Abstract
Amyloids are highly ordered protein aggregates that are associated with both disease (including PrP prion, Alzheimer’s, and Parkinson’s) and biological function. The amyloid structure is composed of the cross-β-sheet entity, which is an almost indefinitely repeating two-layered intermolecular β-sheet motif. The three-dimensional (3D) structure is unique among protein folds because it folds only upon intermolecular contacts (for a folding to occur, only short sequences of amino acid residues are required), and the structure repeats itself at the atomic level (i.e., every 4.7 Å). As a consequence of this structure, among others, it can grow by recruiting corresponding amyloid peptide/protein and thus has the capacity to be an infectious protein (i.e., a prion). Furthermore, its repetitiveness can translate what would be a nonspecific activity as monomer into a potent one through cooperativity. Because of these and other properties, the activities of amyloids are manifold and include peptide storage, template assistance, loss of function, gain of function, generation of toxicity, membrane binding, infectivity, and more. This review summarizes the structural nature of the cross-β-sheet motif on the basis of a few high-resolution structural studies of amyloids in the context of potential biological activities.
Amyloids consist of a β-sheet motif that repeats almost indefinitely. This and other unique properties bestow on these aggregates many biological activities (e.g., template assistance, membrane binding, and infectivity).
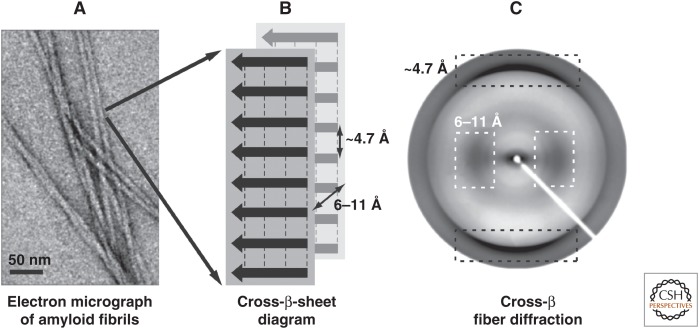
The term “amyloid” has been introduced to describe macroscopic tissue abnormalities that exhibit a pale, waxy appearance and produce a positive iodine-staining reaction (Sipe and Cohen 2000). Consequently, the classic histological definition of an amyloid is an extracellular protein deposit that binds Congo red and produces a yellow-green birefringence under polarized light (Westermark et al. 1999). Because X-ray fiber diffraction of aligned amyloids yields a characteristic diffraction pattern with a meridional reflection at 4.7 Å and an equatorial reflection (i.e., 90° off the other reflection) at ∼8–11 Å, the structural organization of the amyloid has been proposed to be the cross-β-sheet motif (Fig. 1) (Sunde and Blake 1997; Sunde et al. 1997). The cross-β-sheet motif was first described by Astbury in 1935 when measuring an X-ray diffraction pattern from poached, stretched egg white, also a protein aggregate (Astbury et al. 1935). Based on this aforementioned pattern, he suggested that the protein chains of the egg white pack as two intermolecular β-sheets with the individual strands of each β-sheet perpendicular to the fibril axis (4.7 Å spacing), whereas the two β-sheets (∼8- to 11-Å spacing) are parallel to the fibril axis (i.e., 90° off the β-sheet axis) (Fig. 1). This packing results in an ordered repetitive arrangement of thousands of copies of a peptide/protein (Astbury et al. 1935) that with electron microscopy (EM) appear as long (up to several microns in length), nonbranched filaments with diameters from 6 to 12 nm. They are usually called amyloid fibrils (Fig. 1) (Sunde and Blake 1997; Sunde et al. 1997). Astbury further suggested that any protein may aggregate into amyloids (Astbury et al. 1935). It took, however, several decades thereafter until the amyloid proteins associated with the several dozens of amyloid diseases were discovered (Glenner et al. 1969; Glenner and Wong 1983) and another few decades until functional amyloids were discovered (Cousteau et al. 1997; Fowler et al. 2006). It is the aim of this review to discuss the structural nature of the cross-β-sheet motif in detail and put its potential actions into perspective.
Figure 1.
Structural organization of the amyloid. (A) Amyloid fibrils are composed of long filaments that are visible in negatively stained transmission electron micrographs. (B) The schematic diagram of the cross-β-sheets in a fibril, with the backbone hydrogen bonds represented by dashed lines, indicates the repetitive spacings that give rise to (C) the typical fiber diffraction pattern with a meridional reflection at ∼4.7 Å (black dashed boxes) and an equatorial reflection at ∼6–11 Å (white dashed boxes). (From Greenwald and Riek 2010; reprinted, with permission, from Elsevier © 2010.)
