Abstract
TP53 missense mutations produce a mutant p53 protein that cannot activate the p53 tumor suppressive transcriptional response, which is the primary selective pressure for TP53 mutation. Specific codons of TP53, termed hotspot mutants, are mutated at elevated frequency. Hotspot forms of mutant p53 possess oncogenic properties in addition to being deficient in tumor suppression. Such p53 mutants accumulate to high levels in the cells they inhabit, causing transcriptional alterations that produce pro-oncogenic activities, such as increased pro-growth signaling, invasiveness, and metastases. These forms of mutant p53 very likely use features of wild-type p53, such as interactions with the transcriptional machinery, to produce oncogenic effects. In this review, we discuss commonalities between wild-type and mutant p53 proteins with an emphasis on transcriptional processes.
Certain mutations in the DNA-binding domain of p53 are particularly common. These accumulate to high levels in cells and promote oncogenesis, likely by using various transcriptional processes of wild-type p53.
TP53 is the most frequently mutated gene found in human cancers (Olivier et al. 2010). Wild-type p53 is a sequence-specific transcription factor that when activated by various stresses, such as DNA damage, oncogenic signaling or nutrient depletion, promotes cellular outcomes, such as cell arrest, cell death, senescence, metabolic changes, and others, depending on the extent and context of the stress (Vousden and Prives 2009). In human cancer, p53 primarily sustains missense mutations in its conserved DNA-binding domain. The small number of residues (∼5 to 6) within this region that are mutated with extraordinarily high frequency are termed hotspot mutations. These mutations can be loosely divided into two categories, the contact mutants (e.g., R273H), which remain well folded but whose mutated residues fail to make specific contact with elements within the DNA-binding site and conformational mutants (e.g., R175H) that are partly unfolded leading to loss of zinc coordination and general DNA binding (Cho et al. 1994; Joerger et al. 2005, 2006). p53 hotspot mutant proteins have been reported to associate with chromatin and alter a cell’s transcriptional profile, leading to oncogenic cellular changes (Di Agostino et al. 2006; Stambolsky et al. 2010; Do et al. 2012; Freed-Pastor et al. 2012; Cooks et al. 2013; Pfister et al. 2015; Zhu et al. 2015).
p53 BACKGROUND AND DISCOVERY
p53 was initially thought to be a cellular oncogene, because early experiments showed cooperation of p53 with SV40 or H-Ras to transform cells (DeLeo et al. 1979; Lane and Crawford 1979; Linzer and Levine 1979; Eliyahu et al. 1984; Parada et al. 1984; Hinds et al. 1989). Histology supported the hypothesis that p53 is an oncogene, as high levels of p53 are observed in transformed cells, whereas low levels of p53 are observed in normal tissues (Rotter 1983; Cattoretti et al. 1988; Bartek et al. 1990a). However, it was eventually realized that transforming versions of p53 were derived from cancer cell lines, whereas p53 cloned from normal cells did not show such oncogenic properties and in fact was able to suppress transformation by other oncogenes. Coupled with the fact that the 17p13.1 chromosomal region containing the TP53 gene is frequently lost in tumors that contain point mutations in the other allele, p53 became firmly established as a tumor suppressor (Baker et al. 1989, 1990; Nigro et al. 1989). Mutant p53 is a dominant-negative inhibitor of wild-type p53 (Bargonetti et al. 1992; Kern et al. 1992) and wild-type p53 confers G1 checkpoint control to cells lacking functional p53 (Kuerbitz et al. 1992; Yin et al. 1992) in a dose-dependent manner, which is mediated by p21 (Waldman et al. 1995). Mechanistically, wild-type p53 binds DNA in a sequence-specific manner to mediate its functions, whereas mutant p53 fails to bind to a wild-type p53 consensus sequence (Bargonetti et al. 1991, 1993; Kern et al. 1991; el-Deiry et al. 1992; Funk et al. 1992). Critically, p53 null mice are predisposed to acquiring tumors, firmly establishing p53 as a tumor suppressor in mice (Donehower et al. 1992). These data provided a framework to understand Li–Fraumeni syndrome, in which germline mutations in TP53 predispose affected individuals to breast cancer, sarcomas, lymphomas, and other neoplasms (Malkin et al. 1990; Srivastava et al. 1990; Varley et al. 1997; Wong et al. 2006).
TP53 MUTATIONS IN HUMAN CANCERS
On average, TP53 is mutated in ∼50% of all human tumors although frequencies vary enormously with tumor type ranging from ∼1% in papillary thyroid cancer (Cancer Genome Atlas Research Network 2014) to 95% in serous ovarian cancer (Cancer Genome Atlas Research 2011 Network, cbioportal.org). Importantly, TP53 is unusual for a tumor suppressor in that it is not frequently deleted. Instead, this gene primarily sustains various other types of mutations, the most frequent of which are missense mutations in the p53 DNA-binding domain. Different datasets may have varying frequencies for TP53 mutation within the same cancer type. Using breast cancer as an example, published TP53 mutation rates range from 27.2% for breast cancers unstratified by subtype (Banerji et al. 2012) to 53.8% (35/65 samples) in triple-negative breast cancer (Shah et al. 2012). TP53 mutation is an independent prognostic indicator (correlating with worse prognosis) in breast cancer (Olivier et al. 2006; Petitjean et al. 2007), stage I non-small-cell lung cancer (Ahrendt et al. 2003), lymphomas (Levine and Vosburgh 2008; O’Shea et al. 2008; Young et al. 2008), and with certain hotspot mutations in colon cancer (Samowitz et al. 2002). TP53 mutations occur at multiple stages in the progression of a tumor, and the stage in which TP53 mutation occurs may affect the malignancy of the tumor (reviewed in Rivlin et al. 2011).
ESSENTIAL SELECTIVE ADVANTAGES OF TP53 MUTATIONS IN TUMORS
With few exceptions, mutations within the TP53 DNA-binding domain either reduce, alter, or preclude interaction of p53 with its consensus DNA-binding sequence (Bargonetti et al. 1991; Kern et al. 1991). The p53 consensus sequence (el-Deiry et al. 1992; Funk et al. 1992; Riley et al. 2008) is a cis-regulatory element in the promoter of canonical p53 target genes, such as MDM2 (Barak et al. 1993), NOXA (Oda et al. 2000), PUMA (Nakano and Vousden 2001), and p21 (CDKN1A) (el-Deiry et al. 1993). Abrogation of DNA binding by mutation in the p53 DNA-binding domain dysregulates the induction of p53-mediated cellular arrest (through p21) and apoptosis (through PUMA and NOXA), and leads to the accumulation of elevated levels of the mutant form of p53 in part because of the impairment of inducing p53’s primary negative regulator, the E3 ubiquitin ligase MDM2 (reviewed in Freed-Pastor and Prives 2012). Loss of p53 function enables a cell to evade cell-arrest mechanisms that would allow adequate repair of damaged DNA and allow a cell to evade apoptosis, which would have eliminated a cell that sustained DNA damage. Hence, abrogation of p53 function is a critical step in oncogenesis (reviewed in Vousden and Prives 2009). Indeed, even when TP53 is not directly affected, its negative regulators MDM2 and MDMX are often overexpressed (reviewed in Toledo and Wahl 2006). Loss of wild-type p53 function is the primary selective advantage conferred by mutation in TP53.
TP53 HOTSPOT MUTATIONS
The spectrum of TP53 missense mutations suggests an oncogenic role of TP53 mutations in promoting tumorigenesis. Six codons have been designated as “hotspots” because of increased selection for these mutants: codons 175, 245, 248, 249, 273, and 282 (Olivier et al. 2010). Each of these codons occurs within the DNA-binding domain of p53, the location of the overwhelming majority of p53 mutations, with other known mutations less frequently occurring outside this domain (Soussi et al. 2005; Petitjean et al. 2007). p53 hotspot mutants are classified as either a DNA-contact mutant (e.g., codon R248, codon R273, codon R282) or a conformational mutant (e.g., codon R175, codon G245) based on the mechanism of alteration of the DNA-binding domain, with conformational mutants being more structurally abnormal, and each class leading to abrogation of sequence-specific DNA binding (Bartek et al. 1990b; Gannon et al. 1990; Bargonetti et al. 1993; Cho et al. 1994; Legros et al. 1994).
p53 DOMAIN STRUCTURE
Structurally, p53 is composed of a bipartite amino-terminal transactivation domain (TAD), a proline-rich domain (PRD), a central DNA-binding domain, an oligomerization domain, and a carboxy-terminal regulatory domain (CTD) (Joerger and Fersht 2008). p53 is a dimer of dimers that through hydrophobic interactions between leucines 344 and 348 in the oligomerization domain form the functional p53 tetramer (Jeffrey et al. 1995; Joerger and Fersht 2008). The p53 protein should, thus, be thought of as a tetramer, allowing for four distinct binding sites per structural element within each tetramer.
The amino-terminal TAD is subdivided into two subdomains, TAD1 within the first 40 amino acids and TAD2 within amino acids 41–61 (Chang et al. 1995; Walker and Levine 1996). TAD1 functionally requires residues 22 (leucine) and 23 (tryptophan) and TAD2 functionally requires residues 53 (tryptophan) and 54 (phenylalanine). These residues can be mutated to polar amino acids (mTAD1 into L22Q/W23S and mTAD2 into W53Q/F54S), leading to abrogation or reduction in p53 transactivation of many target genes (Lin et al. 1994, 1995; Candau et al. 1997; Zhu et al. 1998; Venot et al. 1999; Yan and Chen 2010). Mouse models of p53 transactivation mutants revealed that TAD1 is required for activation of the acute DNA damage response but is dispensable for tumor suppression, whereas simultaneous TAD1 and TAD2 mutations also impairs p53 tumor suppression (Johnson et al. 2005; Brady et al. 2011). The PRD (amino acids 64 to 92) is likely necessary for efficient Pin1 prolyl isomerase-dependent loading of WT p53 on target promoters (Mantovani et al. 2007). On the other hand, the very same domain is required for mutant p53 conformation and function (Girardini et al. 2011), and this domain can be deleted to study PRD-dependent effects (Yan and Chen 2010). The CTD (residues ∼363 to 393) has been shown to be necessary for certain mutant p53 target gene activation but dispensable or even inhibitory for mutant p53 pro-proliferation phenotypes, and this domain can be deleted to study CTD-dependent effects (Frazier et al. 1998; Yan and Chen 2010).
The amino and carboxyl termini of p53 are involved in the majority of described protein–protein interactions with p53 (Boehme and Blattner 2009). The CTD and oligimerization domain (OD, residues 326–356) are required for wild-type p53 interaction with Sp1 and Sp3, as well as other transcriptional proteins (Koutsodontis et al. 2005; Kamada et al. 2011). The CTD contains numerous lysines that are modified to regulate p53, and these residues can be mutated to investigate mutant p53 CTD interactions. A significant challenge to studying CTD interaction is that many studies co-delete the OD along with the CTD, which destabilizes the quaternary structure of p53 and changes its binding interface so it cannot be clearly determined whether an interaction requires the CTD specifically or other aspects of the p53 tetramer. Indeed, specific mutant p53 domains are either required or dispensable to mutant p53 target gene activation, and the mechanisms of this regulation are incompletely defined (Table 1). It is plausible that the charged amino- and carboxy-terminal domains of p53 retain protein–protein interactions in the DNA-binding domain mutants, even though the genomic localization of these mutants is altered. Not only the TAD and CTD but also the DNA-binding domain form interactions with cellular proteins. For example, mutant p53 interactions with members of p63 and p73 families involve DNA-binding domain interactions (Li and Prives 2007). It will be critical to investigate domain-specific p53 interactions to mechanistically define mutant p53 gene regulation.
Table 1.
