SUMMARY
The nucleoskeleton is an important structural feature of the metazoan nucleus and is involved in the regulation of genome expression and maintenance. The nuclear lamins are intermediate filament proteins that form a peripheral nucleoskeleton in concert with other lamin-associated proteins. Several other proteins normally found in the cytoskeleton have also been identified in the nucleus, but, as will be discussed here, their roles in forming a nucleoskeleton have not been elucidated. Nevertheless, mutations in lamins and lamin-associated proteins cause a spectrum of diseases, making them interesting targets for future research.
The peripheral lamina is the only bona fide nucleoskeleton described to date. Several other cytoskeletal proteins (e.g., actins and myosins) are also found in nuclei, but their roles in the nucleoskeleton have not been identified.
1. INTRODUCTION
Over the past 30 years, the organization of the eukaryotic nucleus has been extensively investigated, leading to our current understanding of the nuclear interior as a highly organized collection of chromosomes, transcription products, and protein assemblies (Austin and Bellini 2010). Numerous nuclear subcompartments, or nuclear bodies, have been identified, and the roles of some of these in nuclear processes such as gene expression and genome maintenance have been well described and characterized (Sleeman and Trinkle-Mulcahy 2014). The roles for cytoskeletal structures in organizing the cytoplasm and regulating cellular functions such as mechanics, cellular locomotion, and distribution of cytoplasmic organelles are well known and have been extensively reviewed. In contrast, the possible existence of a nuclear skeleton, or nucleoskeleton, which could provide mechanical support and functional organization to the nucleus, emerged as a fully formed hypothesis only as recently as the 1970s.
As long ago as the 1940s, it was observed that the extraction of nuclei with high-salt solutions to remove chromatin left behind a residual protein structure. A subsequent study in the 1960s identified a nuclear ribonucleoprotein network in the nuclei of Walker tumor cells that had been extracted with high salt (Pederson 2000). Further refinement of these salt extraction methods to produce residual nuclear protein fractions led to the concept of a “nuclear matrix” in the 1970s, which, it was suggested, was responsible for organizing the genome in the nuclear volume and regulating gene expression (Berezney and Coffey 1974). However, as this nuclear matrix was the product of rather harsh extraction conditions that could induce artifactual protein or ribonucleoprotein precipitation, its very existence has proven to be controversial. The nuclear matrix has been described as an extensive and highly branched anastomosing network of fine filaments when viewed by electron microscopy of critical-point-dried matrices in whole-mount specimens. However, numerous studies on the topography of chromosomes and the wide array of nuclear bodies in the nucleus support the idea that each chromosome occupies a unique “territory,” with little intertwining of different chromosomes. The chromatin fibers themselves occupy a volume of the nucleus, and the spaces in between chromatin fibers form a sinusoidal interconnected network, the boundaries of which are formed by the chromatin itself. This network—the interchromatin space—is filled with ribonucleoproteins and other protein complexes forming the nuclear bodies. Therefore, it is difficult to imagine how a dense, highly branched nuclear matrix could accommodate the observed structures of chromosomes, chromatin fibers, and nuclear bodies in the nucleus.
The only bona fide nucleoskeleton present in animal cells lies at the periphery of the nucleus at the outer boundary of the chromatin mass. The nucleus is bordered by a complex organelle called the nuclear envelope (NE), which physically and functionally separates the nuclear and cytoplasmic compartments. Macromolecular movement between the cytoplasm and nucleus is restricted by a double-membrane bilayer structure—the nuclear membrane—the outer nuclear membrane is continuous with the endoplasmic reticulum (ER), whereas the inner nuclear membrane closely surrounds the chromatin. Punctuating the nuclear membranes are large protein structures known as nuclear pore complexes (NPCs), which are the sites of most macromolecular traffic between the nucleus and cytoplasm. The nuclear lamina is a complex protein network comprising the nuclear lamins and associated proteins, and lying between the chromatin and the inner membrane to form a peripheral nucleoskeleton. The lamina provides a structural framework to support nuclear shape and mechanics and to position NPCs in the NE. The nucleus also is linked to different elements of the cytoskeleton through transmembrane complexes in the NE, thus linking the cytoskeleton to the nucleoskeleton (Stancheva and Schirmer 2014).
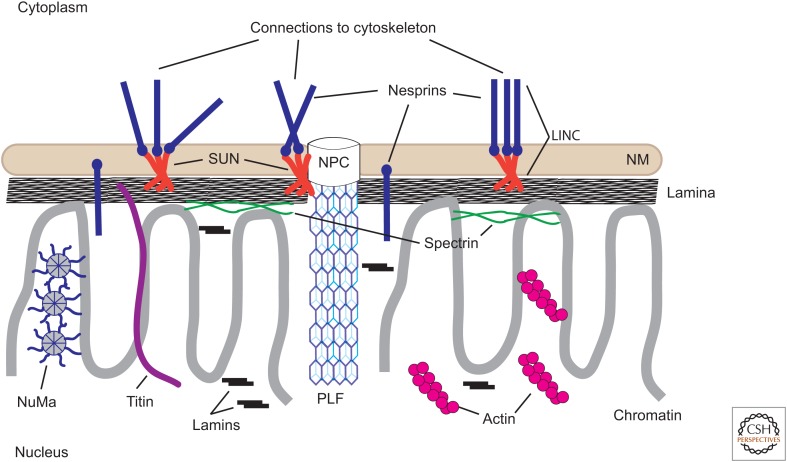
In contrast to the peripheral nucleoskeleton, the existence of an internal nucleoskeleton has remained a controversial idea. Over the past 40 years, advances in molecular methods and microscopy systems have allowed for the identification of several cytoskeletal proteins in the nucleus involved in organizing the genome and regulating gene expression. This review discusses the potential roles for these proteins as internal nucleoskeletal structures and their interactions with the peripheral nuclear lamina (Fig. 1). One should keep in mind that the identification of potential nucleoskeletal proteins has included many different cell types or metazoan model systems. The expression or nuclear localization of some of these proteins might be restricted to certain cell types to form cell type–specific nucleoskeletons.
Figure 1.
Schematic diagram of the nucleoskeleton and its relationships to the nuclear envelope and cytoskeleton. The proposed major components of the vertebrate nucleoskeleton are illustrated. The lamina comprises polymers of A- and B-type lamins. A fraction of lamins are also present in the nuclear interior, possibly associated with chromatin, but not demonstrably in a polymerized form. Short filaments of actin are illustrated, although monomeric actin might also be present. Nuclear spectrin is indicated as associated primarily with the nuclear envelope/lamina. A nuclear isoform of titin also occurs in most eukaryotic cell nuclei and associates with both chromosomes and the lamina. Pore-linked filaments (PLFs) associated with nuclear pore complexes (NPCs) might interact with chromatin and other nucleoskeletal components. The nucleoskeleton is linked to the cytoplasmic skeletal components through LINC (links the nucleoskeleton and cytoskeleton) complexes comprising SUN (Sad1p–UNC-84)-domain proteins and nesprins (nuclear envelope spectrin-repeat proteins). NM, nuclear membrane (nuclear envelope).