THE THREE-DIMENSIONAL STRUCTURES OF PEPTIDE AMYLOIDS
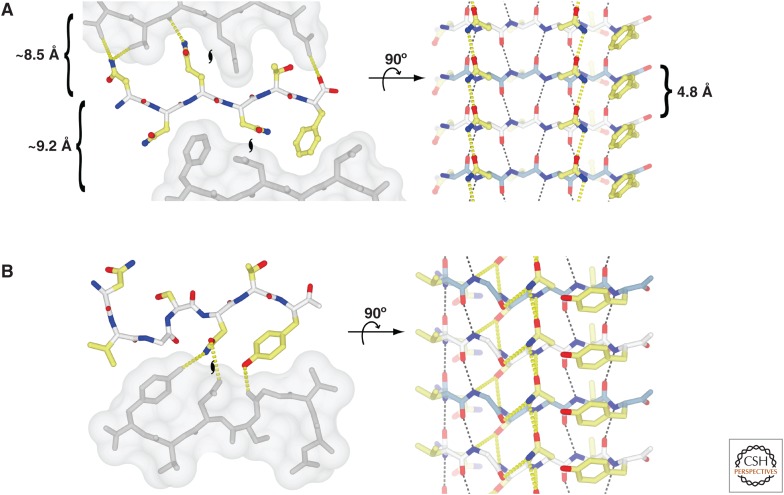
Eisenberg et al. (2006) determined the three-dimensional (3D) structures of short fibril-forming peptide segments of amyloid proteins (such as PrP, Sup35, insulin, Aβ, tau, and amylin) from microcrystals by X-ray crystallography (Nelson et al. 2005; Nelson and Eisenberg 2006a,b; Sawaya et al. 2007). The soluble peptides are able to form microcrystals as well as amyloid fibrils. The microcrystals and fibrils grow in the same condition and coexist in the same solution. Some fibrils grow from the tips of crystals, and the microcrystals can seed the growth of amyloid fibrils, indicating that the peptide conformation in the microcrystals is identical to the fibril structure. All of the structures are composed of intermolecular β-sheets, which are perpendicular to the long axis of the microcrystal reminiscent of the cross-β-sheet motif (Fig. 1). The β-sheets are packed in the crystal with two distinct interfaces, termed “dry” and “wet” (Fig. 2). The dry interface consists of side-chain interdigitation of complementary side chains via van der Waals interactions and hydrogen bonds such as Asn/Gln ladders (Nelson et al. 2005). This structural motif is called the “steric zipper.” In contrast to the dry interface, the wet interface is similar to intermolecular contacts in protein crystals. This finding suggests that the stable structural unit of the microcrystals is a pair of β-sheets forming a minimal cross-β-sheet motif.
Figure 2.
Steric zippers, hydrogen bonding, and van der Waals packing in atomic-resolution amyloid crystal structures. The interactions found in cross-β fibrils are exemplified by the structures of two peptides: (A) NNQNTF (PDB code: 3FVA), and (B) NVGSNTY (PDB code: 3FTL). The left panels show the views looking down the axes of the microcrystal “fibrils,” with the neighboring dry interfaces depicted as a solvent-accessible surface (gray) to show the tight interdigitation of the side chains. The intersheet hydrogen bonds are shown as yellow dashed lines (only a unique set is shown, not symmetry equivalent bonds). The coloring scheme is white for main-chain carbon, yellow for side-chain carbon, blue for nitrogen, and red for oxygen. The positions of the twofold screw axes that relate the individual molecules in the fibril are indicated with black hurricane symbols. The right panels show the view from the side of the “fibrils” showing only the β-sheet corresponding to the colored strand from the left panel. The intrasheet hydrogen bonds are shown (black for main-chain-only bonds, and yellow for any side-chain interactions), and the coloring of the main-chain carbons of individual strands alternates blue and white. The front face of the sheet in A has two Asn ladders and a Phe ladder. The peptide in B is actually composed of two short β-strands with a kink between them. The kink allows the Asn in the ladder to make a hydrogen bond to the main-chain oxygen of residue three (Gly) within the same molecule. In A, the spacing between the repetitive structures is indicated (the left panel has only the average spacing between main-chain atoms because the intersheet distances are not related by crystallographic translations along unit cells). (From Greenwald and Riek 2010; reprinted, with permission, from Elsevier © 2010.)
Although all of the structures determined are composed of the cross-β-sheet motif, (1) the β-sheets are either of a parallel or an antiparallel nature, (2) they may pack either with the same (“face to face”) or to different (“face to back”) surfaces adjacent to one another, and (3) the two closely packed β-sheets can be oriented either in the same direction (“up–up”) or in the opposite direction (“up–down”). Combinations of these three structural arrangements give eight theoretically possible classes of steric zippers.
Although the crystal structures may indicate that the structural complexity of amyloid fibrils is limited, it is noteworthy that only peptides (and not entire proteins) have been crystallized so far in an amyloid-like conformation, and, hence, one should not underestimate the possible structural complexity of amyloid fibrils. In particular, a complex structure might be derived from a combination of steric-zipper-like structures composed by several peptide segments within a single polypeptide chain, as exemplified by the 3D structure of the HET-s(218-289) amyloid fibrils discussed in the following section.