Studies exploring mutant p53 domain-specific effects on gene expression
| Gene | Cell line | Mutant | Method | Study |
|---|---|---|---|---|
| MDR1 | NIH3T3, Saos-2, Caco-2, BHK | 175H, 281G, requires TAD residues 14/19 for gene expression; requires ETS-1 site; Del22/23 blocks Ets1 binding, Del360CTD still has Ets binding to 281G to 143A, 175H, 248W, 273H; 213Q and 234H no effect on MDR1; 281G mTAD1 ineffective | Overexpression—CAT reporter assay, reporter assay | Chin et al. 1992; Lin et al. 1995; Strauss and Haas 1995; Candau et al. 1997; Sampath et al. 2001 |
| dUTPase | SK-OV-3 and 10(1) | 175H, 248W, 273H mTAD1 | Overexpression, qPCR/northern | Pugacheva et al. 2002 |
| TIM50 (Ets-1, CREB ChIP) | 1299, SKBR3, MDA-MB-468, Saos-2 | 175H, 273H, 281G, mTAD1 281G ineffective | Overexpression, siRNA, reporter assay, ChIP | Sankala et al. 2011 |
| NF-κB2 | H1299, 21PT, Saos-2 | 175H, 273H, 281G mTAD1-281G; 175H has increased NF-κB activity | Gene expression array with ectopic expression, qPCR, reporter assay, EMSA | Scian et al. 2005 |
| EBAG9, ITGA6, E2F5, MCM6, C-SYN | H1299 | 281G but not mTAD1-281G effective in gene expression | Gene expression array with ectopic expression, qPCR, | Scian et al. 2005 |
| VDR gene (protein), VDR promoters RNA (IGFBP3, CYP24A1) | SKBR3, H1299, SW480, MDA-MB-231 | 175H (VDR motif overrepresented), 273H; mTAD1-175H no effect on reporter assay | ChIP-on-chip, overexpression, Southwestern blot, reporter assay | Scian et al. 2005 |
| EGR1 | H1299, PC3, SKBR3, HeLa | 175H, 248W, 273H, 281G, but not mTAD1-175H or 179E or wt p53 increase gene expression | Overexpression, gene expression array, ChIP, reporter assay, VEGF-induction by EGR1 | Weisz et al. 2004 |
| CXCL1 (GRO1) | SW480, MIA-PaCa-2, HCT116 | 273H/309S, 248W, 175H but not wt p53, mTAD1, mTAD2, PRD required in 245S and 248W, CTD inhibitory 248W, no effect 245S | siRNA, ectopic expression, ChIP | Yan and Chen 2009, 2010 |
| Id2 (inhibition by mutant p53) | SW480, MIA-PaCa-2, HCT116 | 273H/309S, 248W, 175H but not wt p53, TAD1, TAD2, PRD required in 245S and 248W, CTD not required and perhaps inhibitory | siRNA, ectopic expression, ChIP | Yan et al. 2008; Yan and Chen 2010 |
| c-Myc (mTAD1 and CTD required) | Cx3Ras (rat), 10(1) mouse cells, Saos-2, SK-OV-3 and 10(1) | 143A, 281G, 175H, 273H, 248; mTAD1 and CTD with intermediate phenotype with 281G; 175H, 248W, 273H activated c-myc but mTAD1 does not | Overexpression, reporter assay, qPCR/northern | Frazier et al. 1998; Pugacheva et al. 2002 |
| c-Myc, apoptosis suppression | M1/2 myeloid cells | 143A, mTAD1 required for c-myc and apoptosis suppression | Overexpression | Matas et al. 2001 |
| Induction of invasion/metastasis or p63/p73 inactivation | H1299 | 175H, 273H; TAD not required for transactivation | Overexpression | Adorno et al. 2009; Oren and Rotter 2010 |
| Spindle checkpoint control | Li–Fraumeni fibroblasts | 281G; TAD not required for transactivation | Overexpression | Gualberto et al. 1998 |
| Apoptosis suppression, G2 arrest suppression | M1/2 myeloid cells | 135V; CTD required for apoptosis suppression | Overexpression | Sigal et al. 2001 |
| CXCL1 | WI-38, Ras expressing | 175H, 179R; TAD1 not required for gene expression | Overexpression | Solomon et al. 2012 |
| IL-1β | WI-38, Ras expressing | 175H, 179R; TAD1 not required | Overexpression | Solomon et al. 2012 |
| MMP3 | WI-38, Ras expressing | 175H, 179R; TAD1 not required | Overexpression | Solomon et al. 2012 |
| TGF-β receptor 2 | H1299 | 175H; mTAD1 required for expression | Overexpression, reporter assay | Kalo et al. 2007 |
| Axl | H1299, H1437, H1048 | 175H, 267P, 273C, 273H, 273P, 281G; utilized mTAD1 mutants | siRNA, qPCR, +/−ChIP | Vaughan et al. 2012 |
Summary of known studies that have investigated domain-specific effects of mutant p53 on transcription. The cell lines in which the stated gene was found to be regulated, the p53 mutants that were used, the experiment system that was used, and the associated references are listed.
TGF, Transforming growth factor; wt p53, wild-type p53.
TRANSCRIPTIONAL ACTIVATION BY WILD-TYPE p53
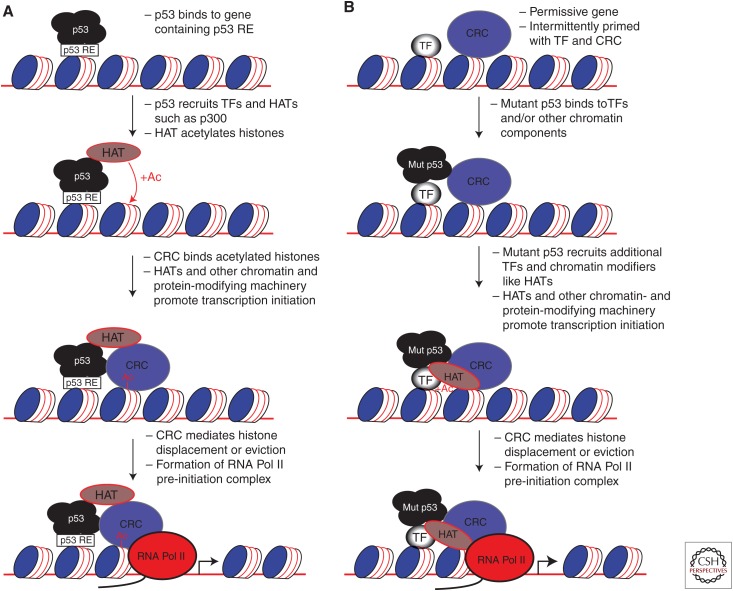
The following section describes the most well-understood mechanisms of how wild-type p53 initiates transcription at genes containing a p53 response element (reviewed in Beckerman and Prives 2010). It must be stated from the outset that the precise mechanisms by which p53 initiates transcription at its target genes is incompletely understood and undoubtedly more complex than discussed here (Espinosa 2008). Although somewhat of an oversimplification, the basic model for activation of a p53 target gene from promoter engagement through transcription initiation occurs in two distinct phases: (1) p53 recognition and association with its DNA response element, followed by (2) recruitment of transcriptional modifiers that culminate in the formation of the RNA polymerase II pre-initiation complex (Fig. 1A) (Laptenko and Prives 2006).
Figure 1.
Transcriptional model for wild-type and mutant p53. (A) Transcriptional model for wild-type p53. Wild-type p53 is modified at specific amino acids in response to cellular stress signals, such as DNA damage, and accumulates at the protein level in part because of decreased degradation by MDM2. p53 has an intact DNA-binding domain and thus recognizes its cognate p53 response element (RE), which it preferentially binds. p53 then recruits other transcriptional regulatory components, such as transcription factors (TFs) and histone acetyltransferases (HATs) such as p300. HATs acetylate (+Ac) p53 as well as chromatin, promoting additional steps toward transcription, such as the stable recruitment of chromatin remodeling complexes (CRCs) such as SWI/SNF. CRCs recognize histone tails that are primed with histone modifications at specific residues, which promotes ATP-dependent histone displacement or eviction by the CRC. RNA polymerase II and other general TFs bind to the open promoter, forming the pre-initiation complex, followed by steps leading to transcriptional elongation. For a more detailed explanation, see the text. (B) Transcriptional model for mutant p53 (Mut p53). Mutant p53 is accumulated at the protein level because of impairment of MDM2 induction and deregulated posttranslational modifications, among other mechanisms. The mutant p53 DNA-binding domain does not recognize the p53 RE, so mutant p53 interaction with chromatin is primarily through protein–protein interactions with other transcriptional regulators such as transcription factors. Because mutant p53 is not restricted through specific binding to the p53 RE, mutant p53 is associated across the genome in a promiscuous manner. At genes that are primed for transcription, such as by being bound by a pioneering transcription factor as well as a chromatin remodeling complex (see Pfister et al. 2015), mutant p53 may promote transcription. Mutant p53 is recruited through protein–protein interactions (that may be conserved in wild-type p53) to such promoters, followed by mutant p53-dependent recruitment of additional transcriptional regulators such as TFs and HATs. Transcription then proceeds in a manner generally similar to that of wild-type p53. Therefore, a key distinction is that wild-type p53 is capable of independently facilitating CRC recruitment and transcriptional activation, while mutant p53 is dependent on the presence of specific promoter factors such as SWI/SNF. Most importantly, wild-type p53 is restricted to specific genomic sites (at p53 responsive genes) by virtue of its DNA-binding domain, whereas mutant p53 is not restricted to specific genomic sequences.
p53 mediates gene expression changes through interaction with multiple chromatin regulators (Laptenko and Prives 2006). The potential for gene regulatory mechanisms is especially complex when the tetrameric structure of p53 is considered. The functional p53 protein composed of four identical monomers allows for four distinct binding sites for binding partners within each tetramer. This allows for a high degree of complexity in the regulation of transcription, as a single tetramer could bind to multiple transcriptional modifiers at its amino or caboxyl termini (or with its DNA-binding domain).
p53 has been proposed to preferentially associate with genomic regions with a high degree of nucleosomal occupancy (Lidor Nili et al. 2010). Increased DNase I cleavage (termed hypersensitivity) is a hallmark of active genes, corresponding to an open promoter configuration in which nucleosomes are not present (reviewed in Krebs and Peterson 2000). DNase I hypersensitivity is a feature of multiple wild-type p53 target gene promoters including p21 (CDKN1A) (Braastad et al. 2003). Interestingly, the p53 binding site within these promoters occurs in regions that tend to be resistant to DNase I (Braastad et al. 2003). These genomic regions where p53 is bound undergo nucleosomal displacement, which is specific to p53 recruitment as the nucleosomal displacement is reversible on p53 inactivation using a temperature-sensitive p53 mutant (Lidor Nili et al. 2010).
At the p21 promoter, which is the best-studied p53 responsive gene, both distal and proximal p53 response elements contain high levels of nucleosomal occupancy (Laptenko et al. 2011). On p53 activation, two nucleosomes at the distal (5′) response element are rapidly displaced on p53 activation (Laptenko et al. 2011). Nucleosomal displacement is most likely to occur subsequent to p53 DNA binding as it is unlikely that the alternative—that nucleosomes and p53 compete for the same site—occurs, especially considering that p53 can bind to its response element while the response element is engaged by a nucleosome (Sahu et al. 2010; Laptenko et al. 2011). Interestingly, the ability of p53 to bend DNA within the p53–DNA complex is directly correlated with the stability of the p53–DNA interaction (Nagaich et al. 1997). It is conceivable that the intrinsic sequence of the p53 response element (which dictates the degree of DNA bending on p53 binding) facilitates nucleosomal repositioning and impacts subsequent steps in transcriptional activation and resetting of the promoter (promoter turnover) (Nagaich et al. 1997; Laptenko and Prives 2006). As mutant p53 does not bind to DNA in a stable manner like wild-type p53, these initial steps of transcriptional activation likely differ.
Relevant to the above-mentioned findings of its association with nucleosomes in vivo, p53 has been reported to cooperate with nucleosomal remodeling complexes including the Mi-2/NuRD complex (Luo et al. 2000) and multiple members of the SWI/SNF chromatin remodeling complex (Lee et al. 2002). p53 is not yet known to function with the INO80, SWR, or ISWI families of CRC.
The genetic and physical interactions between SWI/SNF components and p53 have been well described. Multiple components of the SWI/SNF complex have been identified as wild-type p53-binding partners including BRG1 (Lee et al. 2002; Naidu et al. 2009), SNF5 (Lee et al. 2002), BAF60A and BAF155 (Oh et al. 2008), ARID1A (Guan et al. 2011), and BRD7 (Burrows et al. 2010). p53 has been described to function with the SWI/SNF complex to mediate transcription from nucleosomal promoters, and p53 interaction with the SWI/SNF complex has been proposed to be mediated, at least in part, through interaction with BAF60A (Oh et al. 2008). BRG1 depletion has been reported to activate p53 signaling (Naidu et al. 2009), whereas SNF5 depletion has been reported to lead to inhibition of p53 mRNA translation (Xu et al. 2010). A proline-rich region of BRG1 is necessary for interaction with p53 (Naidu et al. 2009). CBP (which is closely related to p300) has been reported to dissociate from BRG1 on DNA damage, correlating with decreased CBP and BRG1 levels at the p21 promoter and increased p300 and p53 levels at the p21 promoter (Naidu et al. 2009). As previously mentioned, the p21 promoter undergoes nucleosomal remodeling on p53 binding (Laptenko et al. 2011) and SWI/SNF complex components including BAF180 (Xia et al. 2008), SNF5 (Lee et al. 2002), and BRG1 (Lee et al. 2002; Xu et al. 2007) are required for p21 expression, so the relationship with p53 and SWI/SNF components is intricate.
The Mi-2/NuRD complex is interesting because a component of this complex, PID (also known as metastasis-associated protein 2, MTA2), significantly restricts a p53 response by preventing acetylation of p53. The Mi-2/NuRD complex is an atypical chromatin-remodeling complex in that it has both histone deacetylase and chromatin remodeling activities in the same complex (Denslow and Wade 2007). The Mi-2/NuRD complex is composed of the HDAC1 and HDAC2 histone deacetylases and the CHD3 (Mi-2α) and CHD4 (Mi-2β) chromatin remodeling components, among other proteins (Denslow and Wade 2007). The CHD3 and CHD4 ATPases are chromodomain-containing proteins in the SNF2 family of proteins, which includes the SWI/SNF ATPases BRG1 and BRM (Eisen et al. 1995; Woodage et al. 1997; Denslow and Wade 2007). The PID component of the Mi-2/NuRD complex binds to the p53 amino terminus between amino acids 1–80, and binding is abolished when the first TAD of p53 (amino acids 22, 23) is inactivated (Luo et al. 2000). This finding is intriguing because theoretically the Mi-2/NuRD complex could be recruited by p53 to remodel chromatin and then deacetylate p53 to terminate the p53 transcription activation cycle.
p53 is known to direct transcription through interaction with the Mediator complex (Zhang et al. 2005; Meyer et al. 2010). The Mediator complex was initially identified as a group of thyroid hormone receptor–associated proteins (TRAPs) that function as gene-specific transcriptional coactivators (Fondell et al. 1996). The pre-initiation complex is composed of RNA polymerase II, the Mediator complex, and the general TFs TFII-A, -B, -D, -E, -F, and -H (Fondell et al. 1996; Esnault et al. 2008). The human Mediator complex was suggested to interact directly with TFIID in the process of forming the pre-initiation complex (Johnson et al. 2002). In yeast, Med11 is required to recruit TFIIH and TFIIE to the pre-initiation complex that leads to serine 5 phosphorylation of the RNA Pol II CTD (Esnault et al. 2008). Mediator has also been described to interact directly with the unmodified RNA Pol II CTD, which causes the Mediator complex to adopt a specific CTD-bound conformation (Naar et al. 2002). p53 has been reported to interact with Mediator components (Gu et al. 1999), including Med17 (TRAP80) (Gu et al. 1999) and MED1 (RB18A) (Drane et al. 1997; Meyer et al. 2010). Med17 interacts with p53 TAD1 (Gu et al. 1999) and Med1 interacts with the p53 CTD (mapped to residues 363–393) (Meyer et al. 2010). Interestingly, increasing titrations of Med1 lead to decreased p53-dependent p21 expression and increased p53-dependent Bax expression (Frade et al. 2000) and increased MDM2 expression (Frade et al. 2002). Notably, the D5 domain of Med1 has been reported to interact with mutant p53 in Raji lymphoma cells (R213Q, Y234H) (Lottin-Divoux et al. 2005).