2. THE NUCLEAR LAMINS
The nuclear lamina was originally observed as an electron-dense layer or fibrous lamina underlying the inner nuclear membrane in electron photomicrographs of vertebrate cells (Fawcett 1966). Subsequent biochemical fractionation of rat liver nuclei into a pore-complex–lamina fraction with NPCs linked by a protein network structure led to the identification of three major proteins—the nuclear lamins—and several minor proteins (Gerace et al. 1978). Numerous other nonskeletal proteins have also been shown to localize to or associate with the lamina in most cell types (Simon and Wilson 2013). In addition, other proteins that have structural roles in the cytoskeleton or at the plasma membrane have also been suggested to interact with the lamina to form a nucleoskeleton (Zhong et al. 2010).
The lamins are classified as type V intermediate filament proteins, based on biochemical analysis and structural studies (for further information, see Herrmann and Aebi 2016). They are the only nuclear localized members of the intermediate protein family and are present only in metazoans. Lamins are classified as A- or B-type based on their amino acid sequences and biochemical characteristics. In mammalian cells, four A-type lamin proteins can be produced from the LMNA gene by alternative splicing: the predominant isoforms lamin A (LA) and lamin C (LC), a minor isoform LAD10, and a germ-cell-specific isoform LC2. Two B-type lamin proteins are produced from separate genes: lamin B1 (LB1) from LMNB1, and lamin B2 (LB2) from LMNB2. There are no known splice isoforms of the B-type lamins. In invertebrates, only a single lamin gene is present, a B-type lamin, with the exception of the fruitfly Drosophila, which expresses a single B-type gene (lamin Dm0) and a single A-type lamin (LC). Lamin isoforms are not uniformly expressed in all cell types. Most cells express both A- and B-type lamins, but some cells, for example, some T and B cells, express only B-type lamins. Interestingly, LC is expressed in the brain at high levels, but LA is expressed very poorly if at all. Embryonic stem cells express both B-type lamins, but A-type lamins are only expressed as the cells begin to differentiate (Adam and Goldman 2012). The differential expression of lamins in certain cell types suggests that there are unique roles for each lamin isoform. Indeed, the creation of LMNB1 and LMNB2 knockout mice has identified nonredundant roles for each B-type lamin in brain development (Lee et al. 2014).
Similar to all cytoskeletal IF proteins (reviewed in Herrmann and Aebi 2016), lamins have an α-helical central rod domain flanked by short globular amino-terminal “head” and long carboxy-terminal “tail” domains (Parry et al. 1986). In vitro, lamins form coiled-coil dimers through interactions of their central rod domains, which further associate end-to-end to form polar head-to-tail polymers (Heitlinger et al. 1991; Stuurman et al. 1996). These head-to-tail polymers associate laterally in a half-staggered antiparallel fashion to form apolar tetrameric filaments that ultimately interact to form paracrystalline arrays (Heitlinger et al. 1991; Stuurman et al. 1996; Ben-Harush et al. 2009). Although in vitro assembly experiments have provided important information related to the possible mechanisms of lamin assembly and lamin structure, paracrystalline arrays of lamins are not observed in cells, and the physical structure of lamins in cells has yet to be determined. The amount of each lamin present in a lamina could have important implications for the mechanical properties of the structure (Funkhouser et al. 2013; Koster et al. 2015). Lamins A and C are thought to be responsible for the stiffness of the nucleus, and the loss of their expression makes nuclei more susceptible to damage on mechanical stress (Sullivan et al. 1999; Lammerding et al. 2006). In contrast, changing the levels of B-type lamins has little or no effect on the mechanical properties of the nucleus.
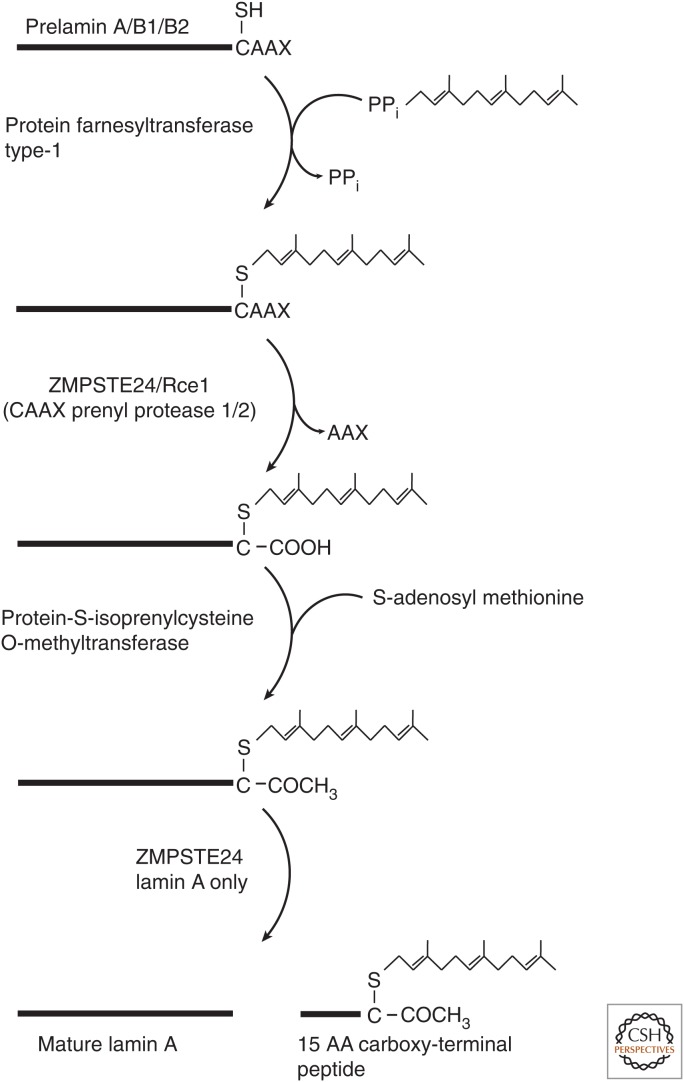
Lamins are unique among the intermediate filament protein family in that they are posttranslationally modified by the addition of a 15-carbon lipid—farnesyl—at their carboxyl terminus by the enzyme farnesyltransferase 1 (protein farnesyltransferase type 1) (Fig. 2). LA, LB1, and LB2 are translated as preproteins with a carboxy-terminal CAAX motif that is a consensus motif for the addition of farnesyl to the proteins. LC lacks a CAAX sequence and is not further processed after translation. Following addition of farnesyl to the cysteine of the CAAX motif, the –AAX residues are proteolytically removed from pre-LA by the zinc metalloprotease ZMPSTE24 (CAAX prenyl protease 1 homolog) and from pre-LB1 and pre-LB2 by the endopeptidase Rce1 (CAAX prenyl protease 2). The free-peptide backbone carboxyl of the farnesyl-cysteine is methylated by isoprenyl carboxymethyltransferase (protein-S-isoprenylcysteine O-methyltransferase) to complete the processing of the CAAX motif. The B-type lamins remain farnesylated for the life of the protein, but the terminal 15 amino acids, including the farnesyl-cysteine, are removed from pre-LA by the protease Zmpste24/FACE1 to produce mature LA (Adam and Goldman 2012). Farnesylation of the lamins is not required for their assembly into the lamina, but might affect the structure of the lamina (Adam et al. 2013; Jung et al. 2013). Interestingly, mice expressing nonfarnesylatable pre-LA or pre-LB2 are normal, but mice expressing pre-LB1 with a mutated CAAX motif die shortly after birth owing to abnormal brain development. Lamins are also posttranslationally modified by several types of modifications, including phosphorylation, acetylation, and sumoylation (Simon and Wilson 2013).
