, many of the resulting complex traits can legitimately be viewed either as novel (in their current context) or as recapitulations of traits that are normally functional for the multicellular organism in different cell types or at different stages of development.
Also increasing the potential for cancer cell adaptation is the fact that cancer cells have access to a large toolbox of adaptations and traits from the human genome that are available for their use, alteration, and repurposing. For example, the normal conditional response of changing cell behavior and migrating away from regions of depleted resources is already built into the human genome. Hypoxia can trigger cell motility via HIF1α (Semenza 2012). A neoplastic cell need only activate and repurpose this pathway to acquire the cell motility behavior that is central to tissue invasion and metastasis (Chen et al. 2011; Aktipis et al. 2012). We suggest that the large size and complexity of the human genome imparts substantial evolvability through repurposing for cell-level fitness advantage.
Also related to the finite human life span, some investigators have mistakenly argued that the potential for adaptation is limited by the fact that most cancers will die with their host (Davies and Lineweaver 2011; Arnal et al. 2015). In fact, this does not preclude adaptations in a neoplasm during the lifetime of the host. Even if those cancer adaptations will cause the cancer’s extinction, that does not prevent the mechanics of natural selection or otherwise impinge on cancer adaptation. The idea that cancer cells or the mutated genes that determine their behavior are not involved in an evolutionary process because they are destined to rapid extinction because of the host’s death (caused by themselves) is demonstrably wrong. So-called “evolutionary suicide” through adaptive evolution is in fact a well-understood phenomenon with many examples (Ibrahim 2014). Adaptive cellular evolution is notoriously “shortsighted” regarding later consequences, including in particular harm to the host individual (Levin and Bull 1994).
IS ADAPTATION MORE OR LESS LIKELY IN CANCER CELLS BECAUSE OF DIFFERENCES BETWEEN ORGANISMAL AND SOMATIC EVOLUTION?
In addition to the finite life span of the host that is the “environment” of somatic cells, there are other important differences between somatic cellular versus organismal evolution, and understanding these differences can help us better answer the question of whether cancer cells can evolve adaptations via natural selection. Natural selection among multicellular organisms is typically a very slow process because of long generation times (compared with somatic cells) and small population sizes (compared with billions of cells in a growing neoplasm).
Factors that favor adaptation in multicellular organisms include their large diploid genomes and sexual reproduction. Sexual reproduction involves genomic recombination, which can contribute to genetic variation among individuals and remove deleterious mutations. In this regard, somatic cell evolution is more similar to evolution among single-cell organisms. Like somatic cells, many unicellular eukaryotes reproduce asexually and have large population sizes and short generation times. In contrast, somatic cells within a human host have huge diploid genomes that may allow them to evolve over shorter periods of time (15–60 years) (Meza et al. 2008; Yachida et al. 2010), despite the fact that there is currently little evidence of genomic recombination among cancer cells.
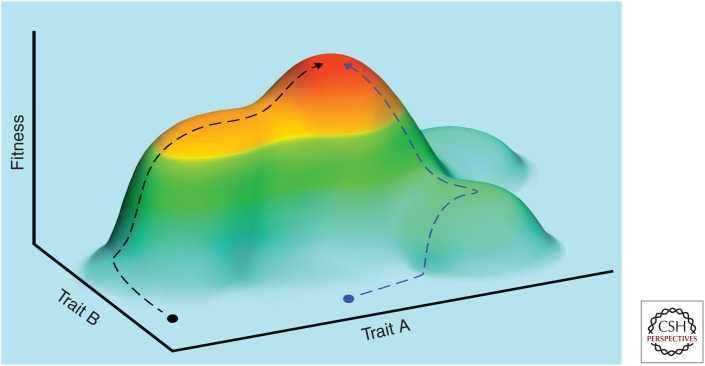
Another fundamental difference increasing the likelihood of adaptation by cancer cells lies in the initial state of the cell. Organisms have typically evolved to an adaptive fitness peak (as shown in Fig. 1), so that most random mutations are detrimental, especially loss-of-function mutations. In contrast, somatic cells start with many built-in constraints on cell survival and reproduction from the legacy of selection among multicellular organisms. In the abnormal context of selection among somatic cells, this puts them in a fitness pit rather than on a peak on the adaptive landscape. Random (epi)-mutations in a neoplastic cell can easily be beneficial, at least early in carcinogenesis, and can allow the neoplastic cell to quickly climb out of its fitness pit. For example, loss-of-function mutations that break the evolved cellular machinery that normally suppresses cellular proliferation, or supports apoptosis, would provide immediate fitness benefit to a somatic cell. The same reasoning applies to most of the cancer “hallmark” traits. This difference between organismal and somatic evolution should lead to more rapid early accumulation of adaptations in somatic evolution than in organismal evolution.
To sum up, some of the differences between somatic evolution and organismal evolution make the evolution of complex adaptations less likely in cancer than in multicellular organisms, including limits on the total number of cell generations and the difficulty of exchanging genetic material between cell lineages. In contrast, other differences, such as the large genome size, large population size, and rapid turnover, make the evolution of complex adaptations in cancer more likely.