Arginine methyltransferases PRMT1 and CARM1 have been implicated in p53 transcriptional activation (An et al. 2004). PRMT1 and CARM1 bind directly to the p53 amino terminus (in a region encompassing TAD1) and carboxyl terminus (OD and CTD), respectively, and cooperate with p300 to stimulate transcription of the p53 target gene GADD45A (An et al. 2004). Histone methylation marks may serve dual roles to recruit co-activators that recognize those sequences and by indirectly enhancing acetylation by blocking histone deacetylases (Nishioka et al. 2002) including the NuRD complex (Zegerman et al. 2002). PRMT1 has been described to modify histones in a manner that permits them to be subsequently modified to transcriptionally active modification modes (Huang et al. 2005), and PRMT1 is known to cooperate with CARM1 to mediate gene expression (Hassa et al. 2008).
The amino terminus of p53 (involving the TAD residues 22 and 23) interacts with p300 (Gu et al. 1997). p300 is a histone acetyltransferase that serves to regulate p53 through direct acetylation (Avantaggiati et al. 1997; Gu and Roeder 1997; Lill et al. 1997). Acetylation of the p53 CTD correlates with increased acetylation of histones H3/H4 and increases the interaction of p53 with the p300 homolog CBP (Barlev et al. 2001). Interestingly, at the p21 promoter it has been reported that p300 does not function in transcriptional coactivation through acetylation of p53 (Espinosa and Emerson 2001). Rather, p53 was found to recruit p300 to acetylate nucleosomal histones to mediate transcriptional activation (Espinosa and Emerson 2001; see also An et al. 2004). Acetylated histone lysine residues are known binding and activating marks for other transcriptional components, especially those that possess bromodomains, including SWI/SNF (Agalioti et al. 2002), which functions in an ATP-dependent manner to reorganize chromatin to allow the binding of TFs (Kwon et al. 1994).
It can be surmised that at the p21 locus p53 binds to its response element and recruits an acetyltransferase, such as p300, to acetylate key histone residues that promote nucleosomal remodeling complex activity to reposition nucleosomes, which facilitates the recruitment of other transcriptional components that culminate in the formation of the RNA Pol II preinitiation complex. Although this is a simplified model, these key ideas—specifically, that wild-type p53 binds to its response element, recruits chromatin-modifying proteins that modify both p53 and/or neighboring histones and chromatin-associated proteins to stimulate subsequent transcriptional processes, such as additional coactivator recruitment—will serve as a guide to understand mutant p53 gain-of-function, whereby one of the most critical actions of mutant p53 may be the recruitment of transcriptional regulators to specific promoters to impact transcription.
MUTANT p53 GAIN OF FUNCTION
It is firmly established that there is a pro-oncogenic gain-of-function role for mutant p53. Li–Fraumeni patients with missense mutations are associated with a 9-year earlier tumor onset than patients with other mechanisms of p53 haploinsufficiency (Bougeard et al. 2008). Mice engineered with haploinsufficient mutant p53 (p53+/mut) have accelerated tumor growth, increased tumor count, altered tumor spectrum, and increased metastases than mice with true haploinsufficiency (p53+/–) (Dittmer et al. 1993; Liu et al. 2000; Lang et al. 2004; Olive et al. 2004; Caulin et al. 2007). Critically, p53mut/– mice spontaneously acquire a variety of additional carcinomas in addition to the tumors associated with p53−/– mice, defining an in vivo gain-of-function phenotype for mutant p53 (Olive et al. 2004).
It is generally appreciated that the majority of mutant p53 gain-of-function effects derive from the ability of mutant p53 to affect transcription of a variety of genes. Indeed, mutant p53 transcriptional effects have been shown to lead to increased cell proliferation (Preuss et al. 2000; Strano et al. 2002; Scian et al. 2004; Bossi et al. 2006, 2008; Yan et al. 2008; Haupt et al. 2009; Yan and Chen 2009; Freed-Pastor et al. 2012), resistance to apoptosis (Bossi et al. 2008; Lim et al. 2009), which can be mediated through mutant p53 interaction with Ets-2 (Do et al. 2012), increased migration (Adorno et al. 2009; Weissmueller et al. 2014), increased invasion through Matrigel (Muller et al. 2009, 2013), increased tumor inflammation (Scian et al. 2005), and increased metastases (Strano et al. 2002; Weissmueller et al. 2014).
In mouse models, mutant p53 is associated with increased rates of metastasis (Pohl et al. 1988; Hsiao et al. 1994; Heinlein et al. 2008; Adorno et al. 2009; Weissmueller et al. 2014). This effect was initially identified in 1988 (Pohl et al. 1988) and was defined to be missense mutant specific by using xenotransplants of leukemia cells expressing different missense p53 mutations (Hsiao et al. 1994). Leukemia cells (Be-13 cells) expressing p53 R175H, R248Q, and R213Q were able to disseminate to distant sites and induce further hematological disease, whereas the same cells expressing Y234H and R273C were unable to metastasize (Hsiao et al. 1994). The missense mutations that resulted in disseminated disease correlated with decreased survival, and these mice underwent rapid death following the mutant p53-mediated metastases (Hsiao et al. 1994). In a mouse model of invasive breast cancer, the mouse equivalent of p53 R273H (R270H) increases the rate of lung metastases (Heinlein et al. 2008). R248Q and G245S have been shown in mouse models to confer different abilities to hasten tumorigenesis, with the R248Q mutant having accelerated tumor onset and shorter survival compared with G245S mice (Hanel et al. 2013).
Mutant p53 has been shown in mouse models to impair p63- and p73-mediated transcription (Lang et al. 2004). Two more recent studies describe how mutant p53 can impede p63 (Adorno et al. 2009) or p73 function to promote metastasis (Weissmueller et al. 2014). Contact mutant p53 R280K (endogenously expressed in MDA-231 breast cancer cells) were studied in xenotransplants. Twenty-one out of 22 mice with unaltered levels of mutant p53 had lymph node positivity, whereas 12 out of 22 mice showed lymph node positivity when mutant p53 was depleted (Adorno et al. 2009). This correlated with the number of micrometastases to the lung, as depletion of mutant p53 resulted in 75%–90% (depending on the clone) reduction in micrometastases (Adorno et al. 2009). Furthermore, when R175H was overexpressed in the presence of depleted R280K, the number of lung micrometastases was increased back to normal levels, demonstrating that both contact (R280K) and conformational (R175H) mutants can mediate this metastatic phenotype (Adorno et al. 2009). This study further describes how Smad3 (a component of transforming growth factor [TGF]-β signaling) cooperates with mutant p53 to oppose p63 action (which impedes metastatic spread to the lung) (Adorno et al. 2009). A second study defined how a p73/NF-Y complex repressed PDGFRβ expression in a mouse model of pancreatic cancer (Weissmueller et al. 2014). Pancreatic cancer cell lines were derived from mice engineered with the mouse equivalent of p53 R175H (R172H). These cell lines have been previously defined and also express constitutively active K-Ras under tissue specific control (Hingorani et al. 2005). These cells were analyzed in the presence and absence of p53 R172H. Following orthotopic injection into the pancreas, metastasis to the lung and liver are significantly reduced when mutant p53 is depleted (Weissmueller et al. 2014). Mutant p53 interaction with p73 was found to inhibit p73 interaction with NF-YB, allowing NF-Y to activate PDGFRβ expression that is critical to the metastatic phenotype of mutant p53 in these cells (Weissmueller et al. 2014). Hence, it is well established that mutant p53 promotes metastasis in mouse models, which is consistent with the observations that mutant p53 correlates with worse survival in human cancers (Elledge et al. 1993; Olivier et al. 2006; Langerod et al. 2007; Petitjean et al. 2007; Alsner et al. 2008).
Mutant p53 is known to promote NF-κB signaling through tumor necrosis factor (TNF)-α (Weisz et al. 2007). This observation was especially interesting because mutant p53 is documented to impact the expression of genes in the NF-κB pathway that lead to increased cell growth and survival (Scian et al. 2005). Mutant p53 has recently been shown through cytoplasmic interaction with DAB2IP to alter TNF-α downstream signaling by promoting NF-κB and dampening ASK1/JNK signaling, respectively, suggesting a role for mutant p53 in harnessing the inflammatory effects of the immune system to promote tumor development and metastases (Di Minin et al. 2014). Hotspot p53 mutants have also been reported to contribute to chronic inflammation that underlies the development of colorectal cancer by cooperation with NF-κB (Cooks et al. 2013). This suggests that early p53 mutation can promote tumor development in early stages of tumor progression (Cooks et al. 2013) as well as promote invasiveness and metastases in later stages (Di Minin et al. 2014). Inflammation has a multifaceted role in cancer (reviewed in Grivennikov et al. 2010), and it is worth considering that novel immunomodulatory agents, such as PD1, PDL1, or CTLA4 checkpoint inhibitors (reviewed in Pardoll 2012), could be effective in mutant p53-containing tumors, especially in conjunction with radiotherapy.
These studies identify a variety of mechanisms through which mutant p53 promotes oncogenesis. As each tumor is unique in its development, it should be considered at this point that mutant p53 may be a promiscuous transcription factor that is used by the tumor cell–based on its specific mutations, dominant signaling pathways, and interaction with the microenvironment—in a manner that is selectively advantageous. The next section will summarize what is known about mutant p53-mediated transcription with a focus on specific mutant p53 interacting partners and transcriptional targets.
TRANSCRIPTIONAL ACTIVATION BY MUTANT p53
It remains to be fully understood how mutant p53 mechanistically affects transcription. Mutant p53 does not directly bind to a consensus DNA sequence (Bargonetti et al. 1991, 1993; Kern et al. 1991; el-Deiry et al. 1992). Rather, it is likely that mutant p53 through its individual domains associates with other transcription factors, histone-modifying machinery, or the transcription initiation complex to promote transcription (Fig. 1B; summarized in Table 2).
Table 2.
Transcription factors (TFs) that interact with mutant p53
| Protein | Interaction with wild-type p53 | Mutant | Study |
|---|---|---|---|
| Ets-1 | Yes | V143A, D281G, CTD may be required | Sampath et al. 2001; Kim et al. 2003; Kim and Deppert 2007; Strano et al. 2007; Do et al. 2012 |
| Ets-2 | Yes | Do et al. 2012 | |
| Sp1 | Yes | V134A, R175H, R249S, R273H, CTD/OD required for wt p53 interaction | Gualberto and Baldwin 1995; Bargonetti et al. 1997; Chicas et al. 2000; Lee et al. 2000; Torgeman et al. 2001; Koutsodontis et al. 2005; Hwang et al. 2011 |
| NF-Y | Yes | R175H, R273H, R273C, CTD required for binding to wt p53 | Imbriano et al. 2005; Di Agostino et al. 2006; Liu et al. 2011 |
| VDR | Yes | R175H, interaction does not occur in 1–292 amino acid mutant | Stambolsky et al. 2010 |
| SMADs (2/3, maybe 4) | Unclear | 175H, 273H, TAD likely required | Wilkinson et al. 2008; Adorno et al. 2009; Ji et al. 2015 |
| E2F1 | Yes | E2F1 binds wt p53 carboxyl terminus, 175H and perhaps 280K recruit E2F1 to CDE consensus sequence | Fogal et al. 2005; Fontemaggi et al. 2009 |
| TBP | Yes | mTAD1 and 2 required for wt p53 | Seto et al. 1992; Ragimov et al. 1993; Chang et al. 1995; Lee et al. 2000 |
| p63 | No | R175H, Y220C, R248W, R273H (not D281G), interaction may not require TAD | Davison et al. 1999; Gaiddon et al. 2001; Strano et al. 2002; Adorno et al. 2009 |
| p73 | No | R175H, Y220C, V143A, R248W (not R273H), interaction may not require TAD | Davison et al. 1999; Di Como et al. 1999; Marin et al. 2000; Gaiddon et al. 2001; Bensaad et al. 2003; Di Agostino et al. 2008; Oren and Rotter 2010 |
| MED1 | Yes | R213Q, Y234H | Drane et al. 1997; Lottin-Divoux et al. 2005; Meyer et al. 2010 |
| p300 | Yes | R175H | Avantaggiati et al. 1997; Di Agostino et al. 2006; Ali et al. 2013 |
Summary of known transcription factors (TFs) or chromatin modifiers for which there is a reported interaction with mutant p53. Whether or not wild-type (wt) p53 is known to interaction with the same factor is listed. Known domain-specific TF interactions with wild-type p53 may predict domain interactions with mutant p53. Note that NF-κB may interact with mutant p53 (Solomon et al. 2012). The table is an extension of a similar table prepared by Freed-Pastor and Prives (2012).
Mutant p53 has been shown to interact with the following transcription factors: p53 homologs p63 (Davison et al. 1999; Gaiddon et al. 2001; Strano et al. 2002; Adorno et al. 2009) and p73 (Davison et al. 1999; Di Como et al. 1999; Marin et al. 2000; Gaiddon et al. 2001; Bensaad et al. 2003; Di Agostino et al. 2008; Oren and Rotter 2010), Sp1 (Gualberto and Baldwin 1995; Bargonetti et al. 1997; Chicas et al. 2000; Lee et al. 2000; Torgeman et al. 2001; Koutsodontis et al. 2005; Hwang et al. 2011), Smad2 (Adorno et al. 2009), Smad3 (Adorno et al. 2009; Ji et al. 2015), NF-Y (Imbriano et al. 2005; Di Agostino et al. 2006; Liu et al. 2011), E2F1 (Fontemaggi et al. 2009), Ets-1 (Sampath et al. 2001; Kim et al. 2003; Kim and Deppert 2007; Strano et al. 2007; Do et al. 2012), Ets-2 (Do et al. 2012), and the Med1 component of the Mediator complex (Drane et al. 1997; Lottin-Divoux et al. 2005; Meyer et al. 2010) (see Table 2). It is presumed that mutant p53 depends on these interactions (and others not yet identified) to mediate the transcription of numerous genes to mediate its gain-of-function effects.