SOLID-STATE NUCLEAR MAGNETIC RESONANCE: THE 3D STRUCTURE OF HET-s(218-289) AMYLOID FIBRILS
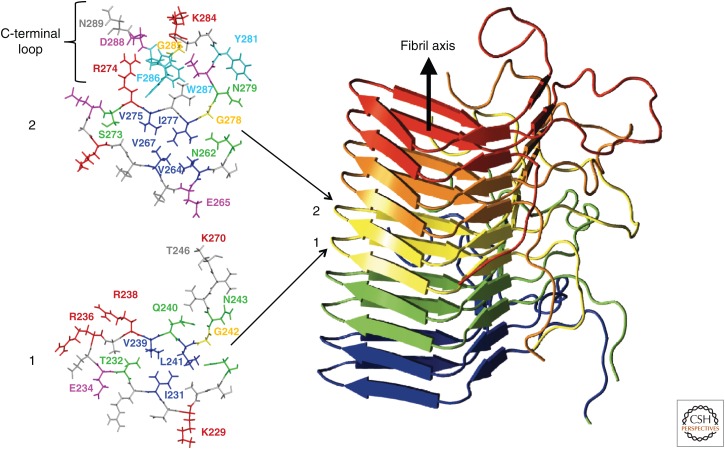
The 3D structure of HET-s(218-289) amyloid fibrils has been determined by solid-state nuclear magnetic resonance (NMR) on the basis of an extensive set of experimental restraints (Fig. 3) (Wasmer et al. 2008). The 3D structure is composed of a left-handed β-solenoid, with each molecule forming two helical windings connected by an unstructured 15-residues-long segment. The four β-strands β1–β4 are linked by two short loops (β1 to β2 and β3 to β4) (Fig. 3). This β-solenoid structure is built up of structural entities observed in the crystal structures of the amyloid peptides (see above). In particular, the four β-sheets of HET-s(218-289) fibrils are parallel and in-register. The interactions between three strands are of a hydrophobic nature and are accompanied at the arches by several hydrogen bonds and an Asn-ladder consisting of residues N226/N262. The surface residues are mostly charged and form several salt bridges. At the C-terminus of the domain, a C-terminal loop folds back onto the core region and forms a semihydrophobic pocket, which can be seen as an extension of the hydrophobic core (van Melckebeke et al. 2010). These structural arrangements result in an overall β-helix characteristic fold. This fold is of much higher structural complexity than the structures of the short peptide fibrils, a complexity that is typically observed in soluble protein folds. Indeed, the three-stranded triangular hydrophobic core superimposes well with the β-solenoid structures of soluble proteins, such as hemagglutinin. Interestingly, a recent mutagenesis study of HET-s(218-289) revealed that its 3D structure is much less sensitive to side-chain alterations than usually soluble proteins, and the C-terminal semihydrophobic loop that folds back to the core region is critical for prion activation (Daskalov et al. 2014).
Figure 3.
The β-solenoid motif of HET-s. The right view is a ribbon representation of the HET-s prion-forming domain fibril, with individual peptide chains colored individually. On the left, a top view of layers 1 and 2 with all the side chains is shown. A one-letter amino acid residue code is used for the identification of the residues. In addition, the C-terminal loop in layer 2 is indicated. (From Wasmer et al. 2008; reprinted, with permission, from the American Association for the Advancement of Science © 2008.)
STRUCTURAL POLYMORPHISM OF AMYLOIDS
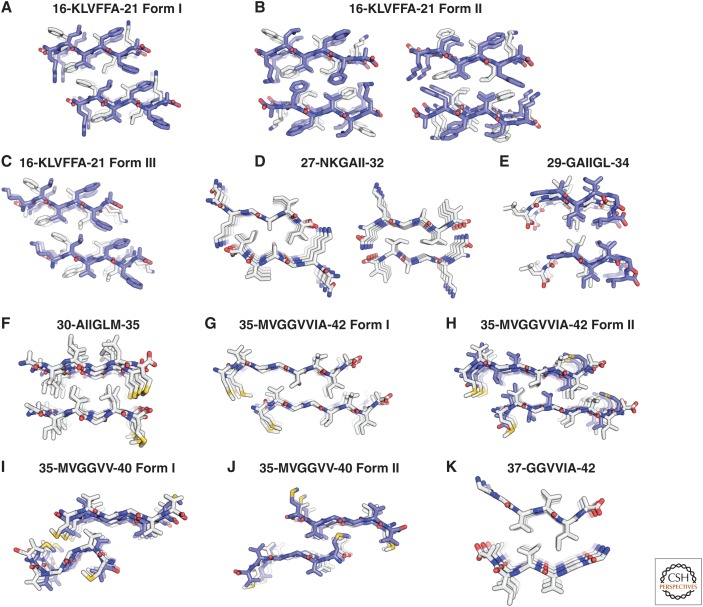
Biophysical investigations suggest the presence of heterogeneous conformations of amyloids within a single sample, which are termed “polymorphisms.” On the mesoscopic scale, EM can differentiate polymorphic fibrils based on the degree of twisting, the number of filaments per fibril, and the diameter or mass per length of the fibrils (Fändrich et al. 2009). Structural heterogeneity is also observed in solid-state NMR spectra, either as multiple NMR signals per chemical shift resonance (Petkova et al. 2005; Bousset et al. 2013) or as very broad resonance lines, indicating that the origin of the macroscopically observed polymorphisms in distinct structures is (at least in part) at the atomic level. This notion is further strengthened by the high-resolution X-ray crystallography studies of short amyloid peptides that form different microcrystals comprising different classes of steric zippers. For example, the fragment of Aβ comprising residues 35-42 crystallized in two forms with both parallel and antiparallel β-sheet stacking, as shown in Figure 4 (Colletier et al. 2011).
Figure 4.
Polymorphisms in amyloids. Crystal structures of Aβ segments, shown in projection down the fiber axes. The Aβ segments are packed as pairs of interdigitated β-sheets, generally with a dry interface between them (termed “steric zippers”), forming the basic unit of the fibril (see main text). The view in this figure looks down the fibril axis, showing only four layers of β-strands in each β-sheet; actual fibrils can contain more than 100,000 layers. Each panel is labeled with the amino acid sequence of each segment and the starting and ending residue numbers. Molecules are shown as sticks with noncarbon atoms colored by atom type. Antiparallel strands forming β-sheet structures (A–C,E,H–J) are alternately colored with carbons colored white and blue. In structures with β-sheets composed of parallel strands (D,F,G,K), the carbons are in white. The closest partners across the dry interface share the same color. Some of the panels are split in two halves; each half represents a different dry interface within the same crystal structure. (From Colletier et al. 2011; reprinted, with permission, from The National Academy of Sciences © 2011.)