EVIDENCE FOR ADAPTATIONS IN CANCER
Evolutionary adaptation occurs in any situation in which the conditions for natural selection are met (Bell 1997, p. 25). This is true for free-living organisms and for cancer cells alike. Natural selection results from the combination of (1) trait variation in a reproducing population (e.g., of cells), (2) inheritance of that variation across (cell) generations, and (3) variation in (cell) fitness correlated with heritable trait variation. All three of these sufficient conditions are met in neoplastic cells (Merlo et al. 2006). Evolution of adaptation through natural selection follows as a logical necessity. This is a precise description of the process, not merely a metaphor (pace Arnal et al. 2015).
In organismal biology, there has long been controversy over the power and limitations of the “adaptationist program” of searching for Darwinian adaptation as a way to generate hypotheses and guide research (Gould and Lewontin 1979; Mayr 1983), and some of this has parallels in cancer biology (Arnal et al. 2015). Does somatic evolution produce complex novelties, or merely remove or activate cell functions that are already encoded in the constitutive human genome? In general, we have information on the mutations and alterations that appear during somatic evolution, but know very little about the tempo and dynamics of their evolution.
Some of the convergent hallmark types of cancer only require a simple loss of molecular function, whereas others are complex and probably require multiple adaptations or extensive repurposing. For example, metastasis is a complex suite of cell behaviors, requiring detachment from the primary tumor, tissue invasion, intravasation into the blood vessels or lymphatic system, survival in that new environment, attachment to a new location, extravasation out of the blood vessels, growth in the new environment of that tissue with a different hormonal and cytokine profile, and finally successful angiogenesis in that environment (Fidler 2003). Although the human genome includes the capacity to express all of those cell traits, their rediscovery and coordination may require selection for multiple genetic or epigenetic alterations.
Some of the clearest examples of adaptation in cancer come from acquired therapeutic resistance. All cytotoxic therapies and targeted therapies select for drug resistance (Pepper 2011, 2012, 2016). As with the hallmarks, the molecular changes underlying acquired resistance often differ between tumors (Gottesman 2002), but the convergent trait of resistance is consistent across many cancer types and therapeutic agents (e.g., Azam et al. 2003; Engelman et al. 2007; Murugaesu et al. 2015; reviewed by Pepper 2016).
Further evidence for evolutionary adaptation in cancer comes from the predictive value of cell genetic diversity, or “mosaicism,” in cancer progression. If there was no cell selection in neoplasms, if all somatic evolution was neutral, then the amount of diversity in a neoplasm would not be predictive of anything. It would only be informative of the past natural history of the neoplasm (Sottoriva et al. 2015; Williams et al. 2016). However, under somatic cell selection, diversity is critical, because it is the basis of fitness selection (Fernandez et al. 2016). Empirically, higher genetic diversity within neoplasms does in fact predict both faster progression to malignancy and reduced patient survival time (Merlo et al. 2006, 2010; Mroz et al. 2015; Andor et al. 2016). The current interpretation of this pattern is that genetic diversity correlates with the likelihood that an adaptive mutation will appear, driving neoplastic progression (Merlo et al. 2006; Merlo and Maley 2010).
Although neutral evolution can generate diversity and may produce cells with the potential for adaptation, those cells become relevant only when they undergo clonal expansion in response to selective pressures from the microenvironment. For example, neoplastic cell populations may adapt only if they contain enough diversity to meet the challenges of metastasis or of therapy (Nguyen et al. 2016).
It is a well-known phenomenon in organismal evolution that mutations that are initially neutral become functionally relevant when the environment changes (Kimura 1983). Similarly, in somatic cell evolution, diversity predicts response to selection, thus drives the evolution first of malignancy, and later of acquired resistance to therapy (Andor et al. 2016; Nguyen et al. 2016).
Recent molecular evidence for cancer cell adaptation comes from examination of spatial heterogeneity in breast cancers (Lloyd et al. 2016). Evolutionary theory predicted that spatial variation of phenotypes could result from local variations in environmental factors that select for different phenotypic properties. For example, regions of low blood flow, which are commonly observed in tumor imaging, would select for tumors that are adapted to such conditions as reduced availability of substrate and blood-derived growth factors. Detailed analysis of spatial molecular heterogeneity in 10 clinical breast cancers showed a consistent regional distribution meeting this prediction, which supported the adaptive hypothesis (Lloyd et al. 2016).
CONVERGENT CELLULAR EVOLUTION IS WHAT MAKES “CANCER” A MEANINGFUL (AND LETHAL) DISEASE CATEGORY
In biology, generally, some of the strongest evidence for adaptation comes from convergent evolution, in which different populations in similar environments evolve similar phenotypes as a result of similar selective pressures. Convergent evolution in cave fish is one classic example with recognized parallels to cancer cell evolution (Gatenby et al. 2011). Around the world, lakes inside caves hold hundreds of different populations of fish adapted to this specialized ecological niche by a consistent set of unusual traits we could call the “hallmarks of cave fish,” including loss of skin pigment and reduced eyesight (Fig. 2). Despite the trait similarities among these different species, genetic analysis reveals that each species evolved these “hallmarks of cave fish” independently, from genetically different founding populations, and that they still retain their genetic uniqueness. Such convergent trait evolution can only arise from a shared adaptive response to the same selective pressures, and such an adaptive response can only be driven by natural selection. Thus, convergent evolution in independent populations is a strong and reliable signature of adaptation through natural selection. This principle holds equally well both for populations of organisms and for populations of somatic cells becoming cancer in different hosts (Gatenby et al. 2011).