The list of mutant p53-regulated genes, most of which have shown a functional purpose, is extensive and includes ABCB1 (MDR) (Chin et al. 1992; Lin et al. 1995; Strauss and Haas 1995; Sampath et al. 2001; Bush and Li 2002), the GRO1 chemokine (Yan and Chen 2009), PCNA (Deb et al. 1992), the ID2 transcription regulator (Yan et al. 2008), the ID4 transcription regulator (Fontemaggi et al. 2009), the hsMAD1 mitotic spindle checkpoint protein (Iwanaga and Jeang 2002), 15-lipoxygenase (Kelavkar and Badr 1999), the galectin-3 antiapoptotic protein (Lavra et al. 2009), insulin-like growth factor II (Lee et al. 2000), insulin-like growth factor receptor I (Werner et al. 1996), epidermal growth factor receptor (Ludes-Meyers et al. 1996), c-Fos (Preuss et al. 2000), NF-κB2 (Scian et al. 2005; Weisz et al. 2007; Cooks et al. 2013), c-Myc (Frazier et al. 1998), the EGR1 transcription factor (Weisz et al. 2004), mitogen-activated protein kinase kinase 3 (Gurtner et al. 2010), asparagine synthetase (Scian et al. 2004), human telomerase reverse transcriptase (Scian et al. 2004), the stathmin microtubule-destabilizing protein (Singer et al. 2007), matrix metalloproteinase 13 (Sun et al. 2000), Pla2g16 phospholipase (Xiong et al. 2014), genes of the mevalonate pathway (Freed-Pastor et al. 2012), genes of nucleotide metabolism (Kollareddy et al. 2015), as well as numerous others (Table 3).
Table 3.
Known mutant p53 target genes
| Gene | Cell line | Mutant | Method | Study |
|---|---|---|---|---|
| RhoGDI α | H1299, SKBR3, HT29 | 175H, 175H, 273H | Overexpression, qPCR, siRNA | Bossi et al. 2008 |
| RANGAP1 | H1299, SKBR3, HT29 | 175H, 175H, 273H | Overexpression, qPCR, siRNA | Bossi et al. 2008 |
| RAB6KIFL | H1299, SKBR3, HT29 | 175H, 175H, 273H | Overexpression, qPCR, siRNA | Bossi et al. 2008 |
| Seladin1 (DHCR24) | H1299, SKBR3, HT29, MDA-MB-468 | 175H, 175H, 273H | Overexpression, qPCR, siRNA | Bossi et al. 2008; Freed-Pastor et al. 2012 |
| MAP2K3 (no TATA) | H1299, SKBR3, HT29/also MDA-MB-468, MDA-MB-231 | 175H, 175H, 273H/ 280K, regulated by NF-κB, NFY | Overexpression, qPCR, siRNA/overexpression (175/273), reporter assay, siRNA, ChIP | Bossi et al. 2008; Gurtner et al. 2010 |
| IGFR1 | H1299, SKBR3, HT29, Saos-2, RD, HeLa | 175H, 175H, 273H, 143A, but wild-type (wt) p53 suppresses | Overexpression, qPCR, siRNA, reporter assay | Werner et al. 1996; Bossi et al. 2008 |
| Paxillin β | SKBR3, HT29 | 175H, 273H | qPCR, siRNA | Bossi et al. 2008 |
| BCL2L1 | SKBR3, HT29 | 175H, 273H | qPCR, siRNA | Bossi et al. 2008 |
| MDR1 | NIH3T3, Saos-2, Caco-2, BHK | 175H, 281G, requires TAD residues 14/19; requires ETS-1 site; Del22/23 blocks Ets binding, Del360CTD still have Ets binding to 281G | Overexpression-CAT reporter assay, reporter assay | Chin et al. 1992; Lin et al. 1995; Strauss and Haas 1995; Candau et al. 1997; Sampath et al. 2001 |
| PCNA | HeLa, Saos-2 | V143A, R175H, R248W, R273H, D281G | Overexpression-CAT reporter assay | Deb et al. 1992 |
| CCNA2 | SKBR3, HT29, SW480, H1299 | 175H, 273H, 273H/309S; induction by adriamycin inhibited | WB, ChIP, reporter assay | Di Agostino et al. 2006 |
| CCNB1 | SKBR3, HT29, SW480 | 175H, 273H, 273H/309S; induction by adriamycin inhibited | WB, ChIP | Di Agostino et al. 2006 |
| CCNB2 | SKBR3, HT29, SW480, H1299-281G | 175H, 273H, 273H/309S, 281G; induction by adriamycin inhibited | WB, ChIP, overexpression, reporter Assay | Di Agostino et al. 2006 |
| CDK1 | SKBR3, HT29, SW480 | 175H, 273H, 273H/309S; induction by adriamycin inhibited | WB, ChIP | Di Agostino et al. 2006 |
| CDC25C | SKBR3, HT29, SW480, H1299-281G | 175H, 273H, 273H/309S, 281G; induction by adriamycin inhibited | WB, ChIP, overexpression, reporter assay | Di Agostino et al. 2006 |
| ID4 | H1299, SKBR3 | 175H, 273H, Sp1 and NF-κB implicated | Overexpression, gene array, ChIP, EMSA | Fontemaggi et al. 2009 |
| c-Myc | Cx3Ras (rat), (10)1 mouse, Saos-2 | 143A, 281G, 175H, 273H | Overexpression, reporter assay | Frazier et al. 1998 |
| CXCL5, CXCL8, CXCL12 | H1299, MDA-MB-231, MDA-MB-435 | 175H, 273H, 281G | Overexpression, siRNA | Yeudall et al. 2012 |
| ACAT2, HMGCS1, HMGCR, PMVK, MVD, IDI1, FDPS, SQLE, LSS, CYP51A1, SC4MOL, DHCR7 | MDA-MB-468 | 273H | siRNA, qPCR, +/−ChIP |
Freed-Pastor et al. 2012 |
| MVK, FDFT1, TM7SF2, NSDH | MDA-MB-468, MDA-MB-231 | 273H, 280K | siRNA, qPCR, +/−ChIP |
Freed-Pastor et al. 2012 |
| TGF-β receptor 2 | H1299 | 175H | Overexpression, reporter Assay | Kalo et al. 2007 |
| EPB41L4B, BUB1, MIS18A (C21orf45), NCAPH, CENPA, FAM64A, DEPDC1, CCNE2 | MDA-MB-231, MDA-MB-468 | 280K, 273H | siRNA, qPCR, ChIP | Girardini et al. 2011 |
| CPSF6, WDR67 | MDA-MB-231 | 280K | siRNA, qPCR, ChIP | Girardini et al. 2011 |
| hsMAD1 | HeLa, HCT116 | 281G, not 143A | Overexpression, reporter assay | Iwanaga and Jeang 2002 |
| Galectin-3 | Saos-2, SW-1736, ARO | 273H | Overexpression | Lavra et al. 2009 |
| EGFR | Saos-2 | V143A, R175H, R248W, R273H, D281G, and also wt p53 | Overeexpression, reporter assay | Ludes-Meyers et al. 1996 |
| Fos | Saos-2 | C174Y | Reporter assay | Preuss et al. 2000 |
| dUTPase (DUT) (TAD1 required) | SK-OV-3 and 10(1) | 175H, 248W, 273H | Overexpression, qPCR/northern | Pugacheva et al. 2002 |
| TIM50 (Ets-1, CREB target) | H1299, SKBR3, MDA-MB-468, Saos-2 | 175H, 273H, 281G | Overexpression, siRNA, reporter assay, ChIP | Sankala et al. 2011 |
| Asparagine synthetase | H1299, 10(3), Saos-2 | 143A, 157F, 163C, 175H, 179Y, 194R, 273H, 281G, 282W | Overexpression, reporter assay, ChIP-273H | Scian et al. 2004 |
| hTERT | H1299, 10(3), Saos-2 | 143A, 157F, 163C, 175H, 179Y, 194R, 273H, 281G, not 282W | Overexpression, reporter assay, ChIP-273H | Scian et al. 2004 |
| NF-κB2 | H1299, 21PT, Saos-2 | 175H, 273H, 281G, 175H increased NF-κB activity | Gene expression array with ectopic expression, qPCR, reporter assay, EMSA | Scian et al. 2005 |
| EBAG9, ITGA6, E2F5, MCM6, C-SYN | H1299 | 281G | Gene expression Array with ectopic expression, qPCR | Scian et al. 2005 |
| Stathmin | Huh-7, HepG2, U138-MG | 213Q, 220C, not wt p53 | Knockdown, WB | Singer et al. 2007 |
| IGFBP3 and CYP24A1 (VDR target genes) | SKBR3, H1299, SW480, MDA-MB-231 | 175H, 273H | ChIP-on-chip, overexpression, Southwestern blot, reporter assay | Stambolsky et al. 2010 |
| hMMP-13 | Saos-2 | 175H, 281G | Overexpression, WB, reporter assay | Sun et al. 2000 |
| EGR1 | H1299, PC3, SKBR3, HeLa | 175H, 179E, 248W, 273H, 281G, not wt p53 | Overexpression, gene expression array, ChIP, reporter assay, VEGF-induction by EGR1 | Weisz et al. 2004 |
| CXCL1 (GRO1) | SW480, MIA-PaCa-2, HCT116 | 273H/309S, 248W, 175H, not wt p53 | siRNA, ectopic expression, ChIP | Yan and Chen 2009, 2010 |
| Id2 (inhibition by mutant p53) | SW480, MIA-PaCa-2, HCT116 | 273H/309S, 248W, 175H, not wt p53 | siRNA, ectopic expression, ChIP | Yan et al. 2008; Yan and Chen 2010 |
| REGγ | H1299, UMSCC-1 | 175H, 248W, 273H; showed mutant p53 cooperation with p300 and negative regulation of Smad3 | Ectopic expression, reporter assays, ChIP | Ali et al. 2013 |
| Axl | H1299, H1437, H1048 | 175H, 267P, 273C, 273H, 273P, 281G; used mTAD1 mutants | siRNA, qPCR, +/−ChIP |
Vaughan et al. 2012 |
| VEGFR2 | MDA-MB-231, MDA-MB-468 | 175H, 245S, 248W, 273H, 280K | siRNA, qPCR, +/−ChIP |
Pfister et al. 2015 |
| PDGFRβ | KPC mouse cells, MiaPaCa-2, BXPC3, CFPAC, A2.1, SW620, H1975, MBA-MB-231 | 155P, 172H (mouse), 220C, 242R, 248R, 280K | sIRNA, qPCR, +/−ChIP |
Weissmueller et al. 2014 |
| Nucleotide metabolism genes | HCC38, BT549, MDA-MB-231, MiaPaCa-2, H1299 | 175H, 248W, 249S, 273L, 280K | Ectopic expression, siRNA, qPCR, +/–ChIP |
Kollareddy et al. 2015 |
| Pla2g16 | H76 (mouse), Saos-2 | 175H, 179R, 245S, 248Q, 273H | Ectopic expression, siRNA, qPCR, +/–ChIP |
Xiong et al. 2014 |
Listing of known mutant p53-regulated genes (primarily resulting in increased expression), the cell lines in which the gene was found to be regulated, the p53 mutants that were used, the experiment system that was used, and the associated references. Note mutant p53 represses other genes, such as CD95 (Fas receptor) (Gurova et al. 2003; Zalcenstein et al. 2003), ATF3 (a CREB TF family protein) (Buganim et al. 2006), TGF-β receptor 2 (Kalo et al. 2007), caspase 3 (Wong et al. 2007), Id2 (Yan et al. 2008), and wild-type p53 target genes p21, GADD45, PERP, and PTEN (Vikhanskaya et al. 2007).
Mutant p53 has been reported to stimulate expression of multiple receptor tyrosine kinases and other signaling components. It is well described that receptor tyrosine kinases promote pro-proliferative signaling (Lemmon and Schlessinger 2010). Common signaling nodes can be engaged by multiple receptor tyrosine kinases, even though the output is different; for example, the same signaling pathway can shift from promoting a differentiated state (common to normal cells) to a pro-proliferative state (Marshall 1995; Lemmon and Schlessinger 2010). It is conceivable that mutant p53 may affect cell signaling pathways to promote dedifferentiation of tumor cells. Mutant p53 has been reported to stimulate multiple receptor tyrosine kinases, including EGFR (Ludes-Meyers et al. 1996), IGF1R (Werner et al. 1996), Axl (Vaughan et al. 2012), MET (Muller et al. 2013), PDGFRβ (Weissmueller et al. 2014), and VEGFR2 (Pfister et al. 2015), all of which promote pro-proliferative signaling. In the case of EGFR and MET, this activation has been shown to be dependent on Rab-coupling protein, which increases recycling of these receptor tyrosine kinases to enhance their signaling outputs (Muller et al. 2009, 2013). It is worth considering that as a tumor forms, acquisition of a hotspot mutation in TP53 may facilitate transcriptional plasticity, whereby tumor cells increase capacity for gene expression changes and, therefore, undergo selection for the greatest pro-proliferative transcriptional program for the particular tumor context. This hypothesis explains, for instance, why such a wide array of genes and pathways has been reported to mediate mutant p53 gain of function.
Mutant p53 regulation of all of these genes varies based on the specific p53 mutant and the cell line. Notably, many of these genes were studied by overexpressing mutant p53 in p53-null cell backgrounds (indicated in the table as overexpression) and often with reporter assays (Table 3). Reporter assays are inadequate to study mutant p53-dependent transcription because they lack the full complexity of chromatin, and mutant p53 does not bind to a DNA response element. Although overexpressing mutant p53 in p53-null cell backgrounds may be adequate to study mutant p53 gain-of-function effects because there is no impact of wild-type p53, this approach is imperfect because the cell lines underwent selective changes without regard to the selective advantages conferred by mutant p53. Such selective changes—the tumor evolving to harness and depend on mutant p53 for its proproliferative or other capacities—must impact how mutant p53 functions in a particular tumor. Therefore, we suggest that cell culture studies involving mutant p53 be performed using cell lines that express single-point mutations in one allele of p53 and have undergone loss of heterozygosity (for example, see Freed-Pastor et al. 2012; Pfister et al. 2015).