Figure 2.
Carboxy-terminal processing of lamins. The flow diagram illustrates the sequential processing steps necessary to convert prelamin A, prelamin B1, and prelamin B2 to mature proteins. Each successive step requires the completion of the previous modification. Lamins B1 and B2 do not undergo the final protease cleavage step and remain permanently farnesylated and carboxymethylated. The fate of the 15-amino-acid farnesylated peptide removed from prelamin A is unknown, but the peptide is thought to be degraded.
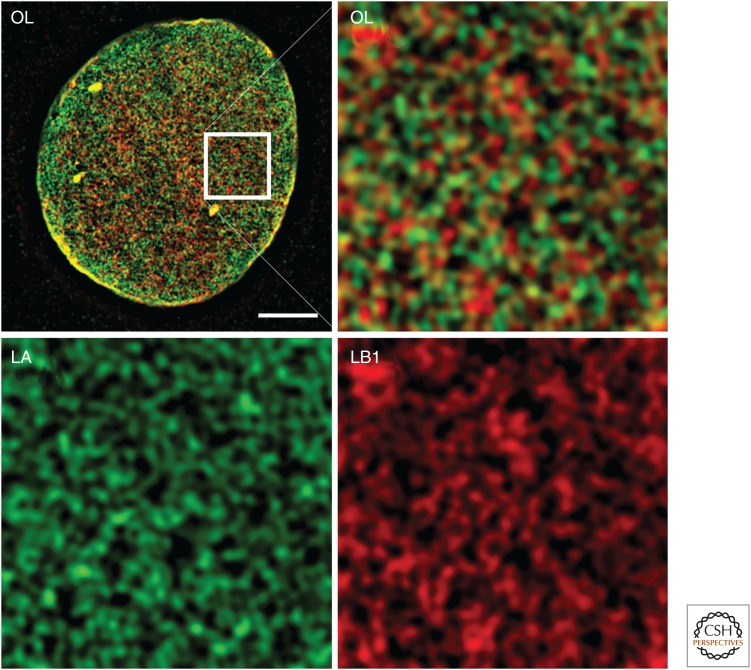
The lamins form a dense structure in the lamina comprising meshworks created by the apparent homotypic polymerization of each lamin isoform, although some degree of heteropolymer formation cannot be excluded based on available data (Delbarre et al. 2006; Kolb et al. 2011). Using superresolution microscopy, lamin meshworks formed by the four major lamin isoforms have been visualized, revealing that each lamin forms a distinct meshwork (Fig. 3) (Shimi et al. 2015). Owing to the resolution limitations of light microscopy, it has not been determined whether these meshworks comprise individual 10-nm lamin fibers or bundles of fibers. Also, because of the high density of chromatin, lamins, and other proteins at the inner face of the NE, it has not been possible to obtain high-resolution images of the endogenous lamin filaments by using electron microscopy in somatic cells. However, using manually isolated NEs from Xenopus oocyte germinal vesicles, regions of the lamina have been visualized by whole-mount EM. In these cells, the lamina appears as a network of 10-nm filaments, with small areas of the lamina arranged in an orthogonal lattice or meshwork pattern (Aebi et al. 1986). In somatic cells, the lamin meshworks are quite stable, with little if any subunit exchange into and out of the lamin polymers and very slow or small-scale diffusion of the meshworks within the plane of the NE. Although the meshworks appear to be largely separate, they must interact in some fashion because the introduction of a mutant lamin, or loss of expression for one type of lamin, affects the structure of the other meshworks (Goldman et al. 2004; Muchir et al. 2004; Shimi et al. 2008). A small population of each lamin also exists in the nuclear interior, or nucleoplasm. The A-type lamins in the nuclear interior are fairly mobile, suggesting that either they do not form extensive polymeric structures or any structures are much more transient than the lamin polymers in the lamina. In contrast, the B-type lamins show very slow mobility in the nuclear interior, but no filamentous structures containing A- or B-type lamins have yet been detected by light or electron microscopy (Shimi et al. 2008).
Figure 3.
Visualization of the lamin meshworks in the lamina of mouse embryo fibroblasts. Lamin A (LA) and lamin B1 (LB1) were detected by indirect immunofluorescence and visualized using three-dimensional structured illumination microscopy (Shimi et al. 2015). The LA and LB1 proteins form separate structures, seen by the lack of overlap between the red and blue colors. The upper left panel is an image of the bottom surface of a fibroblast nucleus. The white box is magnified in the image to the right, and the contribution of each lamin is shown in the lower two panels. Scale bar, 5 µm. OL, overlay.
Differences in the A- and B-type lamins are especially evident during mitosis. At the onset of mitosis, the nuclear lamina disassembles along with the other components of the NE, coincident with their phosphorylation by mitotic kinases (Fields and Thompson 1995). Disassembled A-type lamins diffuse throughout the cell. In contrast, the disassembled B-type lamins are dispersed with the nuclear membranes into the ER, presumably because their attached farnesyl lipid allows them to be retained at the membrane. This idea is supported by the observation that a permanently farnesylated form of LA that is mutated in the premature-aging disease Hutchinson–Gilford progeria (see following section) also partitions into the ER during mitosis (Dechat et al. 2007). A fraction of B-type lamins also accumulate in a poorly defined collection of membranes that cluster within and around the mitotic spindle (Zheng and Tsai 2006). This structure, known as the spindle matrix, is essential for the correct orientation of microtubules in the spindle. Although the exact roles of lamins in the spindle matrix remain to be resolved, LB has been shown to bind, and possibly regulate the activity of, at least one mitotic kinesin and dynein (Ma et al. 2009). At the conclusion of mitosis during telophase, the B-type lamins along with the nuclear membranes associate with the chromosome mass before assembly of A-type lamins into the reforming NE. In some cell lines, A-type lamins only accumulate appreciably in the reformed NE after the chromosomes are completely enclosed by the NE (Moir et al. 2000).
