Abstract
Prions are alternatively folded, self-perpetuating protein isoforms involved in a variety of biological and pathological processes. Yeast prions are protein-based heritable elements that serve as an excellent experimental system for studying prion biology. The propagation of yeast prions is controlled by the same Hsp104/70/40 chaperone machinery that is involved in the protection of yeast cells against proteotoxic stress. Ribosome-associated chaperones, proteolytic pathways, cellular quality-control compartments, and cytoskeletal networks influence prion formation, maintenance, and toxicity. Environmental stresses lead to asymmetric prion distribution in cell divisions. Chaperones and cytoskeletal proteins mediate this effect. Overall, this is an intimate relationship with the protein quality-control machinery of the cell, which enables prions to be maintained and reproduced. The presence of many of these same mechanisms in higher eukaryotes has implications for the diagnosis and treatment of mammalian amyloid diseases.
In yeast, prion propagation is controlled by the same Hsp104/70/40 chaperone machinery that is involved in protection against proteotoxic stress. Similar mechanisms may be present in higher eukaryotes.
Recent developments have led to the expansion of the prion concept from one specific group of infectious diseases to a variety of self-perpetuating protein states associated with both pathogenic and potentially adaptive processes. Yeast prions, typically represented by amyloid isoforms of various yeast proteins, manifest as protein-based heritable elements and are therefore amenable to high-resolution genetic analysis. A significant fraction of the data reviewed below has been obtained by using the Sup35/[PSI+] prion system. Sup35 is a translational termination factor, which includes the N-terminal prion domain ([PrD] or Sup35N), the middle linker domain (Sup35M), and the C-proximal domain (Sup35C) functioning in translation termination (Liebman and Chernoff 2012; Nizhnikov et al. 2014b). Detailed information about other yeast prions, respective proteins, and their functions is discussed elsewhere (Liebman and Chernoff 2012; Wickner 2016). Because of the simplicity of yeast cultivation and the availability of the powerful tools of yeast genetics and cell biology, yeast prions provide an excellent model for studying the fundamental mechanisms of prion formation and propagation. General rules uncovered from yeast prion studies can be extended to mammalian and human prion and amyloid phenomena, including neurodegenerative diseases.
A connection between prions and chaperones was first established using a yeast model. Additionally, our knowledge of the cellular control of prion formation and propagation has since been expanded in significant part because of experiments with yeast. This review summarizes the overall progress and recent breakthroughs in understanding the connections between prions and cellular-protein quality control in yeast.
CONTROL OF PRION PROPAGATION BY THE Hsp104/70/40 CHAPERONE MACHINERY IN YEAST
The connection between chaperones and prions was first established by demonstrating that propagation of [PSI+], the prion form of the translation termination factor Sup35, requires an intermediate level of the chaperone Hsp104 (Chernoff et al. 1995). Hsp104 is a hexameric ATPase of the AAA+ superfamily (Glover and Lindquist 1998) and is involved in protein disaggregation (Glover and Lum 2009); its prokaryotic counterpart is ClpB (Weibezahn et al. 2004; Glover and Lum 2009; Winkler et al. 2012a). Hsp104 is also required for all other known amyloid-based cytosolic yeast prions (Moriyama et al. 2000; Du et al. 2008; Alberti et al. 2009; Patel et al. 2009; Rogoza et al. 2010; Liebman and Chernoff 2012). Two yeast prions that are independent of Hsp104 include [ISP+], presumably located in the nucleus (Rogoza et al. 2010), and the membrane-associated [GAR+], which is not yet proven to show amyloid-like aggregation (Brown and Lindquist 2009; Jarosz et al. 2014). ATPases of the Hsp70 chaperone family (Verghese et al. 2012) and their partners from the Hsp40 family (Cyr and Ramos 2015) are involved with Hsp104 in the process of prion propagation (for reviews, see Liebman and Chernoff 2012; Chernova et al. 2014).