Tumor-specific genetic alterations, cellular metabolism, chromatin landscape, and the availability of specific TFs affect mutant p53 function (Kim and Deppert 2003, 2004; Strano et al. 2007; Li et al. 2008; Adorno et al. 2009; Haupt et al. 2009; Dell’Orso et al. 2011; Rodriguez et al. 2012). It follows that one p53 mutant may be observed to behave differently than another p53 mutant based on changes in conformation, binding partners, cellular localization, and transactivation capability. Indeed, mutant p53 gain of function depends on cell and tumor context. Note that posttranslational modifications, nucleosome state, quarter-site orientation and spacer length of the p53 response element, and cofactors affect wild-type p53 gene regulation (Riley et al. 2008). Posttranslational modifications, nucleosome state, and cofactors likely impact mutant p53 transcription as well.
These observations (in addition to the fact that mutant p53 does not bind to the p53 response element) may explain why wild-type p53 and mutant p53, despite binding to many of the same TFs (Table 2) differentially affect gene expression. Mutant p53 can even lead to diametric outcomes of gene expression, even though mutant p53 can still cooperate with wild-type p53 coactivators, such as the histone acetyltransferase p300 (Avantaggiati et al. 1997; Di Agostino et al. 2006). For instance, mutant p53, when located at genes that wild-type p53 activates, such as p21, GADD45, PERP, and PTEN, leads to their repression (Vikhanskaya et al. 2007). It is not immediately obvious why mutant p53 and wild-type p53 lead to diametric gene expression outcomes when they can interact with similar subsets of TFs and coactivators. One possibility is that wild-type p53 has interactions with the same transcription factors that mutant p53 binds, but when overexpressed or activated, wild-type p53 retains preference to its response element while mutant p53 is brought to different promoters (or in the case of wild-type p53-regulated genes, different sites within the promoter) by the transcription factor interactions leading to gene dysregulation.
In the following paragraphs, we speculate on possible mechanisms of mutant p53 transcriptional activity. Mutant p53 likely mediates transcription by co-opting sets of TFs to initiate gene activation at the transcription factor’s location, as it is likely the transcription factor is DNA-bound. Coactivators recruited by the transcription factor or mutant p53 then stimulate gene expression. The extent that mutant p53 co-opts individual TFs (or other chromatin modulators) for target gene activation is unclear and may be dependent on the specific mutation in p53 and the active cell signaling pathways leading to subsets of active TFs in the cell. It is also possible that mutant p53, following recruitment by a transcription factor or chromatin modulator, recruits additional factors that can stimulate the function of the initial recruiting factor. It is possible that there are mechanisms for mutant p53 to change the state of chromatin, and these mechanisms may rely on mutant p53 recruitment to DNA through transcription factor binding followed by a change in chromatin architecture by known and unknown chromatin-modifying machinery that are recruited by the mutant p53–TF complex.
TFs may not need to be active to be recruited by mutant p53, although mutant p53 may need to be modified in a specific manner. Mutant p53 engagement may lead to conformational change of an inactive TF to a conformation that can bind DNA. In this mechanism, the presence of mutant p53 in a cell leads to transcription factor engagement by mutant p53. If these TFs were inactive and cytoplasmic, mutant p53 presence would shift their localization to the nucleus, forming a complex with mutant p53, the engaged transcription factor, and a coactivator such as p300. Indeed, cell context, perhaps through mutant p53 modifications, can alter mutant p53-coactivator binding (Di Agostino et al. 2006). Promoters known to be engaged by NF-Y and mutant p53 shift from containing the repressive HDAC1 to the activating p300 on DNA damage by doxorubicin (Di Agostino et al. 2006). Additionally, serine-6 and serine-9 phosphorylations are required for mutant p53-Smad binding (Cordenonsi et al. 2007; Adorno et al. 2009). Mutant p53 contact mutants can cooperate through NF-κB signaling to increase mutant p53 target gene activation (Solomon et al. 2012). It should also be considered that mutant p53 may refine transcription factor-binding specificities, a process termed latent specificity (a cofactor-induced change in DNA recognition) (Slattery et al. 2011). Significantly, mutant p53 impacts the expression of transcription factors such as NF-κB2 (Scian et al. 2005; Weisz et al. 2007; Cooks et al. 2013), c-Fos (Preuss et al. 2000), c-Myc (Frazier et al. 1998), EGR1 (Weisz et al. 2004), ID2 (Yan et al. 2008), ID4 (Fontemaggi et al. 2009), as well as chromatin modulators such as the MLL1 and MLL2 histone methyltransferases (Zhu et al. 2015) and the MOZ histone acetyltransferase (Zhu et al. 2015), all of which could influence mutant p53 transcriptional processes.
We recently showed that mutant p53 promotes SWI/SNF chromatin-remodeling complex activity (Pfister et al. 2015). Further, our results show that mutant p53 and SWI/SNF function to mediate nucleosomal remodeling at the VEGFR2 promoter (Pfister et al. 2015). Mutant p53 and SWI/SNF coordinately repress or increase transcription at approximately half of mutant p53 target genes (Pfister et al. 2015). As mutant p53 and wild-type p53 often mediate opposing effects on their interacting partners, in theory, mutant p53 could dysregulate normal SWI/SNF complex functions that wild-type p53 requires for transcriptional activities (Lee et al. 2002; Xu et al. 2007) by affecting its activity, interaction with other proteins, or chromosomal positioning. Mutant p53 has been proposed to facilitate transcriptional plasticity (Quante et al. 2012), and functional interaction of mutant p53 with a chromatin-remodeling complex like SWI/SNF that has broad genomic distribution (Euskirchen et al. 2011) may explain the ability of mutant p53 to mediate gene expression at multiple loci (see Fig. 1B). Because nucleosomal positioning is a critical factor in gene regulation, promoting or inhibiting transcription by regulating access to DNA-binding proteins (Wilson and Roberts 2011), mutant p53 interaction with SWI/SNF could mediate gene activation and repression (Pfister et al. 2015).
It must be emphasized that the mechanism of mutant p53-transcription factor and mutant p53-coactivator interaction is not well understood, and it is these interactions culminating in transcriptional changes that likely represent the majority of mutant p53 gain-of-function effects (versus cytoplasmic or other nontranscriptional mechanisms). This supposition is predicated on experimental data and that the majority of wild-type p53 effects are transcriptional. To understand mutant p53 gain-of-function transcriptional effects, the mechanisms for mutant p53 interactions with transcription factors, coactivators, and other chromatin regulators required for gene expression changes must be delineated. Novel coactivators and corepressors must be identified that could account for mutant p53 transcriptional outcomes. Future drug discovery targets rely entirely on the previous point—if a factor is found that is required for mutant p53 gene transactivation, such as a novel coactivator, then a drug could be developed that would be highly specific for the mutant p53–coactivator complex that would only exist in cells expressing mutant p53 gain-of-function mutants.
CONCLUDING REMARKS
We are headed toward molecular medicine whereby specific TP53 mutations, combined with the other characteristics of that tumor, will determine clinical response. Knowing whether a tumor is TP53 wild type or mutated is incomplete information. TP53 wild-type tumors may or may not predictably function as if they expressed wild-type p53 depending on other aspects of that tumor. Likewise, all mutant p53 proteins are not alike. Hotspot mutants affect transcription in ways that are currently unpredictable, although common mechanisms of action could lead to new drug candidates. Pharmaceuticals that can target specific epigenetic machinery (and chemical modifications of such machinery) are likely to be powerful anticancer drugs in the future. Defining tumor-specific protein interactions, such as mutant p53 interactions with other proteins, are likely to be more therapeutically viable as the targeted interaction would only exist within the tumor. Signaling pathways that are activated by mutant p53 are also candidates for anticancer drugs. Defining the transcriptional changes mediated by mutant p53 in tumors is fundamental to the classification and treatment of tumors harboring mutant p53 and provides insight to the mechanism of mutant p53 target gene activation. Ultimately, we hope work in this direction furthers the goal of defining common vulnerabilities in mutant p53-expressing tumors that are common to multiple types of p53 alterations (so treatment options are simplified) that significantly enhance patient survival.
ACKNOWLEDGMENTS
We thank Oleg Laptenko for critically reading this manuscript. We also thank the members of the Prives laboratory as well as members of the p53 research community who have stimulated provoking thoughts about these subjects. We apologize to those whose research we could not discuss because of space constraints. This work is supported by the National Cancer Institute (NCI) Program Project Grant (P01) CA 87497 to C.P.
Footnotes
Editors: Guillermina Lozano and Arnold J. Levine
Additional Perspectives on The p53 Protein available at www.perspectivesinmedicine.org
REFERENCES
- Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, Solari A, Bobisse S, Rondina MB, Guzzardo V, et al. 2009. A mutant-p53/Smad complex opposes p63 to empower TGF-β-induced metastasis. Cell 137: 87–98. [DOI] [PubMed] [Google Scholar]
- Agalioti T, Chen G, Thanos D. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111: 381–392. [DOI] [PubMed] [Google Scholar]
- Ahrendt SA, Hu Y, Buta M, McDermott MP, Benoit N, Yang SC, Wu L, Sidransky D. 2003. p53 mutations and survival in stage I non-small-cell lung cancer: Results of a prospective study. J Natl Cancer Inst 95: 961–970. [DOI] [PubMed] [Google Scholar]
- Ali A, Wang Z, Fu J, Ji L, Liu J, Li L, Wang H, Chen J, Caulin C, Myers JN, et al. 2013. Differential regulation of the REGγ-proteasome pathway by p53/TGF-β signalling and mutant p53 in cancer cells. Nat Commun 4: 2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsner J, Jensen V, Kyndi M, Offersen BV, Vu P, Borresen-Dale AL, Overgaard J. 2008. A comparison between p53 accumulation determined by immunohistochemistry and TP53 mutations as prognostic variables in tumours from breast cancer patients. Acta Oncol 47: 600–607. [DOI] [PubMed] [Google Scholar]
- An W, Kim J, Roeder RG. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117: 735–748. [DOI] [PubMed] [Google Scholar]
- Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. 1997. Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89: 1175–1184. [DOI] [PubMed] [Google Scholar]
- Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y, et al. 1989. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 244: 217–221. [DOI] [PubMed] [Google Scholar]
- Baker SJ, Preisinger AC, Jessup JM, Paraskeva C, Markowitz S, Willson JK, Hamilton S, Vogelstein B. 1990. p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res 50: 7717–7722. [PubMed] [Google Scholar]
- Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L, et al. 2012. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 486: 405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak Y, Juven T, Haffner R, Oren M. 1993. mdm2 expression is induced by wild type p53 activity. EMBO J 12: 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargonetti J, Friedman PN, Kern SE, Vogelstein B, Prives C. 1991. Wild-type but not mutant p53 immunopurified proteins bind to sequences adjacent to the SV40 origin of replication. Cell 65: 1083–1091. [DOI] [PubMed] [Google Scholar]
- Bargonetti J, Reynisdottir I, Friedman PN, Prives C. 1992. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes Dev 6: 1886–1898. [DOI] [PubMed] [Google Scholar]
- Bargonetti J, Manfredi JJ, Chen X, Marshak DR, Prives C. 1993. A proteolytic fragment from the central region of p53 has marked sequence-specific DNA-binding activity when generated from wild-type but not from oncogenic mutant p53 protein. Genes Dev 7: 2565–2574. [DOI] [PubMed] [Google Scholar]
- Bargonetti J, Chicas A, White D, Prives C. 1997. p53 represses Sp1 DNA binding and HIV-LTR directed transcription. Cell Mol Biol (Noisy-le-grand) 43: 935–949. [PubMed] [Google Scholar]
- Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell 8: 1243–1254. [DOI] [PubMed] [Google Scholar]
- Bartek J, Bartkova J, Vojtesek B, Staskova Z, Rejthar A, Kovarik J, Lane DP. 1990a. Patterns of expression of the p53 tumour suppressor in human breast tissues and tumours in situ and in vitro. Int J Cancer 46: 839–844. [DOI] [PubMed] [Google Scholar]
- Bartek J, Iggo R, Gannon J, Lane DP. 1990b. Genetic and immunochemical analysis of mutant p53 in human breast cancer cell lines. Oncogene 5: 893–899. [PubMed] [Google Scholar]
- Beckerman R, Prives C. 2010. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol 2: a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaad K, Le Bras M, Unsal K, Strano S, Blandino G, Tominaga O, Rouillard D, Soussi T. 2003. Change of conformation of the DNA-binding domain of p53 is the only key element for binding of and interference with p73. J Biol Chem 278: 10546–10555. [DOI] [PubMed] [Google Scholar]
- Boehme KA, Blattner C. 2009. Regulation of p53—Insights into a complex process. Crit Rev Biochem Mol Biol 44: 367–392. [DOI] [PubMed] [Google Scholar]
- Bossi G, Lapi E, Strano S, Rinaldo C, Blandino G, Sacchi A. 2006. Mutant p53 gain of function: Reduction of tumor malignancy of human cancer cell lines through abrogation of mutant p53 expression. Oncogene 25: 304–309. [DOI] [PubMed] [Google Scholar]