The close association of chromatin, especially heterochromatin, with the nuclear lamina led to speculation that lamins might regulate chromatin structure, transcription, DNA replication, and DNA repair. A-type lamins interact with sites of active DNA replication and might be involved in stabilizing the replication fork (Singh et al. 2013; Butin-Israeli et al. 2015). Cells expressing permanently farnesylated LA owing to mutations or deletion of the Zmpste24 protease also show defects in the repair of double-stranded DNA lesions (Liu et al. 2005). The partial loss of LB1 expression in tumor cells leads to defects in the elongation phase of DNA replication and in several DNA repair processes, although these effects are likely to be indirect as LB1 does not appear to interact with sites of replication or repair (Butin-Israeli et al. 2015). Lamins have also been suggested to be important organizers of chromatin in the nucleus. Using techniques such as proximity labeling of chromatin and chromatin immunoprecipitation, lamins have been shown to be bound to specific regions of chromatin in association with specific subsets of epigenetic histone modifications (Amendola and van Steensel 2014). LB1 interacts with regions of chromatin that bind genomic domains of 50–100 Mb in size, which typically are gene poor and enriched in heterochromatin. Some of these regions are also associated with LA, but other LA-binding regions of chromatin are more “open” and enriched in active genes (Lund et al. 2015). The differences in LA- and LB1-bound regions of chromatin might in part explain differences in gene-expression profiles in cells that express varied amounts of each lamin. Although it is likely that at least some structural features of the lamins are important for regulating chromatin structure and function, direct evidence for this is currently lacking.
3. NUCLEAR ACTIN
Actin is found in all eukaryotic cells and is one of the most highly conserved components of the cytoskeleton, with roles in many biological processes (for reviews, see Bachir et al. 2016; Pollard 2016; Svitkina 2016; Titus 2016). In mammals, six isoforms of actin are expressed from distinct genes. Four of the isoforms are mainly expressed in muscle cells, whereas the other two—β-actin and γ-actin—are expressed universally. After many years of controversy, roles for nuclear β-actin, actin-related proteins (Arps) and other actin-associated proteins have been established (de Lanerolle and Serebryannyy 2011). Actin shuttles between the cytoplasm and nucleus in complexes with actin-binding proteins, entering the nucleus in a complex with cofilin, and returning to the cytoplasm in a complex with profilin, directed by two nuclear export sequences in actin (Pendleton et al. 2003). One reason why the acceptance of a nuclear role for actin took so long is that nuclear actin does not form conventional actin filaments that can be labeled by phalloidin. Most nuclear actin is probably in the form of G-actin, and only a minor fraction is polymeric, which can be detected with a nuclear-actin-specific monoclonal antibody (Schoenenberger et al. 2005). It has also been speculated that nuclear actin polymers might comprise nonclassical conformations of actin or actin modified covalently with one of the small ubiquitin modifiers (SUMO2 or SUMO3) (Hofmann et al. 2009).
The many roles that have been ascribed to nuclear actin include facilitating the transcription, processing and export of messenger RNAs (mRNAs), as well as chromatin remodeling. Actin has been reported to associate with and stimulate all three RNA polymerases (de Lanerolle and Serebryannyy 2011). A nuclear form of myosin, myosin 1, has also been described. Some evidence suggests that nuclear actin and myosin 1 cooperate to position and move chromosomes in the nucleus to regulate gene expression (de Lanerolle and Serebryannyy 2011). How this occurs is unknown as the nature of actin filaments in the nucleus remains obscure and attachment sites for actin filaments, against which forces could be generated, have not been identified. One intriguing possibility is that actin filaments might connect to the peripheral lamina (Zastrow et al. 2004). Actin-binding sites in lamins have been described, and LA can bundle F-actin in vitro. The inner nuclear membrane protein emerin binds to LA directly and can act as a pointed-end F-actin capping protein in vitro (Bengtsson and Wilson 2004).
4. OTHER NUCLEOSKELETAL CANDIDATES
A number of other cytoskeletal proteins have also been implicated as components of a nucleoskeleton. A family of giant elastic proteins, known as titins, function as molecular springs to provide elasticity to vertebrate striated muscle cells (reviewed by Sweeney and Hammers 2016). A nuclear isoform of titin has been reported to exist in most eukaryotic cell nuclei, and associates with both chromosomes and the lamina (Machado and Andrew 2000; Zastrow et al. 2006). The loss of nuclear titin expression is associated with defects in chromosome condensation at mitosis and missegregation. Notably, titin is also an actin-binding protein and can bind lamins in vitro (Zastrow et al. 2006). A large elastic protein such as titin could provide long-range organization and elastic stability for nuclei, but these possibilities have not been tested.
Nuclear mitotic apparatus protein 1 (NuMa) is a 238-kDa protein that forms long coiled-coil homodimers. NuMa can form three-dimensional space-filling structures in vitro, lending weight to the speculation that it might form a space-filling soft scaffold throughout the nucleus that could organize chromosomes (Harborth et al. 1999). As NuMa is nearly as abundant as the lamins in nuclei, it is potentially an important component of an internal nucleoskeleton. The altered expression of NuMa, or expression of fragments of the protein, leads to changes in chromatin organization and nuclear shape, further supporting its proposed role as an organizer of the nuclear volume (Gueth-Hallonet et al. 1998).
The NPC has a basket structure on its nucleoplasmic side comprising, in part, the conserved nucleoporin proteins nucleoporin 153 (Nup153) and translocated promoter region (TPR) (Hase and Cordes 2003). The latter protein is required to maintain the heterochromatin-free channel near the nuclear pore. In amphibian oocytes, lacy open filament channels extend from the nuclear baskets of the NPC and connect to various nuclear bodies (Arlucea et al. 1998). These “pore-linked filaments,” or PLFs, are thought to contain TPR or a related protein, but it is not clear at this stage what comprises the backbone of the PLFs. It has been suggested that the PLFs form chromatin-free channels to facilitate the diffusion or transport of large macromolecules within the nucleus to the NPC (Arlucea et al. 1998). Filamentous networks containing TPR have also been reported in cultured mammalian tumor cells (Cordes et al. 1993). In Drosophila, the ortholog of TPR, Megator, colocalizes with another large Drosophila-specific protein, EAST, in filamentous structures between chromosomes (Wasser and Chia 2000). In turn, EAST promotes the accumulation of actin in the Drosophila cell nucleus and has been proposed to be part of an expandable interchromosomal matrix. The connections between intranuclear filaments and NPCs might be the metazoan equivalent of the linkage of actively transcribed genes and NPCs seen in yeast (Ibarra and Hetzer 2015).
Spectrins are a family of large flexible proteins that crosslink actin and protein 4.1 to membrane-associated proteins to form a membrane skeleton (see Liem 2016). These networks are highly elastic, allowing for the deformation of the membrane and return to normal shape when the deforming force is removed. Nuclear isoforms of spectrin and protein 4.1 have been identified, leading to the proposal of the existence of a complex cortical skeleton in the nucleus, with lamins being the major components anchoring these other accessory skeletal elements (Sridharan et al. 2006). Complexes containing lamins, spectrin isoforms, protein 4.1, and some of the other proposed nucleoskeletal proteins described here have been isolated biochemically from cells. However, a convincing localization and visualization of a membrane-associated skeleton containing these proteins by microscopy is lacking.
In addition to the structural proteins in the nucleus discussed here that could form a nucleoskeleton, it is possible that nonprotein elements could also provide crucial components of a complete functional nucleoskeleton. For example, the chromatin itself could provide a bridge between small isolated regions of internal nucleoskeleton to form an interacting network. In addition, the identification of expressed long noncoding RNAs with no obvious function opens up the intriguing possibility that RNA could be part of a dynamic and flexible nucleoskeleton (Bergmann and Spector 2014).