There are several types of polymorphisms.
The packing polymorphism is based on a different packing between the β-strands, as shown in Figure 4, for the peptide fragment Aβ(35-42) all having an antiparallel β-sheet arrangement (note that different packings are also possible when both structures have a parallel β-sheet arrangement).
The segmental polymorphism is defined such that distinct segments of an amyloid peptide/protein are involved in the cross-β-sheet conformation. For example, for Aβ(1-42), among others, the segments 16-21, 27-32, or 35-42 all individually form an amyloid (Fig. 4) (Colletier et al. 2011), and each of these segments individually may be part of the core of the amyloid.
In the assembly polymorphism, the packing between protofilaments forming a fibril is different between polymorphs, whereas at the local atomic structural level, the structure could be identical. The presence of the assembly polymorphism is based on the rationale that the tight binding between protofilaments of a fibril is based on weak and nonspecific interactions that are translated into potent interaction by the repeat-induced cooperativity.
Finally, it has been suggested that a polymorph may even appear at the amino acid residue side-chain level (Greenwald and Riek 2010).
Regardless of their molecular origins, all four classes of polymorphism (i.e., segmental, packing, side-chain, and assembly polymorphism) can be propagated by the self-templating nature of amyloids that ensures the persistence of distinct amyloid fibrils, each consisting of a unique conformation or polymorph. However, owing to the stochastic nature of nucleation and the fact that nucleation is often a rate-limiting step in the aggregation process, the Boltzmann distribution of peptide conformations is not necessarily reflected in mixtures of polymorphic amyloid aggregates.
How many polymorphs exist for a given amyloid is an important open question, particularly for prion diseases with their many prion strains (Prusiner 1991; Colby and Prusiner 2011). Based on the four classes of polymorphs discussed and the potential that each of the polymorphs may exist independently, the number of possible polymorphs could be skyrocketing. For example, if an amyloid protein has three segmental, three packing, three assembly, and three side-chain polymorphs, respectively, the number of possible polymorphs for the given theoretical example is 81 (i.e., 34). Although this number is most likely an overestimation in reality, it is noteworthy that the number of prion strains known is in the two-digit region (Prusiner 1991; Colby and Prusiner 2011).
THE CROSS-β-SHEET STRUCTURE FROM AN ACTIVITY POINT OF VIEW
The uniqueness of the cross-β-sheet structure gives rise to a plethora of potential functions—both good and bad for the organism. The structural repetitiveness of the aggregates at subnanomolar resolution and, concomitantly, the close spacing of identical side chains are unique. This repetitiveness can generate specificity and binding affinity where none would exist without a repeating structure. For example, a single amphipathic β-strand would have only little affinity for a membrane, but a fibril made up of this peptide could bind tightly to a lipid membrane (also having a repetitive structure). The same is true of an amyloid composed of basic side chains interacting with DNA (having also a repetitive structure). In addition to supporting cooperative binding, the amyloid structure allows very short peptides (as short as a few residues) to assemble into complexes with regular secondary structure, thus mimicking some features of larger folded proteins (Rufo et al. 2014). Another interesting property is that the folding into the cross-β-sheet entity is concentration-dependent; thus, the formation and, concurrently, the action of the amyloid can be induced and/or controlled by the concentration of the amyloid peptide/protein. Furthermore, the amyloid can grow by recruiting corresponding amyloid peptide/protein. Finally, the two-state nature of the amyloid peptide between a soluble and an insoluble state is noteworthy.
Because of these special structural features and biophysical properties, each amyloid entity can have a diverse set of activities that rivals that of soluble proteins. The growing list of known amyloid-related activities shows that the formation of amyloids may result both in a loss of function of the polypeptide, as described for yeast prions (True and Lindquist 2000; Osherovich and Weissman 2002), or in a gain of function, as in the case of the HET-s prion (Maddelein et al. 2002). Pmel17 amyloids serve as a template for ligand binding (Fowler et al. 2006), whereas other amyloids induce a specific toxic response, as shown for the HET-s prion system (Maddelein et al. 2002). Amyloids may be involved in programmed cell death in fungi (Cousteau et al. 1997; Seuring et al. 2012) as well as in mammals (i.e., necroptosis) (Li et al. 2012). The structural repetitiveness of the amyloid fibril provides an ideal template for making it into a transmissible or infectious fold, as discussed in the following section.