- Bossi G, Marampon F, Maor-Aloni R, Zani B, Rotter V, Oren M, Strano S, Blandino G, Sacchi A. 2008. Conditional RNA interference in vivo to study mutant p53 oncogenic gain of function on tumor malignancy. Cell Cycle 7: 1870–1879. [DOI] [PubMed] [Google Scholar]
- Bougeard G, Sesboue R, Baert-Desurmont S, Vasseur S, Martin C, Tinat J, Brugieres L, Chompret A, de Paillerets BB, Stoppa-Lyonnet D, et al. 2008. Molecular basis of the Li–Fraumeni syndrome: An update from the French LFS families. J Med Genet 45: 535–538. [DOI] [PubMed] [Google Scholar]
- Braastad CD, Han Z, Hendrickson EA. 2003. Constitutive DNase I hypersensitivity of p53-regulated promoters. J Biol Chem 278: 8261–8268. [DOI] [PubMed] [Google Scholar]
- Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, Kenzelmann Broz D, Basak S, Park EJ, McLaughlin ME, et al. 2011. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145: 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Kalo E, Brosh R, Besserglick H, Nachmany I, Rais Y, Stambolsky P, Tang X, Milyavsky M, Shats I, et al. 2006. Mutant p53 protects cells from 12-O-tetradecanoylphorbol-13-acetate-induced death by attenuating activating transcription factor 3 induction. Cancer Res 66: 10750–10759. [DOI] [PubMed] [Google Scholar]
- Burrows AE, Smogorzewska A, Elledge SJ. 2010. Polybromo-associated BRG1-associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proc Natl Acad Sci 107: 14280–14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush JA, Li G. 2002. Cancer chemoresistance: The relationship between p53 and multidrug transporters. Int J Cancer 98: 323–330. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2011. Integrated genomic analyses of ovarian carcinoma. Nature 474: 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2014. Integrated genomic characterization of papillary thyroid carcinoma. Cell 159: 676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candau R, Scolnick DM, Darpino P, Ying CY, Halazonetis TD, Berger SL. 1997. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene 15: 807–816. [DOI] [PubMed] [Google Scholar]
- Cattoretti G, Rilke F, Andreola S, D’Amato L, Delia D. 1988. P53 expression in breast cancer. Int J Cancer 41: 178–183. [DOI] [PubMed] [Google Scholar]
- Caulin C, Nguyen T, Lang GA, Goepfert TM, Brinkley BR, Cai WW, Lozano G, Roop DR. 2007. An inducible mouse model for skin cancer reveals distinct roles for gain- and loss-of-function p53 mutations. J Clin Invest 117: 1893–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Kim DH, Lee SW, Choi KY, Sung YC. 1995. Transactivation ability of p53 transcriptional activation domain is directly related to the binding affinity to TATA-binding protein. J Biol Chem 270: 25014–25019. [DOI] [PubMed] [Google Scholar]
- Chicas A, Molina P, Bargonetti J. 2000. Mutant p53 forms a complex with Sp1 on HIV-LTR DNA. Biochem Biophys Res Commun 279: 383–390. [DOI] [PubMed] [Google Scholar]
- Chin KV, Ueda K, Pastan I, Gottesman MM. 1992. Modulation of activity of the promoter of the human MDR1 gene by Ras and p53. Science 255: 459–462. [DOI] [PubMed] [Google Scholar]
- Cho Y, Gorina S, Jeffrey PD, Pavletich NP. 1994. Crystal structure of a p53 tumor suppressor–DNA complex: Understanding tumorigenic mutations. Science 265: 346–355. [DOI] [PubMed] [Google Scholar]
- Cooks T, Pateras IS, Tarcic O, Solomon H, Schetter AJ, Wilder S, Lozano G, Pikarsky E, Forshew T, Rosenfeld N, et al. 2013. Mutant p53 prolongs NF-κB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell 23: 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordenonsi M, Montagner M, Adorno M, Zacchigna L, Martello G, Mamidi A, Soligo S, Dupont S, Piccolo S. 2007. Integration of TGF-β and Ras/MAPK signaling through p53 phosphorylation. Science 315: 840–843. [DOI] [PubMed] [Google Scholar]
- Davison TS, Vagner C, Kaghad M, Ayed A, Caput D, Arrowsmith CH. 1999. p73 and p63 are homotetramers capable of weak heterotypic interactions with each other but not with p53. J Biol Chem 274: 18709–18714. [DOI] [PubMed] [Google Scholar]
- Deb S, Jackson CT, Subler MA, Martin DW. 1992. Modulation of cellular and viral promoters by mutant human p53 proteins found in tumor cells. J Virol 66: 6164–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo AB, Jay G, Appella E, Dubois GC, Law LW, Old LJ. 1979. Detection of a transformation-related antigen in chemically induced sarcomas and other transformed cells of the mouse. Proc Natl Acad Sci 76: 2420–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Orso S, Fontemaggi G, Stambolsky P, Goeman F, Voellenkle C, Levrero M, Strano S, Rotter V, Oren M, Blandino G. 2011. ChIP-on-chip analysis of in vivo mutant p53 binding to selected gene promoters. OMICS 15: 305–312. [DOI] [PubMed] [Google Scholar]
- Denslow SA, Wade PA. 2007. The human Mi-2/NuRD complex and gene regulation. Oncogene 26: 5433–5438. [DOI] [PubMed] [Google Scholar]
- Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, Blandino G, Piaggio G. 2006. Gain of function of mutant p53: The mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell 10: 191–202. [DOI] [PubMed] [Google Scholar]
- Di Agostino S, Cortese G, Monti O, Dell’Orso S, Sacchi A, Eisenstein M, Citro G, Strano S, Blandino G. 2008. The disruption of the protein complex mutantp53/p73 increases selectively the response of tumor cells to anticancer drugs. Cell Cycle 7: 3440–3447. [DOI] [PubMed] [Google Scholar]
- Di Como CJ, Gaiddon C, Prives C. 1999. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol Cell Biol 19: 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Minin G, Bellazzo A, Dal Ferro M, Chiaruttini G, Nuzzo S, Bicciato S, Piazza S, Rami D, Bulla R, Sommaggio R, et al. 2014. Mutant p53 reprograms TNF signaling in cancer cells through interaction with the tumor suppressor DAB2IP. Mol Cell 56: 617–629. [DOI] [PubMed] [Google Scholar]
- Dittmer D, Pati S, Zambetti G, Chu S, Teresky AK, Moore M, Finlay C, Levine AJ. 1993. Gain of function mutations in p53. Nat Genet 4: 42–46. [DOI] [PubMed] [Google Scholar]
- Do PM, Varanasi L, Fan S, Li C, Kubacka I, Newman V, Chauhan K, Daniels SR, Boccetta M, Garrett MR, et al. 2012. Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev 26: 830–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA Jr, Butel JS, Bradley A. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356: 215–221. [DOI] [PubMed] [Google Scholar]
- Drane P, Barel M, Balbo M, Frade R. 1997. Identification of RB18A, a 205 kDa new p53 regulatory protein which shares antigenic and functional properties with p53. Oncogene 15: 3013–3024. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC. 1995. Evolution of the SNF2 family of proteins: Subfamilies with distinct sequences and functions. Nucleic Acids Res 23: 2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. 1992. Definition of a consensus binding site for p53. Nat Genet 1: 45–49. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75: 817–825. [DOI] [PubMed] [Google Scholar]
- Eliyahu D, Raz A, Gruss P, Givol D, Oren M. 1984. Participation of p53 cellular tumour antigen in transformation of normal embryonic cells. Nature 312: 646–649. [DOI] [PubMed] [Google Scholar]
- Elledge RM, Fuqua SA, Clark GM, Pujol P, Allred DC, McGuire WL. 1993. Prognostic significance of p53 gene alterations in node-negative breast cancer. Breast Cancer Res Treat 26: 225–235. [DOI] [PubMed] [Google Scholar]
- Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, Holstege F, Werner M. 2008. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol Cell 31: 337–346. [DOI] [PubMed] [Google Scholar]
- Espinosa JM. 2008. Mechanisms of regulatory diversity within the p53 transcriptional network. Oncogene 27: 4013–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JM, Emerson BM. 2001. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol cell 8: 57–69. [DOI] [PubMed] [Google Scholar]
- Euskirchen GM, Auerbach RK, Davidov E, Gianoulis TA, Zhong G, Rozowsky J, Bhardwaj N, Gerstein MB, Snyder M. 2011. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet 7: e1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogal V, Hsieh JK, Royer C, Zhong S, Lu X. 2005. Cell cycle-dependent nuclear retention of p53 by E2F1 requires phosphorylation of p53 at Ser315. EMBO J 24: 2768–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondell JD, Ge H, Roeder RG. 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci 93: 8329–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontemaggi G, Dell’Orso S, Trisciuoglio D, Shay T, Melucci E, Fazi F, Terrenato I, Mottolese M, Muti P, Domany E, et al. 2009. The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis. Nat Struct Mol Biol 16: 1086–1093. [DOI] [PubMed] [Google Scholar]
- Frade R, Balbo M, Barel M. 2000. RB18A, whose gene is localized on chromosome 17q12-q21.1, regulates in vivo p53 transactivating activity. Cancer Res 60: 6585–6589. [PubMed] [Google Scholar]
- Frade R, Balbo M, Barel M. 2002. RB18A regulates p53-dependent apoptosis. Oncogene 21: 861–866. [DOI] [PubMed] [Google Scholar]
- Frazier MW, He X, Wang J, Gu Z, Cleveland JL, Zambetti GP. 1998. Activation of c-myc gene expression by tumor-derived p53 mutants requires a discrete C-terminal domain. Mol Cell Biol 18: 3735–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor WA, Prives C. 2012. Mutant p53: One name, many proteins. Genes Dev 26: 1268–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R, Barsotti A, Chicas A, Li W, Polotskaia A, et al. 2012. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 148: 244–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk WD, Pak DT, Karas RH, Wright WE, Shay JW. 1992. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol 12: 2866–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. 2001. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol 21: 1874–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon JV, Greaves R, Iggo R, Lane DP. 1990. Activating mutations in p53 produce a common conformational effect. A monoclonal antibody specific for the mutant form. EMBO J 9: 1595–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E, Capaci V, Jordan L, Quinlan P, Thompson A, et al. 2011. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell 20: 79–91. [DOI] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. 2010. Immunity, inflammation, and cancer. Cell 140: 883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Roeder RG. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90: 595–606. [DOI] [PubMed] [Google Scholar]
- Gu W, Shi XL, Roeder RG. 1997. Synergistic activation of transcription by CBP and p53. Nature 387: 819–823. [DOI] [PubMed] [Google Scholar]
- Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG. 1999. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell 3: 97–108. [DOI] [PubMed] [Google Scholar]
- Gualberto A, Baldwin AS Jr. 1995. p53 and Sp1 interact and cooperate in the tumor necrosis factor-induced transcriptional activation of the HIV-1 long terminal repeat. J Biol Chem 270: 19680–19683. [DOI] [PubMed] [Google Scholar]
- Gualberto A, Aldape K, Kozakiewicz K, Tlsty TD. 1998. An oncogenic form of p53 confers a dominant, gain-of-function phenotype that disrupts spindle checkpoint control. Proc Natl Acad Sci 95: 5166–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B, Wang TL, Shih Ie M. 2011. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res 71: 6718–6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurova KV, Rokhlin OW, Budanov AV, Burdelya LG, Chumakov PM, Cohen MB, Gudkov AV. 2003. Cooperation of two mutant p53 alleles contributes to Fas resistance of prostate carcinoma cells. Cancer Res 63: 2905–2912. [PubMed] [Google Scholar]
- Gurtner A, Starace G, Norelli G, Piaggio G, Sacchi A, Bossi G. 2010. Mutant p53-induced up-regulation of mitogen-activated protein kinase kinase 3 contributes to gain of function. J Biol Chem 285: 14160–14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanel W, Marchenko N, Xu S, Yu SX, Weng W, Moll U. 2013. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ 20: 898–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Covic M, Bedford MT, Hottiger MO. 2008. Protein arginine methyltransferase 1 coactivates NF-κB-dependent gene expression synergistically with CARM1 and PARP1. J Mol Biol 377: 668–678. [DOI] [PubMed] [Google Scholar]
- Haupt S, di Agostino S, Mizrahi I, Alsheich-Bartok O, Voorhoeve M, Damalas A, Blandino G, Haupt Y. 2009. Promyelocytic leukemia protein is required for gain of function by mutant p53. Cancer Res 69: 4818–4826. [DOI] [PubMed] [Google Scholar]
- Heinlein C, Krepulat F, Lohler J, Speidel D, Deppert W, Tolstonog GV. 2008. Mutant p53(R270H) gain of function phenotype in a mouse model for oncogene-induced mammary carcinogenesis. Int J Cancer 122: 1701–1709. [DOI] [PubMed] [Google Scholar]
- Hinds P, Finlay C, Levine AJ. 1989. Mutation is required to activate the p53 gene for cooperation with the ras oncogene and transformation. J Virol 63: 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. 2005. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7: 469–483. [DOI] [PubMed] [Google Scholar]
- Hsiao M, Low J, Dorn E, Ku D, Pattengale P, Yeargin J, Haas M. 1994. Gain-of-function mutations of the p53 gene induce lymphohematopoietic metastatic potential and tissue invasiveness. Am J Pathol 145: 702–714. [PMC free article] [PubMed] [Google Scholar]
- Huang S, Litt M, Felsenfeld G. 2005. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev 19: 1885–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CI, Matoso A, Corney DC, Flesken-Nikitin A, Korner S, Wang W, Boccaccio C, Thorgeirsson SS, Comoglio PM, Hermeking H, et al. 2011. Wild-type p53 controls cell motility and invasion by dual regulation of MET expression. Proc Natl Acad Sci 108: 14240–14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbriano C, Gurtner A, Cocchiarella F, Di Agostino S, Basile V, Gostissa M, Dobbelstein M, Del Sal G, Piaggio G, Mantovani R. 2005. Direct p53 transcriptional repression: In vivo analysis of CCAAT-containing G2/M promoters. Mol Cell Biol 25: 3737–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanaga Y, Jeang KT. 2002. Expression of mitotic spindle checkpoint protein hsMAD1 correlates with cellular proliferation and is activated by a gain-of-function p53 mutant. Cancer Res 62: 2618–2624. [PubMed] [Google Scholar]