Figure 2.
Although tumors do not converge genetically, all typically converge on the same “hallmark” phenotypic traits. Here, we use a fitness landscape to illustrate convergent evolution of tumor phenotypes. The x- and y-axes (horizontal dimensions) represent two different quantitative traits (trait A and trait B), and the z-axis (vertical dimension) represents fitness. Fitness increases vertically and also scales in color from light blue (low fitness) to red (high fitness). We show two different neoplasms starting from different phenotypic states, depicted by the black and blue circles in the blue landscape valley. As neoplastic progression advances, these two tumors may follow different trajectories (indicated by the dotted lines) up the fitness peak. However, both tumors will converge on the same malignant phenotype (red fitness peak) showing the hallmarks of cancer.
Without convergent evolution, there would be no similarities between tumors from different tissues, and so there would be no reason for us to categorize them together as the disease(s) we call “cancer.” In fact, we see the same phenotypes evolving over and over again, in entirely different types of cancers, in different hosts. We call these phenotypes the “hallmarks of cancer” (Hanahan and Weinberg 2011). They include the generation of growth signals, insensitivity to antigrowth signals, stabilization of telomeres, which allows unlimited replication, suppression of apoptosis, neo-angiogenesis, evasion of immune predation, and tissue invasion leading to metastasis. These are cancer hallmarks precisely for the reason that they each increase the fitness (survival and reproduction) of neoplastic cells and so evolve consistently, even from different genetic and phenotypic starting points (Fig. 1). The ecological niche for all these cancers is that of an “endogenous parasite” (Charlton 1996), meaning parasitic cells derived from normal cells but now competing with and exploiting them. The similarity across all cancers of this niche and its selective pressures explains why all cancers tend to generate similar cell traits even though each cancer can be as genetically unique as a species of cave fish (Fig. 2).
Somatic evolutionary adaptation not only is what makes “cancer” a meaningful disease category but also what makes cancer a malignant and deadly disease regardless of what tissue it arises in. Neoplasms that begin as benign growths soon outgrow their local resources, after which the only neoplastic cells that survive and proliferate are those that can acquire resources by invading the surrounding normal tissues. This constitutes a strong selective pressure, with malignant tissue invasion as the adaptive response (Aktipis et al. 2012).
ADAPTATIONIST THINKING PUSHES THE FRONTIERS OF CANCER RESEARCH
Beyond merely explaining why cancer is deadly, and refractory to treatment, adaptationist perspectives are also guiding current medical advances. The central problem in cancer medicine is that, “cancer is continuously adapting” (Willyard 2016), so that initially promising new therapeutics almost invariably fail in the face of acquired therapeutic resistance (Pepper 2016). The problem is not the specific mutations, which are highly variable and heterogeneous. Rather, the problem is adaptive evolution, and recognizing this has led some clinicians to refocus on “the possibility to track and treat evolution” (Willyard 2016), by applying standard tools of evolutionary biology, such as phylogenetic reconstruction of ancestral lineages and evolutionary origins (Kostadinov et al. 2013; Jamal-Hanjani et al. 2014; Willyard 2016) or mathematical models of fitness optimization through evolutionary adaptation (Misale et al. 2015; Enriquez-Navas et al. 2016; Lloyd et al. 2016).
Rather than just providing metaphors for thinking about the problem, adaptationist theory also provides mathematical models, such as evolutionary optimization on fitness landscapes, that can translate into new therapeutic strategies, such as the adaptive therapy regimens that have recently succeeded in animal models by avoiding acquired drug resistance and thereby prolonging progression-free survival (Enriquez-Navas et al. 2016; Schmidt 2016). To succeed, the emerging trend toward precision oncology must incorporate insights from ecology and adaptive evolution (Lloyd et al. 2015).
DO CANCER CELLS EVOLVE NOVEL TRAITS?
One of the classic conceptual challenges to adaptationist thinking that has reappeared in cancer biology (Arnal et al. 2015) is the question of novelty, or the distinction between new adaptations versus repurposed “exaptations” that originally served a different function (Gould and Lewontin 1979). Many traits of cancer are novel for the cell types in which the cancer arises but are normally expressed in other cell types or at other stages of development, from the same normal genome. In neoplasms, genes and traits that originally functioned in multicellularity are often “hijacked,” or repurposed for competition among cells. For example, the seemingly novel cancer phenotypes of rapid proliferation, motility, and tissue invasion often result from activation of functions that are normally expressed only by embryonic stem cells and are built into the normal human genome for that context (Brewer et al. 2009; Chen et al. 2015).
Another dramatic example of a complex cancer cell behavior that appears novel in its repurposed context of cancer progression is cell cannibalism, or consuming and digesting other cells (Lugini et al. 2006; Melendez-Lazo et al. 2015). This cell behavior is normal only for lymphocytes and is presumably enabled by normal human genes that are abnormally expressed in cancer cells.
Many other traits that evolve in cancers also are novel only in their abnormal context, such as shifting to a glycolytic metabolism in the presence of plentiful oxygen (the Warburg effect). Normal cells can shift to a glycolytic metabolism under oxygen deprivation, but constitutive expression (the Warburg effect) is novel to cancer cells (Lunt and Vander Heiden 2011).