THE INFECTIOUS NATURE OF AMYLOIDS
Based on the cross-β-sheet structure with its intermolecular β-strands, the amyloid can, in principle, grow by recruiting the corresponding soluble amyloid protein. Hence, under certain circumstances, amyloid replication can occur in an organism once exposed by the corresponding amyloid (seed). The replication of the amyloid depends thereby on the concentration of the soluble amyloid protein in the host as well as on the concentration of the amyloid, the growth rate of the amyloid, and the rate of breakage of the amyloid fiber into seeds (Tanaka et al. 2004). According to this theoretical framework, manipulation of these variables (such as the concentration of the amyloid protein in the host or the presence of chaperones sequestering large amyloids into smaller seeds) (Chernoff et al. 1995) could transform any amyloid into an entity with infectivity potential (Riek 2006). However, to be a prion, the amyloid needs to have an additional property: It must be transmissible between individuals, as discussed in a review by Aguzzi and Rajendran (2009). To differentiate between an amyloid that is transmissible between cells and a prion, which is by definition transmissible between species, Aguzzi and Rajendran (2009) introduced the term “prionoid,” which is a protein (amyloid) aggregate that replicates in an organism and is transmissible between cells, but not between individuals, and from a medical point of view is not infectious. For example, the Aβ amyloid, which is associated with Alzheimer’s disease, becomes transmissible in a mouse that overexpresses the precursor to Aβ (Kane et al. 2000; Meyer-Luehmann et al. 2006) and thus can be classified as a prionoid. Other amyloids that appear to be prionoids are α-synuclein amyloids associated with Parkinson’s disease (Luk et al. 2012), the tau amyloid associated with Alzheimer’s disease, and others listed in the review by Aguzzi and Rajendran (2009) and Prusiner (2013). It remains to be demonstrated whether and under which circumstances these amyloid species are prions. Because of the extraordinary properties of amyloids (such as high stability and the capacity to bind and penetrate membranes), it is, however, likely that under certain (experimental) conditions, these prionoids may be prions with a very low infectivity titer. For further details, three excellent reviews concentrate on this topic (Aguzzi and Rajendran 2009; Colby and Prusiner 2011; Prusiner 2013).
THE TOXIC NATURE OF AMYLOIDS
The long list of amyloid diseases suggests that amyloids are inherently toxic (Pieri et al. 2012). However, this observation is countered by the existence of functional amyloids (Greenwald and Riek 2010). Thus, toxicity, like other protein activities, may be associated with a particular conformation. Notably, in this context, the functional nontoxic HET-s prion amyloid has a hydrophilic surface typically observed for soluble proteins (HET-s(218-289) overexpression, both in its normal host Podospora anserina and in rat, is not toxic) (Fig. 2) (Winner et al. 2011), whereas the disease-associated Aβ amyloids appear to have significant hydrophobic patches on their surfaces with the potential for nonspecific hydrophobic interactions with other proteins or membranes, which may cause toxicity (Lu et al. 2013; Schütz et al. 2015).
Alternatively, it has been suggested that a folding/aggregation intermediate or an off-pathway oligomer of the amyloid peptide/protein may be the most toxic species (Hardy and Selkoe 2002; Klein et al. 2004; Lazo et al. 2005; Winner et al. 2011). Low-resolution structural studies indicate that (toxic) amyloid oligomers are rich in intermolecular β-sheet secondary structure (Kayed et al. 2003; Chimon et al. 2007; Stroud et al. 2012), suggesting similar structural properties as the amyloids and, concomitantly, potentially similar activities. The presence of intermolecular β-sheet secondary structure in amyloid oligomers is strengthened by the recent structural investigation of a designed toxic oligomer, which has elucidated a new type of fold, termed “cylindrin.” The cylindrin fold is composed of intermolecular β-sheets with a hydrophobic core and a hydrophilic solvent exposed side, respectively. It remains to be demonstrated how this fold exerts its mechanism of toxicity. If such an oligomer acts through membrane permeation as suggested for other oligomers, large conformational changes are necessary to refold the cylindrin inside out (in the membrane state, the hydrophobic amino acid side chains should face outward toward the membrane, and the charged solvent exposed side chains inward, respectively). Interestingly, it has been suggested that this class of oligomers is off-pathway to the amyloid kinetics, because a cylindric β-sheet can only be formed if the individual β-strands are out of register, whereas the flat amyloid fibrils are in general composed of in-register β-sheets (Liu et al. 2012).
More recently, it has been suggested that it is not a specific structural entity that is toxic, but rather the process of amyloid aggregation (Jan et al. 2011). This hypothesis is supported by the finding that, in a model membrane, growing α-synuclein aggregates continuously extract lipids from the membrane (Reynolds et al. 2011), resulting in membrane thinning and, consequently, in a failure of membrane integrity, which eventually triggers a cascade of events that leads to cell death.
THE AMYLOID COMPARED TO OTHER PROTEIN AGGREGATES
There are ordered protein aggregates that are not composed of the cross-β-sheet motif. Folded proteins may aggregate by domain-swapping or by interaction with their surfaces, such as the so-called “end-to-end” stacking (Bennett et al. 2006; Eisenberg et al. 2006; Nelson and Eisenberg 2006a,b). Because such aggregates are based on interactions between folded proteins, there is not a gross rearrangement of the protein fold, and the repetitive nature of the structure is at the suprananomolar level, contrasting the subnanomolar repetitive nature of the amyloid, which gives rise to the many potential activities of the amyloid through cooperativity effects (see above). Examples of nonamyloid protein aggregates are the cytoskeletal complexes of the actins and tubulins as well as the assembly of hemoglobin S (HbS) associated with sickle cell anemia. Another family of protein aggregation occurs in the assembly of the structural proteins like collagen and the keratins, which comprise a variety of folds distinct from other ordered aggregates.
Yet, another class of protein aggregates is arthropod silk. There are large varieties of silks, and many are not yet characterized in detail. However, the more well-studied spider fibroins are composed of a β-sheet-rich structure in which, unlike amyloids, the strands run parallel to the fiber axis (Römer and Scheibel 2008). Furthermore, they are composed of other sections such as the Gly/Pro/Gln segments, which give the elasticity to silk and other protein domains that are important for the induction of the aggregation process (Hagn et al. 2010).