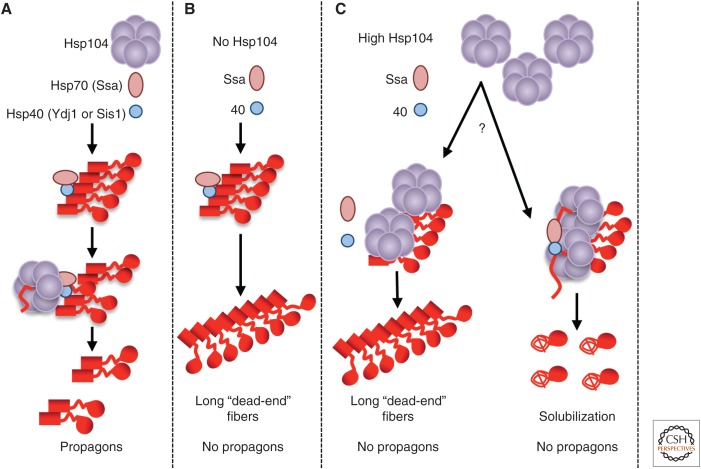
Our current understanding of the role of Hsp in the propagation of cytosolic yeast prions is summarized in Figure 1. In this model, the Hsp70/Hsp40 complex binds to prion polymers first and then binds Hsp104, which initiates polymer fragmentation by extracting individual protein molecules from the middle of an amyloid chain (Winkler et al. 2012b). Polymer fragmentation generates “propagons,” smaller “seeds” that are essential for initiating new rounds of prion propagation as well as prion segregation (Fig. 1A). In the absence of fragmentation, new propagons are not generated, and prions are eventually diluted and lost in the course of cell divisions (Fig. 1B). In the case of the Sup35 protein, chaperone binding is most likely mediated or modulated by the region of oligopeptide repeats present within the PrD (Borchsenius et al. 2001; Osherovich et al. 2004). However, the Hsp machinery also binds other PrDs, including those not containing repeats. The role of Hsps in prion propagation is based on their major evolutionarily conserved enzymatic functions, because Escherichia coli orthologs of yeast—Hsp104, Hsp70-Ssa, and Hsp40 proteins (ClpB, DnaK, and DnaJ, respectively)—can partially propagate yeast prions, but only if the whole bacterial chaperone network (with the addition of the DnaK guanine-exchange factor GrpE) is reassembled in the yeast cell (Miot et al. 2011).
Figure 1.
Role of chaperones in the propagation of the Sup35 prion. (A) Polymer fragmentation and propagon formation at the normal levels of Hsp104, Hsp70, and Hsp40 proteins. (B) Impairment of prion propagation in the absence of Hsp104. (C) Impairment of prion propagation at high levels of Hsp104 (two models are shown). (Red square) Sup35N (PrD) in prion form; (red line) Sup35M linker and Sup35N in a misfolded intermediate form; (red spiral line) Sup35NM region in a nonprion form; (red circle) globular Sup35C domain.
EFFECTS OF Hsp104 LEVELS ON YEAST PRIONS
Interestingly, transient overproduction of Hsp104 also cures yeast of some prions, for example, [PSI+] (Chernoff et al. 1995) and [MOD+] (Suzuki et al. 2012). This agrees with the notion that Hsp104 can bind polymeric Sup35 without the help of Ssa, which leads to prion loss rather than polymer fragmentation (Winkler et al. 2012a,b). Such “nonproductive” binding (Fig. 1B) involves the Hsp104 N-terminus, which is not required for prion fragmentation and propagation (Hung and Masison 2006; Winkler et al. 2012b) and is apparently mediated by a portion of the linker region of Sup35 (Sup35M) (Glover and Lindquist 1998; Glover and Lum 2009; Helsen and Glover 2012a,b). Two possible (and not mutually exclusive) mechanisms proposed for [PSI+] curing by excess Hsp104 include “chopping” monomers from polymers and inhibiting polymer fragmentation and/or segregation in cell divisions (Fig. 1C). Evidence in favor of each mechanism has been reported (Newnam et al. 2011; Liebman and Chernoff 2012; Chernova et al. 2014; Klaips et al. 2014; Park et al. 2014). An Hsp104 derivative lacking the N-terminal region can fragment Sup35NM polymers in vitro but cannot extract monomers from polymers (Sweeny et al. 2015), which is consistent with its inability to eliminate [PSI+] on overproduction in vivo. However, overproduction of full-size Hsp104 leads to an increase in the average size of Sup35 polymers in vivo (Kryndushkin et al. 2003), pointing to the possibility of the fragmentation defect, which results in fiber growth.