5. NUCLEAR MOTOR PROTEINS
One surprising result of recent research on the nucleoskeleton has been the discovery of at least six different nuclear myosin isoforms and four nuclear kinesins, despite the lack of conventional tracks for these motors to move along (Simon and Wilson 2013). The only suggested “motor” activity for any of these motors is the rapid relocation of chromosome territories in interphase cells in conjunction with alterations in gene activity (Mehta et al. 2008). Although the evidence for polymerized actin in the nucleus is scant, it is possible that local polymerization of a small number of unconventional actin filaments could be sufficient for chromosome movement during interphase. It is also possible that some or most of these motors have nonmotor functions in the nucleus as regulatory proteins. Nuclear myosin, along with nuclear actin, interacts with both ribosomal and polymerase-II-transcribed genes and evidence suggests that they are important for regulating transcription from these genes (Simon and Wilson 2013). The potential activity of kinesins in the nucleus is even more elusive. The lack of any tracks for kinesins in the interphase nucleus suggests that their roles there are to regulate the activity of other factors. Currently, the best evidence suggests that some nuclear kinesins interact with various factors to regulate some types of DNA repair, although this is likely distinct from any motor activity (Simon and Wilson 2013).
6. CONNECTING THE NUCLEOSKELETON TO THE CYTOSKELETON
Cells can change their patterns of gene expression when exposed to external mechanical forces from outside the cell, a phenomenon called mechanotransduction. Although some of these changes could be due to the activation of signal transduction pathways arising from mechano-sensitive receptors in the plasma membrane, cytoskeletal connections between the plasma membrane and nucleus can also transmit information to the genome. The transmission of mechanical signals from the cytoskeleton to the nucleoskeleton is mediated by a family of protein complexes anchored in the NE collectively called LINC (links the nucleoskeleton and cytoskeleton) complexes.
Making connections between the cytoskeleton and the nucleoskeleton is complicated by the fact that the NE contains a double-bilayer membrane system that must be bridged. LINC complexes are built from KASH (Klarischt, ANC-1, and SYNE homology) domain-containing proteins called nesprins (nuclear envelope spectrin-repeat proteins) embedded in the outer nuclear membrane, and the SUN (Sad1p–UNC-84) proteins embedded in the inner nuclear membrane. Four full-length nesprins and several shorter isoforms have been identified, although potentially dozens more could be produced by alternative expression (Rajgor and Shanahan 2013). The SUN protein family consists of SUN1 and SUN2. The linkage between the outer-nuclear-membrane-associated nesprin and inner-nuclear-membrane-associated SUN proteins occurs when the carboxy-terminal KASH domain of a nesprin binds to the carboxy-terminal end of a SUN protein within the lumen of the NE. SUN-domain proteins and nesprins can form both homo-oligomers and hetero-oligomers, suggesting that the diversity of the types of connections formed is quite complex and specific to particular cell types. The amino-terminal tails of SUN1 and SUN2 project to the nuclear side of the NE and interact with the lamina, completing the connection of the cytoskeleton (actin, microtubules, and intermediate filaments) to the nucleoskeleton (lamins). Short isoforms of nesprin-1 and nesprin-2—nesprin-1α and nesprin-2β—localize to the inner nuclear membrane and have been proposed to interact with lamins and other lamin-associated proteins. In skeletal and smooth-muscle cells, nesprin-1α is enriched and has been proposed to provide additional structural reinforcement to the lamina structure in cells exposed to higher levels of mechanical stress. The full-length isoforms of nesprin-1 and nesprin-2, also known as giant nesprins, can dimerize with nesprin-3 at the outer nuclear membrane to form a meshwork that surrounds the nucleus. This meshwork could act to buffer mechanical forces on the LINC complexes and the nucleus during periods of exceptional force on the nucleus, such as during nuclear migration, or in cells that undergo large stretching forces.
7. THE NUCLEOSKELETON AND DISEASES
Much of the interest in diseases related to the nucleoskeleton has centered on the lamins and lamin-associated proteins. This interest is due to the seemingly ever-increasing number of mutations identified primarily in the LMNA gene encoding lamins A and C linked to a spectrum of 15 different diseases collectively known as laminopathies. A few rare diseases caused by mutations in LMNB1 or LMNB2 have been identified, including autosomal dominant leukodystrophy for LMNB1 (Giorgio et al. 2015) and acquired partial lipodystrophy for LMNB2 (Hegele et al. 2006). The laminopathies include accelerated-aging disorders (progerias), lipodystrophies, muscular dystrophies affecting striated muscle, and peripheral neuropathies (Box 1) (Maraldi et al. 2011). It should be noted that, although mutated lamins are expressed in most tissues, only those cells of mesenchymal origin appear to be affected. Mutations are found throughout the LMNA gene, with the majority being autosomal-dominant missense mutations, although nonsense mutations, compound-heterozygous recessive mutations, deletions, and mutations causing alternative splicing have also been identified. The presence of mutations in all parts of the protein suggests that individual mutations could affect folding (Bollati et al. 2012), assembly (Bank et al. 2012), or interactions with lamin-associated proteins (Krimm et al. 2002). In-depth discussions on the implications of lamin mutations in many laminopathies have been published elsewhere (Gruenbaum and Foisner 2015), and so this review will consider, in more detail, two of the best-studied diseases, the premature aging ailment known as Hutchinson–Gilford progeria syndrome (HGPS) and Emery–Dreifuss muscular dystrophy (EDMD). These two diseases provide some of the best evidence that structural defects in the nucleoskeleton could be involved in the disease pathology (Davidson and Lammerding 2014).
BOX 1. LAMINOPATHIES ATTRIBUTABLE TO MUTATIONS IN LMNA.