Finally, there is a group of protein aggregates that are classified as amorphous aggregates, a type that is, for example, the precipitates obtained by heat or high concentration treatments. Although this class of aggregates is on the mesoscopic scale and different from the fibrous aggregate, a recent study from our laboratory indicated that amorphous aggregates may also contain a defined cross-β structure, but without the long-range (>μm) order found in fibrils (Wang et al. 2010). This finding, which is in line with the early work by Astbury et al. (1935), suggests that the manner in which a polypeptide backbone achieves a kinetically accessible local-energy minimum might require some local order or repeated structure throughout the aggregate, and concomitantly, the nomenclature “amorphous” aggregate is misleading.
CAN ANY PROTEIN FORM AN AMYLOID?
Based on the aforementioned structural studies, the cross-β fold can be as simple as a single four-residue β-strand that assembles in a repetitive manner. Hence, because of the simplicity of the most basic repeating β-strand unit of the amyloid, one might suggest that any polypeptide that can form a β-strand could eventually form an amyloid. In fact, many proteins aggregate into amyloids or amyloid-like states when the delicate balance between folding and aggregation is disturbed. For example, overexpression in Escherichia coli can cause many proteins to form inclusion bodies that have an amyloid-like substructure (Ventura and Villaverde 2006; Wang et al. 2008). Some proteins can aggregate into amyloid-like structures when heat-denatured, such as in the poached-egg-white studies by Astbury et al. (1935), and some proteins that have stable soluble folds can be transformed into an amyloid state in nonbiological conditions (Guijarro et al. 1998; Chiti et al. 1999). However, amyloid aggregation is highly amino acid sequence specific, as demonstrated by the structures discussed above (Nelson et al. 2005; Nelson and Eisenberg 2006a; Sawaya et al. 2007; Wasmer et al. 2009). The essential involvement of side-chain interactions in the aggregation process is evident from the observed sequence-specific nature of amyloid aggregation (Tjernberg et al. 2002; Zanuy and Nussinov 2003; Margittai and Langen 2006) and from the predictive power of the algorithms for aggregation propensities (Fernandez-Escamilla et al. 2004; Trovato et al. 2006; Tartaglia et al. 2008). The predictive power of the algorithms also indicates that the 3D structure of the cross-β state is less complex than most tertiary structures of soluble proteins, a notion that is supported by the recent mutagenesis study of HET-s(218-289), which showed that the structural influence of side-chain alteration is smaller than observed for soluble proteins (Daskalov et al. 2014). Thus, the complexity of the cross-β fold may be classified somewhere between a secondary and a tertiary structural feature. Returning to the question of this section, because already short peptides are able to form amyloid, it is likely that most proteins contain amyloid-prone segments and thus have the capacity to form amyloids. Indeed, when analyzing the amylome, which is the universe of proteins that are capable of forming amyloid-like fibrils, amyloid-prone segments were found in almost all proteins (Goldschmidt et al. 2010).
It is concluded that the cross-β-sheet entity of the amyloid first described in the poached-egg studies is unique within the protein-fold universe. The concurrent unique properties make the amyloid a fascinating structural entity that may act in many different ways, both in health and in disease.
ACKNOWLEDGMENTS
We thank the Swiss Federal Institute of Technology (ETH) Zurich for its continuous financial support of our work on protein amyloids. Furthermore, some text passages and figures were adopted from other papers that the corresponding author of this review wrote in the past.
Footnotes
Editor: Stanley B. Prusiner
Additional Perspectives on Prion Biology available at www.cshperspectives.org
REFERENCES
- Aguzzi A, Rajendran L. 2009. The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron 24: 783–790. [DOI] [PubMed] [Google Scholar]
- Astbury WT, Dickinson S, Bailey K. 1935. CCLXXIX. The X-ray interpretation of denaturation and the structure of the seed globulins. Biochem J 29: 2351–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MJ, Sawaya MR, Eisenberg D. 2006. Deposition diseases and 3D domain swapping. Structure 14: 811–824. [DOI] [PubMed] [Google Scholar]
- Bousset L, Pieri L, Ruiz-Arlandis G, Gath J, Jensen PH, Habenstein B, Madiona K, Olieric V, Böckmann A, Meier BH, et al. 2013. Structural and functional characterization of two α-synuclein strains. Nat Commun 4: 2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268: 880–884. [DOI] [PubMed] [Google Scholar]
- Chimon S, Shaibat MA, Jones CR, Calero DC, Aizezi B, Ishii Y. 2007. Evidence of fibril-like β-sheet structures in a neurotoxic amyloid intermediate of Alzheimer’s β-amyloid. Nat Struct Mol Biol 14: 1157–1164. [DOI] [PubMed] [Google Scholar]
- Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, Dobson CM. 1999. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc Natl Acad Sci 96: 3590–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby DW, Prusiner SB. 2011. De novo generation of prion strains. Nat Rev Microbiol 9: 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colletier JP, Laganowsky A, Landau M, Zhao M, Soriaga AB, Goldschmidt L, Flot D, Cascio D, Sawaya MR, Eisenberg D. 2011. Molecular basis for amyloid-β polymorphism. Proc Natl Acad Sci 108: 16938–16943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustou V, Deleu C, Saupe S, Begueret J. 1997. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci 94: 9773–9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalov A, Gantner M, Wälti MA, Schmidlin T, Chi CN, Wasmer C, Schütz A, Ceschin J, Clavé C, Cescau S, et al. 2014. Contribution of specific residues of the β-solenoid fold to HET-s prion function, amyloid structure and stability. PLoS Pathog 10: e1004158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Nelson R, Sawaya MR, Balbirnie M, Sambashivan S, Ivanova MI, Madsen AØ, Riekel C. 2006. The structural biology of protein aggregation diseases: Fundamental questions and some answers. Acc Chem Res 39: 568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fändrich M, Meinhardt J, Grigorieff N. 2009. Structural polymorphism of Alzheimer Aβ and other amyloid fibrils. Prion 3: 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. 2004. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol 22: 1302–1306. [DOI] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. 2006. Functional amyloid formation within mammalian tissue. PLoS Biol 4: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenner GG, Wong CW. 1983. Alzheimer’s disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun 120: 885–890. [DOI] [PubMed] [Google Scholar]
- Glenner GG, Cuatrecasas P, Isersky C, Bladen HA, Eanes ED. 1969. Physical and chemical properties of amyloid fibers II. Isolation of unique protein constituting the major component from human splenic amyloid fibril concentrates. J Histochem Cytochem 17: 769–780. [DOI] [PubMed] [Google Scholar]
- Goldschmidt L, Teng PK, Riek R, Eisenberg D. 2010. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci 107: 3487–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald J, Riek R. 2010. Biology of amyloid: Structure, function, and regulation. Structure 18: 1244–1260. [DOI] [PubMed] [Google Scholar]
- Guijarro JI, Sunde M, Jones JA, Campbell ID, Dobson CM. 1998. Amyloid fibril formation by an SH3 domain. Proc Natl Acad Sci 95: 4224–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagn F, Eisoldt L, Hardy J, Vendrely C, Coles M, Scheibel T, Kessler H. 2010. A conserved spider silk domain acts as a molecular switch that controls fibre assembly. Nature 465: 239–242. [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. 2002. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 297: 353–356. [DOI] [PubMed] [Google Scholar]
- Jan A, Adolfsson O, Allaman I, Buccarello AL, Magistretti PJ, Pfeifer A, Muhs A, Lashuel HA. 2011. Aβ42 neurotoxicity is mediated by ongoing nucleated polymerization process rather than by discrete Aβ42 species. J Biol Chem 286: 8585–8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane MD, Lipinski WJ, Callahan MJ, Bian F, Durham RA, Schwarz RD, Roher AE, Walker LC. 2000. Evidence for seeding of β-amyloid by intracerebral infusion of Alzheimer brain extracts in β-amyloid precursor protein-transgenic mice. J Neurosci 20: 3606–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. 2003. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300: 486–489. [DOI] [PubMed] [Google Scholar]
- Klein WL, Stine WB Jr, Teplow DB. 2004. Small assemblies of unmodified amyloid β-protein are the proximate neurotoxin in Alzheimer’s disease. Neurobiol Aging 25: 569–580. [DOI] [PubMed] [Google Scholar]
- Lazo ND, Maji SK, Fradinger EA, Bitan G, Teplow DB. 2005. The amyloid β protein. In Amyloid protein: The β sheet conformation disease (ed. Sipe JD), pp. 385–491. Wiley, Weinheim, Germany. [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, et al. 2012. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 150: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zhao M, Jiang L, Cheng PN, Park J, Sawaya MR, Pensalfini A, Gou D, Berk AJ, Glabe CG, et al. 2012. Out-of-register β-sheets suggest a pathway to toxic amyloid aggregates. Proc Natl Acad Sci 109: 20913–20908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. 2013. Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell 154: 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk KC, Kehm V, Carroll J, Zhang B, O’Brien P, Trojanowski JQ, Lee VM. 2012. Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 338: 949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddelein ML, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B, Saupe SJ. 2002. Amyloid aggregates of the HET-s prion protein are infectious. Proc Natl Acad Sci 99: 7402–7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margittai M, Langen R. 2006. Side-chain-dependent stacking modulates tau filament structure. J Biol Chem 281: 37820–37827. [DOI] [PubMed] [Google Scholar]
- Meyer-Luehmann M, Coomaraswamy J, Bolmont T, Kaeser S, Schaefer C, Kilger E, Neuenschwander A, Abramowski D, Frey P, Jaton AL, et al. 2006. Exogenous induction of cerebral β-amyloidogenesis is governed by agent and host. Science 313: 1781–1784. [DOI] [PubMed] [Google Scholar]
- Nelson R, Eisenberg D. 2006a. Recent atomic models of amyloid fibril structure. Curr Opin Struct Biol 16: 260–265. [DOI] [PubMed] [Google Scholar]
- Nelson R, Eisenberg D. 2006b. Structural models of amyloid-like fibrils. Adv Protein Chem 73: 235–282. [DOI] [PubMed] [Google Scholar]
- Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, Eisenberg D. 2005. Structure of the cross-β spine of amyloid-like fibrils. Nature 435: 773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherovich LZ, Weissman JS. 2002. The utility of prions. Dev Cell 2: 143–151. [DOI] [PubMed] [Google Scholar]
- Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. 2005. Self-propagating, molecular-level polymorphism in Alzheimer’s β-amyloid fibrils. Science 307: 262–265. [DOI] [PubMed] [Google Scholar]
- Pieri L, Madiona K, Bousset L, Melki R. 2012. Fibrillar α-synuclein and huntingtin exon 1 assemblies are toxic to the cells. Biophys J 20: 2894–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. 1991. Molecular biology of prion diseases. Science 252: 1515–1522. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. 2013. Biology and genetics of prions causing neurodegeneration. Annu Rev Genet 47: 601–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds NP, Soragni A, Rabe M, Verdes D, Liverani E, Handschin S, Riek R, Seeger S. 2011. Mechanism of membrane interaction and disruption by α-synuclein. J Am Chem Soc 133: 19366–19375. [DOI] [PubMed] [Google Scholar]
- Riek R. 2006. Cell biology: Infectious Alzheimer’s disease? Nature 444: 429–431. [DOI] [PubMed] [Google Scholar]
- Römer L, Scheibel T. 2008. The elaborate structure of spider silk: Structure and function of a natural high performance fiber. Prion 2: 154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufo CM, Moroz YS, Moroz OV, Stöhr J, Smith TA, Hu X, DeGrado WF, Korendovych IV. 2014. Short peptides self-assemble to produce catalytic amyloids. Nat Chem 4: 303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, et al. 2007. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 447: 453–457. [DOI] [PubMed] [Google Scholar]
- Schütz AK, Vagt T, Huber M, Ovchinnikova OY, Cadalbert R, Wall J, Güntert P, Böckmann A, Glockshuber R, Meier BH. 2015. Atomic-resolution three-dimensional structure of amyloid β fibrils bearing the Osaka mutation. Angew Chem Int Ed 54: 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuring C, Greenwald J, Wasmer C, Wepf R, Saupe SJ, Meier BH, Riek R. 2012. The mechanism of toxicity in HET-S/HET-s prion incompatibility. PLoS Biol 10: e1001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipe JD, Cohen AS. 2000. Review: History of the amyloid fibril. J Struct Biol 130: 88–98. [DOI] [PubMed] [Google Scholar]
- Stroud JC, Liu C, Teng PK, Eisenberg D. 2012. Toxic fibrillar oligomers of amyloid-β have cross-β structure. Proc Natl Acad Sci 109: 7717–7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunde M, Blake C. 1997. The structure of amyloid fibrils by electron microscopy and X-ray diffraction. Adv Protein Chem 50: 123–159. [DOI] [PubMed] [Google Scholar]
- Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, Blake CC. 1997. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol 273: 729–739. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. 2004. Conformational variations in an infectious protein determine prion strain differences. Nature 428: 323–328. [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, Pawar AP, Campioni S, Dobson CM, Chiti F, Vendruscolo M. 2008. Prediction of aggregation-prone regions in structured proteins. J Mol Biol 380: 425–436. [DOI] [PubMed] [Google Scholar]
- Tjernberg L, Hosia W, Bark N, Thyberg J, Johansson J. 2002. Charge attraction and β propensity are necessary for amyloid fibril formation from tetrapeptides. J Biol Chem 277: 43243–43246. [DOI] [PubMed] [Google Scholar]
- Trovato A, Chiti F, Maritan A, Seno F. 2006. Insight into the structure of amyloid fibrils from the analysis of globular proteins. PLoS Comput Biol 2: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True HL, Lindquist SL. 2000. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407: 477–483. [DOI] [PubMed] [Google Scholar]
- Van Melckebeke H, Wasmer C, Lange A, Ab E, Loquet A, Böckmann A, Meier BH. 2010. Atomic-resolution three-dimensional structure of HET-s(218-289) amyloid fibrils by solid-state NMR spectroscopy. J Am Chem Soc 132: 13765–13775. [DOI] [PubMed] [Google Scholar]
- Ventura S, Villaverde A. 2006. Protein quality in bacterial inclusion bodies. Trends Biotechnol 24: 179–185. [DOI] [PubMed] [Google Scholar]
- Wang L, Maji SK, Sawaya MR, Eisenberg D, Riek R. 2008. Bacterial inclusion bodies contain amyloid-like structure. PLoS Biol 6: e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Schubert D, Sawaya MR, Eisenberg D, Riek R. 2010. Multidimensional structure–activity relationship of a protein in its aggregated states. Angew Chem Int Ed Engl 49: 3904–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. 2008. Amyloid fibrils of the HET-s(218-289) prion form a β solenoid with a triangular hydrophobic core. Science 319: 1523–1526. Erratum in 320: 50. [DOI] [PubMed] [Google Scholar]
- Wasmer C, Schütz A, Loquet A, Buhtz C, Greenwald J, Riek R, Böckmann A, Meier BH. 2009. The molecular organization of the fungal prion HET-s in its amyloid form. J Mol Biol 394: 119–127. [DOI] [PubMed] [Google Scholar]
- Westermark GT, Johnson KH, Westermark P. 1999. Staining methods for identification of amyloid in tissue. Methods Enzymol 309: 23–25. [DOI] [PubMed] [Google Scholar]
- Winner B, Jappelli R, Maji SK, Desplats PA, Boyer L, Aigner S, Hetzer C, Loher T, Vilar M, Campioni S, et al. 2011. In vivo demonstration that α-synuclein oligomers are toxic. Proc Natl Acad Sci 108: 4194–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanuy D, Nussinov R. 2003. The sequence dependence of fiber organization. A comparative molecular dynamics study of the islet amyloid polypeptide segments 22-27 and 22-29. J Mol Biol 329: 565–584. [DOI] [PubMed] [Google Scholar]