The mechanisms of abnormal gene expression behind trait repurposing in cancer are gradually being revealed. Recent efforts to characterize the genetic regulatory networks in cancer cells have found extensive rewiring of those networks, relative to normal cells (Li et al. 2015b). Similarly, in organismal evolution, important trait changes have also occurred through changes in the regulation of genes (Wray 2007). In cancer, most of the detailed mechanisms remain to be described, because most cancer sequencing so far has exclusively focused on changes in coding regions, for the simple reason that we do not yet have good ways to identify regulatory regions and predict the consequences of mutations in those regions.
ADAPTATION THROUGH GENETIC NOVELTIES IN CANCER
In addition to abnormal gene expression, novel phenotypes can be generated by genetic (or epigenetic) novelties. Various mechanisms lead to novel genetic constructs and adaptive phenotypes in neoplastic cells. Most point mutations that transform proto-oncogenes into oncogenes are gain-of-function mutations, typically changing a conditional proliferative signal into a constant proliferative signal. Similarly, point mutations can generate adaptations by preventing drugs from binding to their target proteins (Milojkovic and Apperley 2009).
Although the size of the essential genome of human cancer cells is unknown, a comparison between the normal human genome (>20,000 genes) and a unicellular eukaryotic species such as yeast (∼6300 genes) suggests the possibility of many nonessential genes in cancer cells. Those genes could constitute an extensive source of nonessential genes to evolve new functions, which could allow somatic evolution to generate novelties more quickly than in most unicellular evolution. In addition, genetic redundancy may allow cancer cells to tolerate deactivating deleterious genes, keeping active only those genes that benefit the fitness of a cell as a single-cell “endogenous parasite.” Taking into consideration the apparent proportion of nonessential genes in cancer, the proportion of random mutations that are advantageous to cancer cells is likely greater than the proportion of mutations that are advantageous in organismal evolution (Rajagopalan et al. 2003).
Many novel genetic constructs not present in normal cells have been described in cancer, including translocations, rearrangements, and gene fusions. The most famous of these is the BCR-ABL translocation in chronic myeloid leukemia, which fuses a promoter that is constitutively activated with a proliferative signal (Ren 2005). Such structural novelties often drive carcinogenesis (Kumar-Sinha et al. 2008; An et al. 2010; Greuber et al. 2013). An increase in retroposition rate in certain cancer types has been reported (Belancio et al. 2010, Helman et al. 2014; reviewed in Rodic and Burns 2013). This constitutes another potential source of novel genetic constructs, either mutating genes via insertion, changing the regulation of nearby genes, or generating gene duplications. Such a role in somatic cell evolution would parallel the known role of retroposition in organismal evolution (Xiao et al. 2008).
Evolution by gene duplication followed by divergence, or “neofunctionalization” (Ohno 1970) is a classical model that explains the generation of new genetic loci in species evolution. Although new models have arisen since then (reviewed in Conant and Wolfe 2008), all are based on two precepts: It is easier to generate new coding regions from preexisting ones; and relaxation of functional constraints is necessary to evolve new gene functions. Copy number amplification is common and relevant in somatic evolution, varying among cancer types (Zack et al. 2013), among tumors, and even within tumors (Gerlinger et al. 2014). Amplifications are even a predictor of prognosis in some cancers (Hieronymus et al. 2014; Li et al. 2015a). The possible role in somatic evolution of divergence after gene duplication will require further investigation.
Horizontal exchanges of genetic material or organelles may also contribute to the adaptive capacity of cancer cells. There is evidence that cancer cells can acquire new genes through fusion of cancer cells or fusion between cancer and normal cells (Duelli and Lazebnik 2007). In addition, neoplastic cells can incorporate fragments of DNA from neighbors that have undergone apoptosis (Holmgren et al. 1999; Bergsmedh et al. 2001). Cancer cells can also acquire or replace their mitochondria from normal cells (Tan et al. 2015).
Each of these sources of genetic novelty can support the evolution of novel traits.
NEUTRAL EVOLUTION IN CANCER
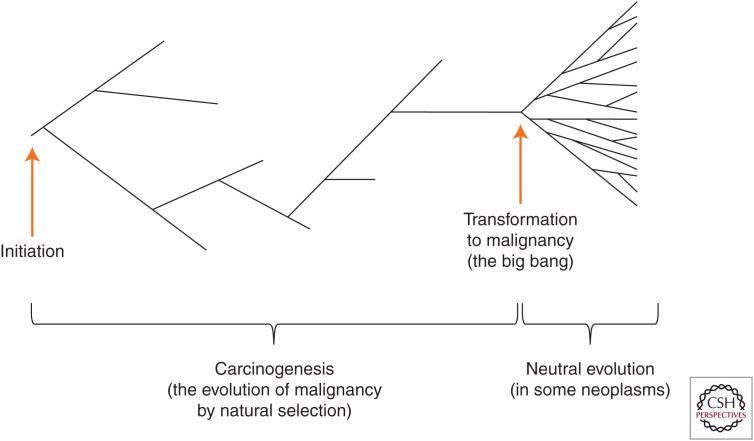
Although we argue here for the central importance of natural selection and adaptation in cancer, selection is not the only mechanism of evolution in cancer. Indeed, both neutral and selected changes have been reported together in some cancers (Shpak et al. 2015). Recent work suggests that many cancers evolve with little evidence of natural selection after transformation to malignancy (Fig. 3) (Sottoriva et al. 2015; Williams et al. 2016). This may be because selection is relaxed after the process of carcinogenesis is complete and most intrinsic limits on cell survival and proliferation from the normal genome have already been removed. One extensive phylogeographic study of a liver tumor found no evidence of selection (Ling et al. 2015). However, it is important to note that this pattern of only neutral evolution is seen only in certain types of cancers. In contrast, evidence of convergent evolution and natural selection (even after transformation in some cases) has been found in many other cancers (Anderson et al. 2011; Gerlinger et al. 2012, 2014; Kovac et al. 2015; Yates et al. 2015).