- Jeffrey PD, Gorina S, Pavletich NP. 1995. Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science 267: 1498–1502. [DOI] [PubMed] [Google Scholar]
- Ji L, Xu J, Liu J, Amjad A, Zhang K, Liu Q, Zhou L, Xiao J, Li X. 2015. Mutant p53 promotes tumor cell malignancy by both positive and negative regulation of the transforming growth factor β (TGF-β) pathway. J Biol Chem 290: 11729–11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger AC, Fersht AR. 2008. Structural biology of the tumor suppressor p53. Annu Rev Biochem 77: 557–582. [DOI] [PubMed] [Google Scholar]
- Joerger AC, Ang HC, Veprintsev DB, Blair CM, Fersht AR. 2005. Structures of p53 cancer mutants and mechanism of rescue by second-site suppressor mutations. J Biol Chem 280: 16030–16037. [DOI] [PubMed] [Google Scholar]
- Joerger AC, Ang HC, Fersht AR. 2006. Structural basis for understanding oncogenic p53 mutations and designing rescue drugs. Proc Natl Acad Sci 103: 15056–15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KM, Wang J, Smallwood A, Arayata C, Carey M. 2002. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev 16: 1852–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TM, Hammond EM, Giaccia A, Attardi LD. 2005. The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nat Genet 37: 145–152. [DOI] [PubMed] [Google Scholar]
- Kalo E, Buganim Y, Shapira KE, Besserglick H, Goldfinger N, Weisz L, Stambolsky P, Henis YI, Rotter V. 2007. Mutant p53 attenuates the SMAD-dependent transforming growth factor β1 (TGF-β1) signaling pathway by repressing the expression of TGF-β receptor type II. Mol Cell Biol 27: 8228–8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada R, Nomura T, Anderson CW, Sakaguchi K. 2011. Cancer-associated p53 tetramerization domain mutants: Quantitative analysis reveals a low threshold for tumor suppressor inactivation. J Biol Chem 286: 252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelavkar UP, Badr KF. 1999. Effects of mutant p53 expression on human 15-lipoxygenase-promoter activity and murine 12/15-lipoxygenase gene expression: Evidence that 15-lipoxygenase is a mutator gene. Proc Natl Acad Sci 96: 4378–4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern SE, Kinzler KW, Bruskin A, Jarosz D, Friedman P, Prives C, Vogelstein B. 1991. Identification of p53 as a sequence-specific DNA-binding protein. Science 252: 1708–1711. [DOI] [PubMed] [Google Scholar]
- Kern SE, Pietenpol JA, Thiagalingam S, Seymour A, Kinzler KW, Vogelstein B. 1992. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science 256: 827–830. [DOI] [PubMed] [Google Scholar]
- Kim E, Deppert W. 2003. The complex interactions of p53 with target DNA: We learn as we go. Biochem Cell Biol 81: 141–150. [DOI] [PubMed] [Google Scholar]
- Kim E, Deppert W. 2004. Transcriptional activities of mutant p53: When mutations are more than a loss. J Cell Biochem 93: 878–886. [DOI] [PubMed] [Google Scholar]
- Kim E, Deppert W. 2007. Interactions of mutant p53 with DNA: Guilt by association. Oncogene 26: 2185–2190. [DOI] [PubMed] [Google Scholar]
- Kim E, Gunther W, Yoshizato K, Meissner H, Zapf S, Nusing RM, Yamamoto H, Van Meir EG, Deppert W, Giese A. 2003. Tumor suppressor p53 inhibits transcriptional activation of invasion gene thromboxane synthase mediated by the proto-oncogenic factor ets-1. Oncogene 22: 7716–7727. [DOI] [PubMed] [Google Scholar]
- Kollareddy M, Dimitrova E, Vallabhaneni KC, Chan A, Le T, Chauhan KM, Carrero ZI, Ramakrishnan G, Watabe K, Haupt Y, et al. 2015. Regulation of nucleotide metabolism by mutant p53 contributes to its gain-of-function activities. Nature Commun 6: 7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsodontis G, Vasilaki E, Chou WC, Papakosta P, Kardassis D. 2005. Physical and functional interactions between members of the tumour suppressor p53 and the Sp families of transcription factors: Importance for the regulation of genes involved in cell-cycle arrest and apoptosis. Biochem J 389: 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs JE, Peterson CL. 2000. Understanding “active” chromatin: A historical perspective of chromatin remodeling. Crit Rev Eukaryot Gene Expr 10: 1–12. [PubMed] [Google Scholar]
- Kuerbitz SJ, Plunkett BS, Walsh WV, Kastan MB. 1992. Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc Natl Acad Sci 89: 7491–7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. 1994. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 370: 477–481. [DOI] [PubMed] [Google Scholar]
- Lane DP, Crawford LV. 1979. T antigen is bound to a host protein in SV40-transformed cells. Nature 278: 261–263. [DOI] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. 2004. Gain of function of a p53 hot spot mutation in a mouse model of Li–Fraumeni syndrome. Cell 119: 861–872. [DOI] [PubMed] [Google Scholar]
- Langerod A, Zhao H, Borgan O, Nesland JM, Bukholm IR, Ikdahl T, Karesen R, Borresen-Dale AL, Jeffrey SS. 2007. TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer. Breast Cancer Res 9: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laptenko O, Prives C. 2006. Transcriptional regulation by p53: One protein, many possibilities. Cell Death Differ 13: 951–961. [DOI] [PubMed] [Google Scholar]
- Laptenko O, Beckerman R, Freulich E, Prives C. 2011. p53 binding to nucleosomes within the p21 promoter in vivo leads to nucleosome loss and transcriptional activation. Proc Natl Acad Sci 108: 10385–10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavra L, Ulivieri A, Rinaldo C, Dominici R, Volante M, Luciani E, Bartolazzi A, Frasca F, Soddu S, Sciacchitano S. 2009. Gal-3 is stimulated by gain-of-function p53 mutations and modulates chemoresistance in anaplastic thyroid carcinomas. J Pathol 218: 66–75. [DOI] [PubMed] [Google Scholar]
- Lee YI, Lee S, Das GC, Park US, Park SM. 2000. Activation of the insulin-like growth factor II transcription by aflatoxin B1 induced p53 mutant 249 is caused by activation of transcription complexes; implications for a gain-of-function during the formation of hepatocellular carcinoma. Oncogene 19: 3717–3726. [DOI] [PubMed] [Google Scholar]
- Lee D, Kim JW, Seo T, Hwang SG, Choi EJ, Choe J. 2002. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J Biol Chem 277: 22330–22337. [DOI] [PubMed] [Google Scholar]
- Legros Y, Meyer A, Ory K, Soussi T. 1994. Mutations in p53 produce a common conformational effect that can be detected with a panel of monoclonal antibodies directed toward the central part of the p53 protein. Oncogene 9: 3689–3694. [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. 2010. Cell signaling by receptor tyrosine kinases. Cell 141: 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Vosburgh E. 2008. P53 mutations in lymphomas: Position matters. Blood 112: 2997–2998. [DOI] [PubMed] [Google Scholar]
- Li Y, Prives C. 2007. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene 26: 2220–2225. [DOI] [PubMed] [Google Scholar]
- Li Y, Guessous F, Kwon S, Kumar M, Ibidapo O, Fuller L, Johnson E, Lal B, Hussaini I, Bao Y, et al. 2008. PTEN has tumor-promoting properties in the setting of gain-of-function p53 mutations. Cancer Res 68: 1723–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidor Nili E, Field Y, Lubling Y, Widom J, Oren M, Segal E. 2010. p53 binds preferentially to genomic regions with high DNA-encoded nucleosome occupancy. Genome Res 20: 1361–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. 1997. Binding and modulation of p53 by p300/CBP coactivators. Nature 387: 823–827. [DOI] [PubMed] [Google Scholar]
- Lim LY, Vidnovic N, Ellisen LW, Leong CO. 2009. Mutant p53 mediates survival of breast cancer cells. Br J Cancer 101: 1606–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Chen J, Elenbaas B, Levine AJ. 1994. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev 8: 1235–1246. [DOI] [PubMed] [Google Scholar]
- Lin J, Teresky AK, Levine AJ. 1995. Two critical hydrophobic amino acids in the N-terminal domain of the p53 protein are required for the gain of function phenotypes of human p53 mutants. Oncogene 10: 2387–2390. [PubMed] [Google Scholar]
- Linzer DI, Levine AJ. 1979. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 17: 43–52. [DOI] [PubMed] [Google Scholar]
- Liu G, McDonnell TJ, Montes de Oca Luna R, Kapoor M, Mims B, El-Naggar AK, Lozano G. 2000. High metastatic potential in mice inheriting a targeted p53 missense mutation. Proc Natl Acad Sci 97: 4174–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Ling S, Lin WC. 2011. TopBP1 mediates mutant p53 gain of function through NF-Y and p63/p73. Mol Cell Biol 31: 4464–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottin-Divoux S, Barel M, Frade R. 2005. RB18A enhances expression of mutant p53 protein in human cells. FEBS Lett 579: 2323–2326. [DOI] [PubMed] [Google Scholar]
- Ludes-Meyers JH, Subler MA, Shivakumar CV, Munoz RM, Jiang P, Bigger JE, Brown DR, Deb SP, Deb S. 1996. Transcriptional activation of the human epidermal growth factor receptor promoter by human p53. Mol Cell Biol 16: 6009–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Su F, Chen D, Shiloh A, Gu W. 2000. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408: 377–381. [DOI] [PubMed] [Google Scholar]
- Malkin D, Li FP, Strong LC, Fraumeni JF Jr, Nelson CE, Kim DH, Kassel J, Gryka MA, Bischoff FZ, Tainsky MA, et al. 1990. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 250: 1233–1238. [DOI] [PubMed] [Google Scholar]
- Mantovani F, Tocco F, Girardini J, Smith P, Gasco M, Lu X, Crook T, Del Sal G. 2007. The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nat Struct Mol Biol 14: 912–920. [DOI] [PubMed] [Google Scholar]
- Marin MC, Jost CA, Brooks LA, Irwin MS, O’Nions J, Tidy JA, James N, McGregor JM, Harwood CA, Yulug IG, et al. 2000. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet 25: 47–54. [DOI] [PubMed] [Google Scholar]
- Marshall CJ. 1995. Specificity of receptor tyrosine kinase signaling: Transient versus sustained extracellular signal-regulated kinase activation. Cell 80: 179–185. [DOI] [PubMed] [Google Scholar]
- Matas D, Sigal A, Stambolsky P, Milyavsky M, Weisz L, Schwartz D, Goldfinger N, Rotter V. 2001. Integrity of the N-terminal transcription domain of p53 is required for mutant p53 interference with drug-induced apoptosis. EMBO J 20: 4163–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Lin SC, Bernecky C, Gao Y, Taatjes DJ. 2010. p53 activates transcription by directing structural shifts in Mediator. Nat Struct Mol Biol 17: 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, Lukashchuk N, Gillespie DA, Ludwig RL, Gosselin P, et al. 2009. Mutant p53 drives invasion by promoting integrin recycling. Cell 139: 1327–1341. [DOI] [PubMed] [Google Scholar]
- Muller PA, Trinidad AG, Timpson P, Morton JP, Zanivan S, van den Berghe PV, Nixon C, Karim SA, Caswell PT, Noll JE, et al. 2013. Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene 32: 1252–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar AM, Taatjes DJ, Zhai W, Nogales E, Tjian R. 2002. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev 16: 1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaich AK, Appella E, Harrington RE. 1997. DNA bending is essential for the site-specific recognition of DNA response elements by the DNA binding domain of the tumor suppressor protein p53. J Biol Chem 272: 14842–14849. [DOI] [PubMed] [Google Scholar]
- Naidu SR, Love IM, Imbalzano AN, Grossman SR, Androphy EJ. 2009. The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells. Oncogene 28: 2492–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Vousden KH. 2001. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 7: 683–694. [DOI] [PubMed] [Google Scholar]
- Nigro JM, Baker SJ, Preisinger AC, Jessup JM, Hostetter R, Cleary K, Bigner SH, Davidson N, Baylin S, Devilee P, et al. 1989. Mutations in the p53 gene occur in diverse human tumour types. Nature 342: 705–708. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D. 2002. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev 16: 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N. 2000. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288: 1053–1058. [DOI] [PubMed] [Google Scholar]
- Oh J, Sohn DH, Ko M, Chung H, Jeon SH, Seong RH. 2008. BAF60a interacts with p53 to recruit the SWI/SNF complex. J Biol Chem 283: 11924–11934. [DOI] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. 2004. Mutant p53 gain of function in two mouse models of Li–Fraumeni syndrome. Cell 119: 847–860. [DOI] [PubMed] [Google Scholar]
- Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, Theillet C, Rodriguez C, Lidereau R, Bieche I, et al. 2006. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res 12: 1157–1167. [DOI] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P. 2010. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2: a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren M, Rotter V. 2010. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol 2: a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea D, O’Riain C, Taylor C, Waters R, Carlotti E, Macdougall F, Gribben J, Rosenwald A, Ott G, Rimsza LM, et al. 2008. The presence of TP53 mutation at diagnosis of follicular lymphoma identifies a high-risk group of patients with shortened time to disease progression and poorer overall survival. Blood 112: 3126–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada LF, Land H, Weinberg RA, Wolf D, Rotter V. 1984. Cooperation between gene encoding p53 tumour antigen and ras in cellular transformation. Nature 312: 649–651. [DOI] [PubMed] [Google Scholar]
- Pardoll DM. 2012. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 12: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. 2007. TP53 mutations in human cancers: Functional selection and impact on cancer prognosis and outcomes. Oncogene 26: 2157–2165. [DOI] [PubMed] [Google Scholar]