Hutchinson–Gilford progeria syndrome (HGPS)
Atypical progerias
Dilated cardiomyopathies
Cardiac conduction defects
Limb girdle muscular dystrophy
Dunnigan-type familial partial lipodystrophy
Mandibuloacral dysplasia
Charcot–Marie–Tooth type 2
Atypical Werner syndrome
Restrictive dermopathy (also mutations in ZMPSTE24)
Autosomal-dominant and autosomal-recessive Emery–Dreifuss muscular dystrophy (EDMA)
Heart–hand syndrome
8. HGPS
Patients with HGPS display a wide array of symptoms, encompassing the spectrum of the laminopathies affecting primarily tissues of mesenchymal origin, including adipose, skin, bone, teeth, and hair. Premature death occurs in the teenage years from heart attack or stroke owing to the formation of atherosclerotic plaques in the coronary and cerebral vasculature. A small number of studies suggests that defective tissue renewal from adult stem cells or increased deposition of extracellular matrix proteins might be responsible for many of the phenotypes in the disease (Prokocimer et al. 2013). HGPS is caused by a dominant silent mutation in exon 11 of LMNA, leading to the activation of a cryptic alternative splice junction. Transcripts with this mutation can splice either to a normal mRNA or to a mRNA missing 150 nucleotides encoded by exon 11. The shorter mRNA encodes prelamin A that cannot be correctly processed, leaving a permanently farnesylated protein, progerin, because the 50-amino-acid segment lost contains the protease cleavage site that is normally used to remove the carboxy-terminal farnesylated peptide during processing. Progerin accumulates at the NE, causes the nucleus to become misshapen (Goldman et al. 2004), and increases the mechanical stiffness of the NE (Dahl et al. 2006). It has been well established that the permanent farnesylation of truncated lamin A is the causative agent in the disease. Expression of progerin in normal cells causes the same changes to the nucleus as it does in the cells of patients, and these effects can be ameliorated by the inhibition of farnesylation using farnesyltransferase inhibitors. In addition, a progeria mouse model that does not express the protease necessary for prelamin A processing—Zmpste24—displays phenotypic features similar to those of several laminopathies, including HGPS (Pendas et al. 2002). Owing to the misshapen appearance of the nucleus and increased mechanical stiffness of nuclei in cells of HGPS patients or in normal cells expressing progerin, it has been suggested that mechanical changes in the lamina might be involved in the disease (Davidson and Lammerding 2014). In this respect, it has been reported that segregation of the lamin isoforms into discrete regions in the lamina can occur in cells expressing progerin (Goldman et al. 2004). In addition, the structure of progerin paracrystals assembled from purified protein in vitro shows subtle, yet important, differences from that of the wild-type protein (Taimen et al. 2009). However, progerin expression also has been reported to result in changes to genome organization, epigenetic modification patterns of histones, gene expression, DNA repair pathways, and telomere maintenance (Prokocimer et al. 2013). Some of these changes have been shown to occur before obvious changes in nuclear shape, and so progerin might express its effects in other ways, such as altered interaction with LA-binding proteins (Dittmer et al. 2014).
9. EDMD
EDMD is the third most common form of muscular dystrophy. Most patients with EDMD are normal at birth, but develop contractures of the elbow or ankle during the second decade of life. A slowly progressive muscle weakening, primarily of the peroneal and humeral muscles, typically follows, along with muscle wasting. Variable in age of onset and severity, some patients eventually lose the ability to walk and have difficulty moving their extremities owing to contractures. In some, but not all patients, variable degrees of cardiomyopathy have also been described. The autosomal-dominant and -recessive forms of EDMD (AD-EDMD and AR-EDMD, respectively) are caused by mutations in LMNA, whereas X-linked EDMD is caused by mutations in the lamin-A-interacting inner nuclear membrane protein emerin (Puckelwartz and McNally 2011). Patients with mutations in the LINC-complex-associated nesprins also display EDMD-like phenotypes, and so it seems that the disease might be related to dysfunction of the NE and its connections to the cytoskeleton (Mejat and Misteli 2010).
The expression of LMNA EDMD mutations in cultured cells causes changes to the lamina structure (Ostlund et al. 2001; Raharjo et al. 2001) that are similar to lamina changes seen in EDMD patient fibroblasts (Muchir et al. 2004). Some mutations also result in the loss of emerin from the NE (Ostlund et al. 2001). However, the only obvious nuclear alterations that have been observed in muscle biopsies of patients are the loss of peripheral heterochromatin in a small percentage of cells (Bonne et al. 1999; Sabatelli et al. 2001). Mouse models expressing mutant lamin A/C to phenocopy the disease must be homozygous rather than the heterozygous state usually found in the human disease. Models expressing different mutations in LMNA show some features of the disease, including muscle wasting, cardiomyopathy, and altered nuclear morphology (Davidson and Lammerding 2014).
Although any of the effects of expressing mutant lamin A/C in HGPS, EDMD, or any other laminopathy could be due to changes in the structure of the lamina nucleoskeleton, any direct evidence for such a linkage is lacking.
10. CONCLUSION
The nucleoskeleton is an important regulator of nuclear structure and functions. The peripheral lamina is the only bona fide nucleoskeleton described to date, but several other cytoskeletal proteins are also found in nuclei and might function as part of the lamina and internal nucleoskeleton. The mechanisms that the nucleoskeleton provides for the regulation of chromatin and gene expression remain to be identified.
Footnotes
Editors: Thomas D. Pollard and Robert D. Goldman
Additional Perspectives on The Cytoskeleton available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Adam SA, Goldman RD. 2012. Insights into the differences between the A- and B-type nuclear lamins. Adv Biol Regul 52: 108–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam SA, Butin-Israeli V, Cleland MM, Shimi T, Goldman RD. 2013. Disruption of lamin B1 and lamin B2 processing and localization by farnesyltransferase inhibitors. Nucleus 4: 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi U, Cohn J, Buhle L, Gerace L. 1986. The nuclear lamina is a meshwork of intermediate-type filaments. Nature 323: 560–564. [DOI] [PubMed] [Google Scholar]
- Amendola M, van Steensel B. 2014. Mechanisms and dynamics of nuclear lamina–genome interactions. Curr Opin Cell Biol 28: 61–68. [DOI] [PubMed] [Google Scholar]
- Arlucea J, Andrade R, Alonso R, Arechaga J. 1998. The nuclear basket of the nuclear pore complex is part of a higher-order filamentous network that is related to chromatin. J Struct Biol 124: 51–58. [DOI] [PubMed] [Google Scholar]
- Austin CM, Bellini M. 2010. The dynamic landscape of the cell nucleus. Mol Reprod Dev 77: 19–28. [DOI] [PubMed] [Google Scholar]
- *.Bachir AI, Horwitz AR, Nelson WJ, Bianchini JM. 2016. Actin-based adhesion modules mediate cell interactions with the extracellular matrix and neighboring cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a023234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank EM, Ben-Harush K, Feinstein N, Medalia O, Gruenbaum Y. 2012. Structural and physiological phenotypes of disease-linked lamin mutations in C. elegans. J Struct Biol 177: 106–112. [DOI] [PubMed] [Google Scholar]