Figure 3.
A schematic illustration of a phylogeny for somatic evolution in neoplastic cells over time (x-axis), starting from tumor initiation (far left). Neoplastic cell lineages accumulate somatic mutations during progression and eventually transformation to malignancy (far right). As described in the text, there may be multiple mechanisms of somatic evolution. Before malignancy, cell lineages may evolve by natural selection, in which certain clones have a fitness advantage. However, after transformation to malignancy, some neoplasms may evolve neutrally, in which clonal expansion is rapid (called the “big bang”) (Sottoriva et al. 2015), and most adaptation to the niche of the “endogenous parasite” is already completed.
CONCLUDING REMARKS
There is evidence for extensive evolutionary adaptation among cancer cells, and this process drives the convergent evolution that not only gives meaning to the category of “cancer,” but also is what makes cancer a malignant and deadly disease.
Despite this central role, other mechanisms of evolution are also at work in cancer. Neutral evolutionary theory provides a set of null models, which have recently been exploited to good effect for analyzing cancer evolution (Sottoriva et al. 2015). However, given all the hallmarks of cancers that evolve during neoplastic progression, and in particular the evolution of malignancy and therapeutic resistance, we believe that adaptationist hypotheses are testable and useful, and also offer a crucial foundation for future medical advances.
Although evolutionary adaptation by somatic cells is different from organismal adaptation, it is central to cancer biology and needs to be understood. Natural selection should remain a key tool for generating hypotheses in cancer biology, and those hypotheses should be tested against alternatives including null models such as genetic drift.
ACKNOWLEDGMENTS
We thank Alex May for help in generating Figure 2, and Marc Tollis for pointing us to the literature on retrotransposons. This work is supported in part by National Institutes of Health (NIH) Grants P01 CA91955, R01 CA149566, R01 CA170595, R01 CA185138, and R01 CA140657, as well as CDMRP Breast Cancer Research Program Award BC132057. J.W.P is supported as an employee of the National Cancer Institute (NCI). The findings, opinions, and recommendations expressed here are those of the authors and not necessarily those of the universities where the research was performed or the NIH.
Footnotes
Editors: Charles Swanton, Alberto Bardelli, Kornelia Polyak, Sohrab Shah, and Trevor A. Graham
Additional Perspectives on Cancer Evolution available at www.perspectivesinmedicine.org
REFERENCES
- Aktipis CA, Maley CC, Pepper JW. 2012. Dispersal evolution in neoplasms: The role of disregulated metabolism in the evolution of cell motility. Cancer Prev Res 5: 266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An X, Tiwari AK, Sun Y, Ding PR, Ashby CR Jr, Chen ZS. 2010. BCR-ABL tyrosine kinase inhibitors in the treatment of Philadelphia chromosome positive chronic myeloid leukemia: A review. Leuk Res 34: 1255–1268. [DOI] [PubMed] [Google Scholar]
- Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, Kempski H, Moorman AV, Titley I, Swansbury J, et al. 2011. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469: 356–361. [DOI] [PubMed] [Google Scholar]
- Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, Ji HP, Maley CC. 2016. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 22: 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araten DJ, Golde DW, Zhang RH, Thaler HT, Gargiulo L, Notaro R, Luzzatto L. 2005. A quantitative measurement of the human somatic mutation rate. Cancer Res 65: 8111–8117. [DOI] [PubMed] [Google Scholar]
- Arnal A, Ujvari B, Crespi B, Gatenby RA, Tissot T, Vittecoq M, Ewald PW, Casali A, Ducasse H, Jacqueline C, et al. 2015. Evolutionary perspective of cancer: Myth, metaphors, and reality. Evol Appl 8: 541–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam M, Latek RR, Daley GQ. 2003. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell 112: 831–843. [DOI] [PubMed] [Google Scholar]
- Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. 2010. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res 38: 3909–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. 1997. Selection: The mechanism of evolution. Chapman and Hall, New York. [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. 2011. The genomic complexity of primary human prostate cancer. Nature 470: 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, et al. 2012. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 485: 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsmedh A, Szeles A, Henriksson M, Bratt A, Folkman MJ, Spetz AL, Holmgren L. 2001. Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc Natl Acad Sci 98: 6407–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozic I, Antal T, Ohtsuki H, Carter H, Kim D, Chen S, Karchin R, Kinzler KW, Vogelstein B, Nowak MA. 2010. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci 107: 18545–18550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BG, Mitchell RA, Harandi A, Eaton JW. 2009. Embryonic vaccines against cancer: an early history. Exp Mol Pathol 86: 192–197. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas. 