Hyperfunctional mutations in Hsp104 facilitate [PSI+] curing and may counteract other amyloid-like aggregates expressed in yeast (Gokhale et al. 2005; Jackrel et al. 2014). Likewise, effects of antiprion mutations in Sup35 are modulated by Hsp104 levels (DiSalvo et al. 2011). On the other hand, chaperone sequestration by some overproduced glutamine/asparagine (Q/N)-rich, aggregation-prone proteins may cause destabilization of the [PSI+] prion (Derkatch and Liebman 2013; Yang et al. 2013).
ROLES OF VARIOUS Hsp70 AND Hsp40 PROTEINS AND THEIR COFACTORS IN PRION PROPAGATION
The yeast cell contains multiple subfamilies of the Hsp70 and Hsp40 families. The major Hsp70 subfamily that plays a role in prion propagation is Ssa, which is composed of four chaperones (Ssa2 produced constitutively; Ssa1 produced during vegetative growth and induced by stress; and two strictly stress-inducible proteins, Ssa3 and Ssa4). Any of these chaperones can work in [PSI+] propagation, although some differences between their specific effects exist (Allen et al. 2005; Sharma and Masison 2008; Liebman and Chernoff 2012). Overproduction of Ssa1 (Newnam et al. 1999) or any other Ssa protein (Allen et al. 2005) allows propagation of the [PSI+] prion, even in the presence of high levels of Hsp104, apparently via counteracting the “nonproductive” interaction of Hsp104 with prion polymers and directing Hsp104 to the fragmentation pathway. Mutational alteration and/or deletion of SSA1 and/or SSA2 destabilizes or cures some variants of [PSI+] (Sharma and Masison 2008; Newnam et al. 2011). In the case of [URE3], Ssa1 and Ssa2 show differential effects (Schwimmer and Masison 2002), and this difference is controlled by a single amino-acid substitution (Sharma and Masison 2011). Ssa proteins can physically bind Sup35 (Allen et al. 2005) and represent the most abundant proteins among those co-isolated together with Sup35 from yeast cells (Bagriantsev et al. 2008).
Several members of the Hsp40 family have been shown to influence prion propagation (for review, see Liebman and Chernoff 2012) in a prion-specific manner (e.g., Reidy et al. 2014), with Ydj1 (Moriyama et al. 2000; Sharma et al. 2009; Hines et al. 2011) and/or Sis1 (Sondheimer et al. 2001; Higurashi et al. 2008; Kirkland et al. 2011), the major Hsp40 partners of Ssa (Rikhvanov et al. 2007), typically playing a major role (Summers et al. 2009). Other chaperone-related proteins involved in the propagation of yeast prions include guanine-exchange factors for Ssa, Fes1, and Sse1 (Jones et al. 2004; Fan et al. 2007; Kryndushkin and Wickner 2007; Moran et al. 2013), and Ssa co-chaperones, Sti1 and Cpr7 (Jones et al. 2004; Wolfe et al. 2013), perhaps acting via modulation of Ssa function. Hsp82 (yeast counterpart of Hsp90) somewhat increases curing of [PSI+] by excess Hsp104 (Moosavi et al. 2010; Reidy and Masison 2011); however, the specific mechanism is unclear.