- Bengtsson L, Wilson KL. 2004. Multiple and surprising new functions for emerin, a nuclear membrane protein. Curr Opin Cell Biol 16: 73–79. [DOI] [PubMed] [Google Scholar]
- Ben-Harush K, Wiesel N, Frenkiel-Krispin D, Moeller D, Soreq E, Aebi U, Herrmann H, Gruenbaum Y, Medalia O. 2009. The supramolecular organization of the C. elegans nuclear lamin filament. J Mol Biol 386: 1392–1402. [DOI] [PubMed] [Google Scholar]
- Berezney R, Coffey DS. 1974. Identification of a nuclear protein matrix. Biochem Biophys Res Commun 60: 1410–1417. [DOI] [PubMed] [Google Scholar]
- Bergmann JH, Spector DL. 2014. Long non-coding RNAs: Modulators of nuclear structure and function. Curr Opin Cell Biol 26: 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati M, Barbiroli A, Favalli V, Arbustini E, Charron P, Bolognesi M. 2012. Structures of the lamin A/C R335W and E347K mutants: Implications for dilated cardiolaminopathies. Biochem Biophys Res Commun 418: 217–221. [DOI] [PubMed] [Google Scholar]
- Bonne G, Di Barletta MR, Varnous S, Becane HM, Hammouda EH, Merlini L, Muntoni F, Greenberg CR, Gary F, Urtizberea JA, et al. 1999. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery–Dreifuss muscular dystrophy. Nat Genet 21: 285–288. [DOI] [PubMed] [Google Scholar]
- Butin-Israeli V, Adam SA, Jain N, Otte GL, Neems D, Wiesmuller L, Berger SL, Goldman RD. 2015. Role of lamin B1 in chromatin instability. Mol Cell Biol 35: 884–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Kohler A, Stuurman N, van Driel R, Franke WW. 1993. Intranuclear filaments containing a nuclear pore complex protein. J Cell Biol 123: 1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl KN, Scaffidi P, Islam MF, Yodh AG, Wilson KL, Misteli T. 2006. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson–Gilford progeria syndrome. Proc Natl Acad Sci 103: 10271–10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson PM, Lammerding J. 2014. Broken nuclei−lamins, nuclear mechanics, and disease. Trends Cell Biol 24: 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Shimi T, Adam SA, Rusinol AE, Andres DA, Spielmann HP, Sinensky MS, Goldman RD. 2007. Alterations in mitosis and cell cycle progression caused by a mutant lamin A known to accelerate human aging. Proc Natl Acad Sci 104: 4955–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lanerolle P, Serebryannyy L. 2011. Nuclear actin and myosins: Life without filaments. Nat Cell Biol 13: 1282–1288. [DOI] [PubMed] [Google Scholar]
- Delbarre E, Tramier M, Coppey-Moisan M, Gaillard C, Courvalin JC, Buendia B. 2006. The truncated prelamin A in Hutchinson–Gilford progeria syndrome alters segregation of A-type and B-type lamin homopolymers. Hum Mol Genet 15: 1113–1122. [DOI] [PubMed] [Google Scholar]
- Dittmer TA, Sahni N, Kubben N, Hill DE, Vidal M, Burgess RC, Roukos V, Misteli T. 2014. Systematic identification of pathological lamin A interactors. Mol Biol Cell 25: 1493–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett DW. 1966. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am J Anat 119: 129–145. [DOI] [PubMed] [Google Scholar]
- Fields AP, Thompson LJ. 1995. The regulation of mitotic nuclear envelope breakdown: A role for multiple lamin kinases. Prog Cell Cycle Res 1: 271–286. [DOI] [PubMed] [Google Scholar]
- Funkhouser CM, Sknepnek R, Shimi T, Goldman AE, Goldman RD, Olvera de la Cruz M. 2013. Mechanical model of blebbing in nuclear lamin meshworks. Proc Natl Acad Sci 110: 3248–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace L, Blum A, Blobel G. 1978. Immunocytochemical localization of the major polypeptides of the nuclear pore complex-lamina fraction. Interphase and mitotic distribution. J Cell Biol 79: 546–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgio E, Robyr D, Spielmann M, Ferrero E, Di Gregorio E, Imperiale D, Vaula G, Stamoulis G, Santoni F, Atzori C, et al. 2015. A large genomic deletion leads to enhancer adoption by the lamin B1 gene: A second path to autosomal dominant adult-onset demyelinating leukodystrophy (ADLD). Hum Mol Genet 24: 3143–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, et al. 2004. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson–Gilford progeria syndrome. Proc Natl Acad Sci 101: 8963–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Foisner R. 2015. Lamins: Nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem 84: 131–164. [DOI] [PubMed] [Google Scholar]
- Gueth-Hallonet C, Wang J, Harborth J, Weber K, Osborn M. 1998. Induction of a regular nuclear lattice by overexpression of NuMA. Exp Cell Res 243: 434–452. [DOI] [PubMed] [Google Scholar]
- Harborth J, Wang J, Gueth-Hallonet C, Weber K, Osborn M. 1999. Self assembly of NuMA: Multiarm oligomers as structural units of a nuclear lattice. EMBO J 18: 1689–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase ME, Cordes VC. 2003. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell 14: 1923–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegele RA, Cao H, Liu DM, Costain GA, Charlton-Menys V, Rodger NW, Durrington PN. 2006. Sequencing of the reannotated LMNB2 gene reveals novel mutations in patients with acquired partial lipodystrophy. Amer J Hum Genet 79: 383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitlinger E, Peter M, Haner M, Lustig A, Aebi U, Nigg EA. 1991. Expression of chicken lamin B2 in Escherichia coli: Characterization of its structure, assembly, and molecular interactions. J Cell Biol 113: 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Herrmann H, Aebi U. 2016. Intermediate filaments: Structure and assembly. Cold Spring Harb Perspect Biol 8: a018242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann WA, Arduini A, Nicol SM, Camacho CJ, Lessard JL, Fuller-Pace FV, de Lanerolle P. 2009. SUMOylation of nuclear actin. J Cell Biol 186: 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A, Hetzer MW. 2015. Nuclear pore proteins and the control of genome functions. Genes Dev 29: 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HJ, Nobumori C, Goulbourne CN, Tu Y, Lee JM, Tatar A, Wu D, Yoshinaga Y, de Jong PJ, Coffinier C, et al. 2013. Farnesylation of lamin B1 is important for retention of nuclear chromatin during neuronal migration. Proc Natl Acad Sci 110: E1923–E1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb T, Maass K, Hergt M, Aebi U, Herrmann H. 2011. Lamin A and lamin C form homodimers and coexist in higher complex forms both in the nucleoplasmic fraction and in the lamina of cultured human cells. Nucleus 2: 425–433. [DOI] [PubMed] [Google Scholar]
- Koster S, Weitz DA, Goldman RD, Aebi U, Herrmann H. 2015. Intermediate filament mechanics in vitro and in the cell: From coiled coils to filaments, fibers and networks. Curr Opin Cell Biol 32: 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimm I, Ostlund C, Gilquin B, Couprie J, Hossenlopp P, Mornon JP, Bonne G, Courvalin JC, Worman HJ, Zinn-Justin S. 2002. The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure 10: 811–823. [DOI] [PubMed] [Google Scholar]
- Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT. 2006. Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem 281: 25768–25780. [DOI] [PubMed] [Google Scholar]