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MF, van Amerongen R, Nijjar T, Cuppen E, Jones PA, Laird PW. 2001. Reduced rates of gene loss, gene silencing, and gene mutation in Dnmt1-deficient embryonic stem cells. Mol Cell Biol 21: 7587–7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton BG. 1996. Senescence, cancer and “endogenous parasites”: A salutogenic hypothesis. J R Coll Physicians Lond 30: 10–12. [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sprouffske K, Huang QH, Maley CC. 2011. Solving the puzzle of metastasis: The evolution of cell migration in neoplasms. PLoS ONE 6: e17933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lin FQ, Xing K, He XL. 2015. The reverse evolution from multicellularity to unicellularity during carcinogenesis. Nat Commun 6: 6367. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. 2008. Probabilistic cross-species inference of orthologous genomic regions created by whole-genome duplication in yeast. Genetics 179: 1681–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PC, Lineweaver CH. 2011. Cancer tumors as Metazoa 1.0: Tapping genes of ancient ancestors. Phys Biol 8: 015001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Monte U. 2009. Does the cell number 10(9) still really fit one gram of tumor tissue? Cell Cycle 8: 505–506. [DOI] [PubMed] [Google Scholar]
- de Nooij-van Dalen AG, van Buuren-van Seggelen VH, Lohman PH, Giphart-Gassler M. 1998. Chromosome loss with concomitant duplication and recombination both contribute most to loss of heterozygosity in vitro. Genes Chromosomes Cancer 21: 30–38. [DOI] [PubMed] [Google Scholar]
- Duelli D, Lazebnik Y. 2007. Cell-to-cell fusion as a link between viruses and cancer. Nat Rev Cancer 7: 968–976. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. 2007. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316: 1039–1043. [DOI] [PubMed] [Google Scholar]
- Enriquez-Navas PM, Kam Y, Das T, Hassan S, Silva A, Foroutan P, Ruiz E, Martinez G, Minton S, Gillies RJ, Gatenby RA. 2016. Exploiting evolutionary principles to prolong tumor control in preclinical models of breast cancer. Sci Transl Med 8: 327ra324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez LC, Torres M, Real FX. 2016. Somatic mosaicism: On the road to cancer. Nat Rev Cancer 16: 43–55. [DOI] [PubMed] [Google Scholar]
- Fidler IJ. 2003. The pathogenesis of cancer metastasis: The “seed and soil” hypothesis revisited. Nat Rev Cancer 3: 453–458. [DOI] [PubMed] [Google Scholar]
- Friberg S, Mattson S. 1997. On the growth rates of human malignant tumors: Implications for medical decision making. J Surg Oncol 65: 284–297. [DOI] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ, Brown JS. 2011. Of cancer and cave fish. Nat Rev Cancer 11: 237–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR, et al. 2014. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 46: 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman MM. 2002. Mechanisms of cancer drug resistance. Annu Rev Med 53: 615–627. [DOI] [PubMed] [Google Scholar]
- Gould SJ, Lewontin RC. 1979. The spandrels of San Marco and the Panglossian paradigm: A critique of the adaptationist programme. Proc R Soc Lond B Biol Sci 205: 581–598. [DOI] [PubMed] [Google Scholar]
- Greuber EK, Smith-Pearson P, Wang J, Pendergast AM. 2013. Role of ABL family kinases in cancer: From leukaemia to solid tumours. Nat Rev Cancer 13: 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger AW, Salmon SE. 1977a. Primary bioassay of human tumor stem cells. Science 197: 461–463. [DOI] [PubMed] [Google Scholar]
- Hamburger A, Salmon SE. 1977b. Primary bioassay of human myeloma stem cells. J Clin Invest 60: 846–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2000. The hallmarks of cancer. Cell 100: 57–70. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: The next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Helman E, Lawrence MS, Stewart C, Sougnez C, Getz G, Meyerson M. 2014. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res 24: 1053–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H, Schultz N, Gopalan A, Carver BS, Chang MT, Xiao Y, Heguy A, Huberman K, Bernstein M, Assel M, et al. 2014. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci 111: 11139–11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren L, Szeles A, Rajnavolgyi E, Folkman J, Klein G, Ernberg I, Falk KI. 1999. Horizontal transfer of DNA by the uptake of apoptotic bodies. Blood 93: 3956–3963. [PubMed] [Google Scholar]
- Ibrahim A. 2014. Invasive cancer as an empirical example of evolutionary suicide. Netw Biol 4: 58–66. [Google Scholar]
- Jamal-Hanjani M, Hackshaw A, Ngai Y, Shaw J, Dive C, Quezada S, Middleton G, de Bruin E, Le Quesne J, Shafi S, et al. 2014. Tracking genomic cancer evolution for precision medicine: The Lung TRACERx Study. Plos Biol 12: e1001906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Chen WD, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, Traulsen A, Nowak MA, Siegel C, Velculescu VE, et al. 2008. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci 105: 4283–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. 2013. Mutational landscape and significance across 12 major cancer types. Nature 502: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1983. The neutral theory of molecular evolution. Cambridge University Press, New York. [Google Scholar]