- Pfister NT, Fomin V, Regunath K, Zhou JY, Zhou W, Silwal-Pandit L, Freed-Pastor WA, Laptenko O, Neo SP, Bargonetti J, et al. 2015. Mutant p53 cooperates with the SWI/SNF chromatin remodeling complex to regulate VEGFR2 in breast cancer cells. Genes Dev 29: 1298–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl J, Goldfinger N, Radler-Pohl A, Rotter V, Schirrmacher V. 1988. p53 increases experimental metastatic capacity of murine carcinoma cells. Mol Cell Biol 8: 2078–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss U, Kreutzfeld R, Scheidtmann KH. 2000. Tumor-derived p53 mutant C174Y is a gain-of-function mutant which activates the fos promoter and enhances colony formation. Int J Cancer 88: 162–171. [DOI] [PubMed] [Google Scholar]
- Pugacheva EN, Ivanov AV, Kravchenko JE, Kopnin BP, Levine AJ, Chumakov PM. 2002. Novel gain of function activity of p53 mutants: Activation of the dUTPase gene expression leading to resistance to 5-fluorouracil. Oncogene 21: 4595–4600. [DOI] [PubMed] [Google Scholar]
- Quante T, Otto B, Brazdova M, Kejnovska I, Deppert W, Tolstonog GV. 2012. Mutant p53 is a transcriptional co-factor that binds to G-rich regulatory regions of active genes and generates transcriptional plasticity. Cell Cycle 11: 3290–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragimov N, Krauskopf A, Navot N, Rotter V, Oren M, Aloni Y. 1993. Wild-type but not mutant p53 can repress transcription initiation in vitro by interfering with the binding of basal transcription factors to the TATA motif. Oncogene 8: 1183–1193. [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. 2008. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9: 402–412. [DOI] [PubMed] [Google Scholar]
- Rivlin N, Brosh R, Oren M, Rotter V. 2011. Mutations in the p53 tumor suppressor gene: Important milestones at the various steps of tumorigenesis. Genes Cancer 2: 466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez OC, Choudhury S, Kolukula V, Vietsch EE, Catania J, Preet A, Reynoso K, Bargonetti J, Wellstein A, Albanese C, et al. 2012. Dietary downregulation of mutant p53 levels via glucose restriction: Mechanisms and implications for tumor therapy. Cell Cycle 11: 4436–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotter V. 1983. p53, a transformation-related cellular-encoded protein, can be used as a biochemical marker for the detection of primary mouse tumor cells. Proc Natl Acad Sci 80: 2613–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu G, Wang D, Chen CB, Zhurkin VB, Harrington RE, Appella E, Hager GL, Nagaich AK. 2010. p53 binding to nucleosomal DNA depends on the rotational positioning of DNA response element. J Biol Chem 285: 1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samowitz WS, Curtin K, Ma KN, Edwards S, Schaffer D, Leppert MF, Slattery ML. 2002. Prognostic significance of p53 mutations in colon cancer at the population level. Int J Cancer 99: 597–602. [DOI] [PubMed] [Google Scholar]
- Sampath J, Sun D, Kidd VJ, Grenet J, Gandhi A, Shapiro LH, Wang Q, Zambetti GP, Schuetz JD. 2001. Mutant p53 cooperates with ETS and selectively up-regulates human MDR1 not MRP1. J Biol Chem 276: 39359–39367. [DOI] [PubMed] [Google Scholar]
- Sankala H, Vaughan C, Wang J, Deb S, Graves PR. 2011. Upregulation of the mitochondrial transport protein, Tim50, by mutant p53 contributes to cell growth and chemoresistance. Arch Biochem Biophys 512: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scian MJ, Stagliano KE, Deb D, Ellis MA, Carchman EH, Das A, Valerie K, Deb SP, Deb S. 2004. Tumor-derived p53 mutants induce oncogenesis by transactivating growth-promoting genes. Oncogene 23: 4430–4443. [DOI] [PubMed] [Google Scholar]
- Scian MJ, Stagliano KE, Anderson MA, Hassan S, Bowman M, Miles MF, Deb SP, Deb S. 2005. Tumor-derived p53 mutants induce NF-κB2 gene expression. Mol Cell Biol 25: 10097–10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E, Usheva A, Zambetti GP, Momand J, Horikoshi N, Weinmann R, Levine AJ, Shenk T. 1992. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc Natl Acad Sci 89: 12028–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. 2012. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486: 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal A, Matas D, Almog N, Goldfinger N, Rotter V. 2001. The C-terminus of mutant p53 is necessary for its ability to interfere with growth arrest or apoptosis. Oncogene 20: 4891–4898. [DOI] [PubMed] [Google Scholar]
- Singer S, Ehemann V, Brauckhoff A, Keith M, Vreden S, Schirmacher P, Breuhahn K. 2007. Protumorigenic overexpression of stathmin/Op18 by gain-of-function mutation in p53 in human hepatocarcinogenesis. Hepatology 46: 759–768. [DOI] [PubMed] [Google Scholar]
- Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, Zhou T, Rohs R, Honig B, Bussemaker HJ, et al. 2011. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147: 1270–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon H, Buganim Y, Pomeraniec L, Beatus T, Assia Y, Kogan-Sakin I, Madar S, Goldstein I, Brosh R, Kalo E, et al. 2012. Various p53 mutant types differently regulate the Ras circuit to induce a cancer-related gene signature. J Cell Sci 125: 3144–3152. [DOI] [PubMed] [Google Scholar]
- Soussi T, Kato S, Levy PP, Ishioka C. 2005. Reassessment of the TP53 mutation database in human disease by data mining with a library of TP53 missense mutations. Hum Mutat 25: 6–17. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Zou ZQ, Pirollo K, Blattner W, Chang EH. 1990. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li–Fraumeni syndrome. Nature 348: 747–749. [DOI] [PubMed] [Google Scholar]
- Stambolsky P, Tabach Y, Fontemaggi G, Weisz L, Maor-Aloni R, Siegfried Z, Shiff I, Kogan I, Shay M, Kalo E, et al. 2010. Modulation of the vitamin D3 response by cancer-associated mutant p53. Cancer Cell 17: 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strano S, Fontemaggi G, Costanzo A, Rizzo MG, Monti O, Baccarini A, Del Sal G, Levrero M, Sacchi A, Oren M, et al. 2002. Physical interaction with human tumor-derived p53 mutants inhibits p63 activities. J Biol Chem 277: 18817–18826. [DOI] [PubMed] [Google Scholar]
- Strano S, Dell’Orso S, Di Agostino S, Fontemaggi G, Sacchi A, Blandino G. 2007. Mutant p53: An oncogenic transcription factor. Oncogene 26: 2212–2219. [DOI] [PubMed] [Google Scholar]
- Strauss BE, Haas M. 1995. The region 3′ to the major transcriptional start site of the MDR1 downstream promoter mediates activation by a subset of mutant P53 proteins. Biochem Biophys Res Commun 217: 333–340. [DOI] [PubMed] [Google Scholar]
- Sun Y, Cheung JM, Martel-Pelletier J, Pelletier JP, Wenger L, Altman RD, Howell DS, Cheung HS. 2000. Wild type and mutant p53 differentially regulate the gene expression of human collagenase-3 (hMMP-13). J Biol Chem 275: 11327–11332. [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. 2006. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat Rev Cancer 6: 909–923. [DOI] [PubMed] [Google Scholar]
- Torgeman A, Mor-Vaknin N, Zelin E, Ben-Aroya Z, Lochelt M, Flugel RM, Aboud M. 2001. Sp1-p53 heterocomplex mediates activation of HTLV-I long terminal repeat by 12-O-tetradecanoylphorbol-13-acetate that is antagonized by protein kinase C. Virology 281: 10–20. [DOI] [PubMed] [Google Scholar]
- Varley JM, Evans DG, Birch JM. 1997. Li–Fraumeni syndrome—A molecular and clinical review. Br J Cancer 76: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan CA, Singh S, Windle B, Yeudall WA, Frum R, Grossman SR, Deb SP, Deb S. 2012. Gain-of-function activity of mutant p53 in lung cancer through up-regulation of receptor protein tyrosine kinase Axl. Genes Cancer 3: 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venot C, Maratrat M, Sierra V, Conseiller E, Debussche L. 1999. Definition of a p53 transactivation function-deficient mutant and characterization of two independent p53 transactivation subdomains. Oncogene 18: 2405–2410. [DOI] [PubMed] [Google Scholar]
- Vikhanskaya F, Lee MK, Mazzoletti M, Broggini M, Sabapathy K. 2007. Cancer-derived p53 mutants suppress p53-target gene expression—Potential mechanism for gain of function of mutant p53. Nucleic Acids Res 35: 2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Prives C. 2009. Blinded by the light: The growing complexity of p53. Cell 137: 413–431. [DOI] [PubMed] [Google Scholar]
- Waldman T, Kinzler KW, Vogelstein B. 1995. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res 55: 5187–5190. [PubMed] [Google Scholar]
- Walker KK, Levine AJ. 1996. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci 93: 15335–15340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmueller S, Manchado E, Saborowski M, Morris JP, Wagenblast E, Davis CA, Moon SH, Pfister NT, Tschaharganeh DF, Kitzing T, et al. 2014. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor β signaling. Cell 157: 382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz L, Zalcenstein A, Stambolsky P, Cohen Y, Goldfinger N, Oren M, Rotter V. 2004. Transactivation of the EGR1 gene contributes to mutant p53 gain of function. Cancer Res 64: 8318–8327. [DOI] [PubMed] [Google Scholar]
- Weisz L, Damalas A, Liontos M, Karakaidos P, Fontemaggi G, Maor-Aloni R, Kalis M, Levrero M, Strano S, Gorgoulis VG, et al. 2007. Mutant p53 enhances nuclear factor κB activation by tumor necrosis factor α in cancer cells. Cancer Res 67: 2396–2401. [DOI] [PubMed] [Google Scholar]
- Werner H, Karnieli E, Rauscher FJ, LeRoith D. 1996. Wild-type and mutant p53 differentially regulate transcription of the insulin-like growth factor I receptor gene. Proc Natl Acad Sci 93: 8318–8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DS, Tsai WW, Schumacher MA, Barton MC. 2008. Chromatin-bound p53 anchors activated Smads and the mSin3A corepressor to confer transforming-growth-factor-β-mediated transcription repression. Mol Cell Biol 28: 1988–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW. 2011. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 11: 481–492. [DOI] [PubMed] [Google Scholar]
- Wong P, Verselis SJ, Garber JE, Schneider K, DiGianni L, Stockwell DH, Li FP, Syngal S. 2006. Prevalence of early onset colorectal cancer in 397 patients with classic Li–Fraumeni syndrome. Gastroenterology 130: 73–79. [DOI] [PubMed] [Google Scholar]
- Wong RP, Tsang WP, Chau PY, Co NN, Tsang TY, Kwok TT. 2007. p53-R273H gains new function in induction of drug resistance through down-regulation of procaspase-3. Mol Cancer Ther 6: 1054–1061. [DOI] [PubMed] [Google Scholar]
- Woodage T, Basrai MA, Baxevanis AD, Hieter P, Collins FS. 1997. Characterization of the CHD family of proteins. Proc Natl Acad Sci 94: 11472–11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W, Nagase S, Montia AG, Kalachikov SM, Keniry M, Su T, Memeo L, Hibshoosh H, Parsons R. 2008. BAF180 is a critical regulator of p21 induction and a tumor suppressor mutated in breast cancer. Cancer Res 68: 1667–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong S, Tu H, Kollareddy M, Pant V, Li Q, Zhang Y, Jackson JG, Suh YA, Elizondo-Fraire AC, Yang P, et al. 2014. Pla2g16 phospholipase mediates gain-of-function activities of mutant p53. Proc Natl Acad Sci 111: 11145–11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhang J, Chen X. 2007. The activity of p53 is differentially regulated by Brm- and Brg1-containing SWI/SNF chromatin remodeling complexes. J Biol Chem 282: 37429–37435. [DOI] [PubMed] [Google Scholar]
- Xu Y, Yan W, Chen X. 2010. SNF5, a core component of the SWI/SNF complex, is necessary for p53 expression and cell survival, in part through eIF4E. Oncogene 29: 4090–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Chen X. 2009. Identification of GRO1 as a critical determinant for mutant p53 gain of function. J Biol Chem 284: 12178–12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Chen X. 2010. Characterization of functional domains necessary for mutant p53 gain of function. J Biol Chem 285: 14229–14238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Liu G, Scoumanne A, Chen X. 2008. Suppression of inhibitor of differentiation 2, a target of mutant p53, is required for gain-of-function mutations. Cancer Res 68: 6789–6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeudall WA, Vaughan CA, Miyazaki H, Ramamoorthy M, Choi MY, Chapman CG, Wang H, Black E, Bulysheva AA, Deb SP, et al. 2012. Gain-of-function mutant p53 upregulates CXC chemokines and enhances cell migration. Carcinogenesis 33: 442–451. [DOI] [PubMed] [Google Scholar]
- Yin Y, Tainsky MA, Bischoff FZ, Strong LC, Wahl GM. 1992. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell 70: 937–948. [DOI] [PubMed] [Google Scholar]
- Young KH, Leroy K, Moller MB, Colleoni GW, Sanchez-Beato M, Kerbauy FR, Haioun C, Eickhoff JC, Young AH, Gaulard P, et al. 2008. Structural profiles of TP53 gene mutations predict clinical outcome in diffuse large B-cell lymphoma: An international collaborative study. Blood 112: 3088–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalcenstein A, Stambolsky P, Weisz L, Muller M, Wallach D, Goncharov TM, Krammer PH, Rotter V, Oren M. 2003. Mutant p53 gain of function: Repression of CD95(Fas/APO-1) gene expression by tumor-associated p53 mutants. Oncogene 22: 5667–5676. [DOI] [PubMed] [Google Scholar]
- Zegerman P, Canas B, Pappin D, Kouzarides T. 2002. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J Biol Chem 277: 11621–11624. [DOI] [PubMed] [Google Scholar]
- Zhang X, Krutchinsky A, Fukuda A, Chen W, Yamamura S, Chait BT, Roeder RG. 2005. MED1/TRAP220 exists predominantly in a TRAP/ Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol Cell 19: 89–100. [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhou W, Jiang J, Chen X. 1998. Identification of a novel p53 functional domain that is necessary for mediating apoptosis. J Biol Chem 273: 13030–13036. [DOI] [PubMed] [Google Scholar]
- Zhu J, Sammons MA, Donahue G, Dou Z, Vedadi M, Getlik M, Barsyte-Lovejoy D, Al-awar R, Katona BW, Shilatifard A, et al. 2015. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature 525: 206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]