- Lee JM, Tu Y, Tatar A, Wu D, Nobumori C, Jung HJ, Yoshinaga Y, Coffinier C, de Jong PJ, Fong LG, et al. 2014. Reciprocal knock-in mice to investigate the functional redundancy of lamin B1 and lamin B2. Mol Biol Cell 25: 1666–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Liem RKH. 2016. Cytoskeletal integrators: The spectrin superfamily. Cold Spring Harb Perspect Biol 8: a018259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Wang J, Chan KM, Tjia WM, Deng W, Guan X, Huang JD, Li KM, Chau PY, Chen DJ, et al. 2005. Genomic instability in laminopathy-based premature aging. Nat Med 11: 780–785. [DOI] [PubMed] [Google Scholar]
- Lund EG, Duband-Goulet I, Oldenburg A, Buendia B, Collas P. 2015. Distinct features of lamin A-interacting chromatin domains mapped by ChIP-sequencing from sonicated or micrococcal nuclease-digested chromatin. Nucleus 6: 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Tsai MY, Wang S, Lu B, Chen R, Yates JR III, Zhu X, Zheng Y. 2009. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nat Cell Biol 11: 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado C, Andrew DJ. 2000. D-Titin: A giant protein with dual roles in chromosomes and muscles. J Cell Biol 151: 639–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraldi NM, Capanni C, Cenni V, Fini M, Lattanzi G. 2011. Laminopathies and lamin-associated signaling pathways. J Cell Biochem 112: 979–992. [DOI] [PubMed] [Google Scholar]
- Mehta IS, Elcock LS, Amira M, Kill IR, Bridger JM. 2008. Nuclear motors and nuclear structures containing A-type lamins and emerin: Is there a functional link? Biochem Soc Trans 36: 1384–1388. [DOI] [PubMed] [Google Scholar]
- Mejat A, Misteli T. 2010. LINC complexes in health and disease. Nucleus 1: 40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Yoon M, Khuon S, Goldman RD. 2000. Nuclear lamins A and B1: Different pathways of assembly during nuclear envelope formation in living cells. J Cell Biol 151: 1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchir A, Medioni J, Laluc M, Massart C, Arimura T, van der Kooi AJ, Desguerre I, Mayer M, Ferrer X, Briault S, et al. 2004. Nuclear envelope alterations in fibroblasts from patients with muscular dystrophy, cardiomyopathy, and partial lipodystrophy carrying lamin A/C gene mutations. Muscle Nerve 30: 444–450. [DOI] [PubMed] [Google Scholar]
- Ostlund C, Bonne G, Schwartz K, Worman HJ. 2001. Properties of lamin A mutants found in Emery–Dreifuss muscular dystrophy, cardiomyopathy and Dunnigan-type partial lipodystrophy. J Cell Sci 114: 4435–4445. [DOI] [PubMed] [Google Scholar]
- Parry DA, Conway JF, Steinert PM. 1986. Structural studies on lamin. Similarities and differences between lamin and intermediate-filament proteins. Biochem J 238: 305–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. 2000. Half a century of “the nuclear matrix”. Mol Biol Cell 11: 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendas AM, Zhou Z, Cadinanos J, Freije JM, Wang J, Hultenby K, Astudillo A, Wernerson A, Rodriguez F, Tryggvason K, et al. 2002. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat Genet 31: 94–99. [DOI] [PubMed] [Google Scholar]
- Pendleton A, Pope B, Weeds A, Koffer A. 2003. Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J Biol Chem 278: 14394–14400. [DOI] [PubMed] [Google Scholar]
- *.Pollard TD. 2016. Actin and actin-binding proteins. Cold Spring Harb Perspect Biol 8: a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokocimer M, Barkan R, Gruenbaum Y. 2013. Hutchinson–Gilford progeria syndrome through the lens of transcription. Aging Cell 12: 533–543. [DOI] [PubMed] [Google Scholar]
- Puckelwartz M, McNally EM. 2011. Emery–Dreifuss muscular dystrophy. Handb Clin Neurol 101: 155–166. [DOI] [PubMed] [Google Scholar]
- Raharjo WH, Enarson P, Sullivan T, Stewart CL, Burke B. 2001. Nuclear envelope defects associated with LMNA mutations cause dilated cardiomyopathy and Emery–Dreifuss muscular dystrophy. J Cell Sci 114: 4447–4457. [DOI] [PubMed] [Google Scholar]
- Rajgor D, Shanahan CM. 2013. Nesprins: From the nuclear envelope and beyond. Expert Rev Mol Med 15: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatelli P, Lattanzi G, Ognibene A, Columbaro M, Capanni C, Merlini L, Maraldi NM, Squarzoni S. 2001. Nuclear alterations in autosomal-dominant Emery–Dreifuss muscular dystrophy. Muscle Nerve 24: 826–829. [DOI] [PubMed] [Google Scholar]
- Schoenenberger CA, Buchmeier S, Boerries M, Sutterlin R, Aebi U, Jockusch BM. 2005. Conformation-specific antibodies reveal distinct actin structures in the nucleus and the cytoplasm. J Struct Biol 152: 157–168. [DOI] [PubMed] [Google Scholar]
- Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, et al. 2008. The A- and B-type nuclear lamin networks: Microdomains involved in chromatin organization and transcription. Genes Dev 22: 3409–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, Jaqaman K, Goldman RD. 2015. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol Biol Cell 26: 4075–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DN, Wilson KL. 2013. Partners and post-translational modifications of nuclear lamins. Chromosoma 122: 13–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Hunt CR, Pandita RK, Kumar R, Yang CR, Horikoshi N, Bachoo R, Serag S, Story MD, Shay JW, et al. 2013. Lamin A/C depletion enhances DNA damage-induced stalled replication fork arrest. Mol Cell Biol 33: 1210–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman JE, Trinkle-Mulcahy L. 2014. Nuclear bodies: New insights into assembly/dynamics and disease relevance. Curr Opin Cell Biol 28: 76–83. [DOI] [PubMed] [Google Scholar]
- Sridharan DM, McMahon LW, Lambert MW. 2006. αII-Spectrin interacts with five groups of functionally important proteins in the nucleus. Cell Biol Int 30: 866–878. [DOI] [PubMed] [Google Scholar]
- Stancheva I, Schirmer EC. 2014. Nuclear envelope: Connecting structural genome organization to regulation of gene expression. Adv Exp Med Biol 773: 209–244. [DOI] [PubMed] [Google Scholar]
- Stuurman N, Sasse B, Fisher PA. 1996. Intermediate filament protein polymerization: Molecular analysis of Drosophila nuclear lamin head-to-tail binding. J Struct Biol 117: 1–15. [DOI] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. 1999. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol 147: 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Svitkina T. 2016. The actin cytoskeleton and actin-based motility. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sweeney HL, Hammers DW. 2016. Muscle contraction. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taimen P, Pfleghaar K, Shimi T, Moller D, Ben-Harush K, Erdos MR, Adam SA, Herrmann H, Medalia O, Collins FS, et al. 2009. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc Natl Acad Sci 106: 20788–20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Titus MA. 2016. Myosin-driven intracellular transport. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a021972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasser M, Chia W. 2000. The EAST protein of Drosophila controls an expandable nuclear endoskeleton. Nat Cell Biol 2: 268–275. [DOI] [PubMed] [Google Scholar]
- Zastrow MS, Vlcek S, Wilson KL. 2004. Proteins that bind A-type lamins: Integrating isolated clues. J Cell Sci 117: 979–987. [DOI] [PubMed] [Google Scholar]
- Zastrow MS, Flaherty DB, Benian GM, Wilson KL. 2006. Nuclear titin interacts with A- and B-type lamins in vitro and in vivo. J Cell Sci 119: 239–249. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Tsai MY. 2006. The mitotic spindle matrix: A fibro-membranous lamin connection. Cell Cycle 5: 2345–2347. [DOI] [PubMed] [Google Scholar]
- Zhong Z, Wilson KL, Dahl KN. 2010. Beyond lamins: Other structural components of the nucleoskeleton. Methods Cell Biol 98: 97–119. [DOI] [PMC free article] [PubMed] [Google Scholar]