- Knudson AG Jr. 1971. Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci 68: 820–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostadinov RL, Kuhner MK, Li X, Sanchez CA, Galipeau PC, Paulson TG, Sather CL, Srivastava A, Odze RD, Blount PL, et al. 2013. NSAIDs modulate clonal evolution in Barrett’s esophagus. PLoS Genet 9: e1003553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovac M, Blattmann C, Ribi S, Smida J, Mueller NS, Engert F, Castro-Giner F, Weischenfeldt J, Kovacova M, Krieg A, et al. 2015. Exome sequencing of osteosarcoma reveals mutation signatures reminiscent of BRCA deficiency. Nat Commun 6: 8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. 2008. Recurrent gene fusions in prostate cancer. Nat Rev Cancer 8: 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzrock R, Giles FJ. 2015. Precision oncology for patients with advanced cancer: The challenges of malignant snowflakes. Cell Cycle 14: 2219–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin BR, Bull JJ. 1994. Short-sighted evolution and the virulence of pathogenic microorganisms. Trends Microbiol 2: 76–81. [DOI] [PubMed] [Google Scholar]
- Li X, Paulson TG, Galipeau PC, Sanchez CA, Liu K, Kuhner MK, Maley CC, Self SG, Vaughan TL, Reid BJ, et al. 2015a. Assessment of esophageal adenocarcinoma risk using somatic chromosome alterations in longitudinal samples in Barrett’s esophagus. Cancer Prev Res (Phila) 8: 845–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Shao T, Jiang C, Bai J, Wang Z, Zhang J, Zhang L, Zhao Z, Xu J, Li X. 2015b. Construction and analysis of dynamic transcription factor regulatory networks in the progression of glioma. Sci Rep 5: 15953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S, Hu Z, Yang Z, Yang F, Li Y, Lin P, Chen K, Dong L, Cao L, Tao Y, et al. 2015. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proc Natl Acad Sci 112: E6496–6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Edgerton SM, Moore DH II, Thor AD. 2001. Measures of cell turnover (proliferation and apoptosis) and their association with survival in breast cancer. Clin Cancer Res 7: 1716–1723. [PubMed] [Google Scholar]
- Lloyd MC, Rejniak KA, Brown JS, Gatenby RA, Minor ES, Bui MM. 2015. Pathology to enhance precision medicine in oncology: Lessons from landscape ecology. Adv Anat Pathol 22: 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd MC, Cunningham JJ, Bui MM, Gillies RJ, Brown JS, Gatenby RA. 2016. Darwinian dynamics of intratumoral heterogeneity: Not solely random mutations but also variable environmental selection forces. Cancer Res 76: 3136–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Lin AW. 2000. Apoptosis in cancer. Carcinogenesis 21: 485–495. [DOI] [PubMed] [Google Scholar]
- Lugini L, Matarrese P, Tinari A, Lozupone F, Federici C, Iessi E, Gentile M, Luciani F, Parmiani G, Rivoltini L, et al. 2006. Cannibalism of live lymphocytes by human metastatic but not primary melanoma cells. Cancer Res 66: 3629–3638. [DOI] [PubMed] [Google Scholar]
- Lunt SY, Vander Heiden MG. 2011. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol 27: 441–464. [DOI] [PubMed] [Google Scholar]
- Martinez P, Timmer MR, Lau CT, Calpe S, del Carmen Sancho-Serra M, Straub D, Baker A, Meijer SL, ten Kate FJW, Mallant-Hent RC, et al. 2016. Dynamic clonal equilibrium and predetermined cancer risk in Barrett’s oesophagus. Nat Commun 7: 12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. 1983. How to carry out the adaptationist program. Am Nat 121: 324–334. [Google Scholar]
- Melendez-Lazo A, Cazzini P, Camus M, Doria-Torra G, Marco Valle AJ, Solano-Gallego L, Pastor J. 2015. Cell cannibalism by malignant neoplastic cells: Three cases in dogs and a literature review. Vet Clin Pathol 44: 287–294. [DOI] [PubMed] [Google Scholar]
- Merlo LM, Maley CC. 2010. The role of genetic diversity in cancer. J Clin Invest 120: 401–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo LM, Pepper JW, Reid BJ, Maley CC. 2006. Cancer as an evolutionary and ecological process. Nat Rev Cancer 6: 924–935. [DOI] [PubMed] [Google Scholar]
- Merlo LM, Shah NA, Li X, Blount PL, Vaughan TL, Reid BJ, Maley CC. 2010. A comprehensive survey of clonal diversity measures in Barrett’s esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prev Res (Phila) 3: 1388–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlos-Suarez A, Barriga FM, Jung P, Iglesias M, Cespedes MV, Rossell D, Sevillano M, Hernando-Momblona X, da Silva-Diz V, Munoz P, et al. 2011. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8: 511–524. [DOI] [PubMed] [Google Scholar]
- Meza R, Jeon J, Moolgavkar SH, Luebeck EG. 2008. Age-specific incidence of cancer: Phases, transitions, and biological implications. Proc Natl Acad Sci 105: 16284–16289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milojkovic D, Apperley J. 2009. Mechanisms of resistance to imatinib and second-generation tyrosine inhibitors in chronic myeloid leukemia. Clin Cancer Res 15: 7519–7527. [DOI] [PubMed] [Google Scholar]
- Misale S, Bozic I, Tong J, Peraza-Penton A, Lallo A, Baldi F, Lin KH, Truini M, Trusolino L, Bertotti A, et al. 2015. Vertical suppression of the EGFR pathway prevents onset of resistance in colorectal cancers. Nat Commun 6: 8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroz EA, Tward AM, Hammon RJ, Ren Y, Rocco JW. 2015. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: Analysis of data from The Cancer Genome Atlas. PLoS Med 12: 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugaesu N, Wilson GA, Birkbak NJ, Watkins TBK, McGranahan N, Kumar S, Abbassi-Ghadi N, Salm M, Mitter R, Horswell S, et al. 2015. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov 5: 821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A, Yoshida M, Goodarzi H, Tavazoie SF. 2016. Highly variable cancer subpopulations that exhibit enhanced transcriptome variability and metastatic fitness. Nat Commun 7:11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Springer, New York. [Google Scholar]
- Pepper JW. 2011. Somatic evolution of acquired drug resistance in cancer. In Targeted therapies: Mechanisms of resistance (ed. Gioeli D), pp. 127–134. Springer, New York. [Google Scholar]
- Pepper JW. 2012. Drugs that target pathogen public goods are robust against evolved drug resistance. Evol Appl 5: 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper JW. 2016. Darwinian strategies to avoid the evolution of drug resistance during cancer treatment. Evolutionary thinking in medicine (ed. Alvergne A), pp. 167–176. Springer International, Cham, Switzerland. [Google Scholar]
- Pepper JW, Dunn BK, Fagerstrom RM, Gohagan JK, Vydelingum NA. 2014. Using systems biology to understand cancer as an evolutionary process. J Evol Med 2: 1–8. [Google Scholar]
- Potten CS, Booth C, Hargreaves D. 2003. The small intestine as a model for evaluating adult tissue stem cell drug targets. Cell Prolif 36: 115–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan H, Nowak MA, Vogelstein B, Lengauer C. 2003. The significance of unstable chromosomes in colorectal cancer. Nat Rev Cancer 3: 695–701. [DOI] [PubMed] [Google Scholar]
- Ren R. 2005. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer 5: 172–183. [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. 2007. Identification and expansion of human colon-cancer-initiating cells. Nature 445: 111–115. [DOI] [PubMed] [Google Scholar]
- Rodic N, Burns KH. 2013. Long interspersed element-1 (LINE-1): Passenger or driver in human neoplasms? PLoS Genet 9: e1003402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C. 2016. Dose schedules inspired by evolutionary concepts lengthen progression-free survival in mice. J Natl Cancer Inst 108: djw157. [DOI] [PubMed] [Google Scholar]
- Semenza GL. 2012. Hypoxia-inducible factors: Mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci 33: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak M, Goldberg MM, Cowperthwaite MC. 2015. Rapid and convergent evolution in the glioblastoma multiforme genome. Genomics 105: 159–167. [DOI] [PubMed] [Google Scholar]
- Sottoriva A, Spiteri I, Shibata D, Curtis C, Tavare S. 2013. Single-molecule genomic data delineate patient-specific tumor profiles and cancer stem cell organization. Cancer Res 73: 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva A, Kang H, Ma Z, Graham TA, Salomon MP, Zhao J, Marjoram P, Siegmund K, Press MF, Shibata D, et al. 2015. A Big Bang model of human colorectal tumor growth. Nat Genet 47: 209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. 2009. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462: 1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PW, Salmon SE. 1972. Kinetics of tumor growth and regression in IgG multiple myeloma. J Clin Invest 51: 1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan AS, Baty JW, Dong LF, Bezawork-Geleta A, Endaya B, Goodwin J, Bajzikova M, Kovarova J, Peterka M, Yan B, et al. 2015. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab 21: 81–94. [DOI] [PubMed] [Google Scholar]
- Tlsty TD, Margolin BH, Lum K. 1989. Differences in the rates of gene amplification in nontumorigenic and tumorigenic cell lines as measured by Luria–Delbruck fluctuation analysis. Proc Natl Acad Sci 86: 9441–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, Vogelstein B, Parmigiani G. 2013. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc Natl Acad Sci 110: 1999–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen L, Morrissey E, van der Heijden M, Nicholson AM, Sottoriva A, Buczacki S, Kemp R, Tavare S, Winton DJ. 2013. Defining stem cell dynamics in models of intestinal tumor initiation. Science 342: 995–998. [DOI] [PubMed] [Google Scholar]
- Wang TL, Rago C, Silliman N, Ptak J, Markowitz S, Willson JK, Parmigiani G, Kinzler KW, Vogelstein B, Velculescu VE. 2002. Prevalence of somatic alterations in the colorectal cancer cell genome. Proc Natl Acad Sci 99: 3076–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MJ, Werner B, Barnes CP, Graham TA, Sottoriva A. 2016. Identification of neutral tumor evolution across cancer types. Nat Genet 48: 238–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willyard C. 2016. Cancer therapy: An evolved approach. Nature 532: 166–168. [DOI] [PubMed] [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet 8: 206–216. [DOI] [PubMed] [Google Scholar]
- Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E. 2008. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319: 1527–1530. [DOI] [PubMed] [Google Scholar]
- Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu BJ, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, et al. 2010. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467: 1114–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, Aas T, Alexandrov LB, Larsimont D, Davies H, et al. 2015. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 21: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang CZ, Wala J, Mermel CH, et al. 2013. Pan-cancer patterns of somatic copy number alteration. Nat Genet 45: 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]