ROLE OF RIBOSOME-ASSOCIATED CHAPERONES IN PRION FORMATION AND PROPAGATION
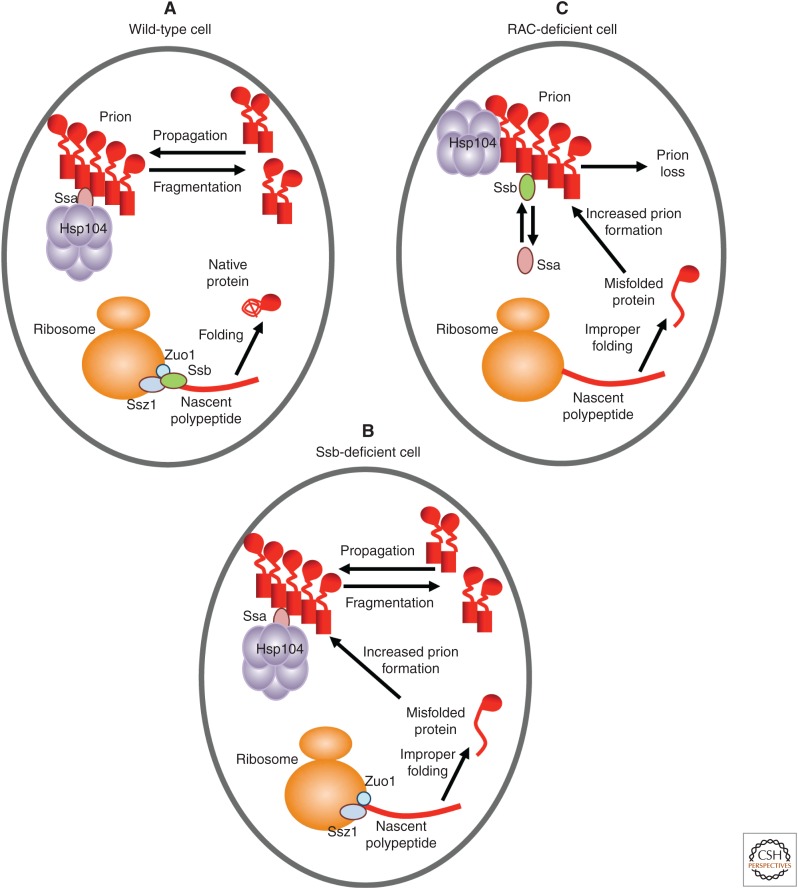
In addition to Ssa, the yeast cell possesses a variety of other Hsp70 subfamilies (Peisker et al. 2010; Liebman and Chernoff 2012). One of them, Ssb, is composed of two nearly identical members, Ssb1 and Ssb2 (termed Ssb protein). Ssb is neither essential for cell viability nor stress-inducible (Nelson et al. 1992). It binds translating ribosomes (Fig. 2A) via the ribosome-associated complex (RAC), composed of the Hsp40 protein Zuo1 and a noncanonical Hsp70 protein Ssz1 (Gautschi et al. 2001; Huang et al. 2005), and is involved in folding of nascent polypeptides (James et al. 1997; Willmund et al. 2013). In the absence of a fully assembled RAC (e.g., in strains lacking one or both RAC component[s]), Ssb is dissociated from the ribosome (Willmund et al. 2013). Depletion of Ssb (Fig. 2B) or its dissociation from the ribosome caused by RAC disruption (Willmund et al. 2013) results in accumulation of misfolded aggregated proteins. Ribosome profiling analysis (Ingolia et al. 2009) indicates that the set of potential Ssb substrates is enriched by long proteins with hydrophobic aggregation-prone regions (Willmund et al. 2013).
Figure 2.
Model for the effects of ribosome-associated chaperones in prion formation and propagation. (A) Wild-type cell. (B) Cell lacking Ssb. (C) Cell lacking Zuo1 and/or Ssz1. Sup35 regions and isoforms are shown as in Figure 1. The cytosolic Hsp40 protein is not shown for simplicity.
Notably, double deletion of both genes coding for Ssb (Chernoff et al. 1999), or deletion of gene(s) coding for one or both RAC components, ZUO1 and/or SSZ1 (Kiktev et al. 2015; Amor et al. 2015), increases de novo formation of [PSI+], possibly because of the accumulation of misfolded Sup35 protein, a substrate for prion formation, on the Ssb-depleted ribosomes (Fig. 2B,C). Ssb consistently acts against prions, as excess Ssb destabilizes some variants of the [PSI+] prion (Kushnirov et al. 2000; Chacinska et al. 2001; Allen et al. 2005), and Ssb overproduction facilitates [PSI+] loss in the presence of excess Hsp104, whereas the double ssb1Δ ssb2Δ deletion (ssb1/2Δ) counteracts it (Chernoff et al. 1999). However, RAC disruption increases [PSI+] curing by excess Hsp104 and destabilizes a “weak” variant of [PSI+] at normal levels of Hsp104 (Kiktev et al. 2015). This effect of RAC disruption is not detected in the ssb1/2Δ background, showing that cytosolic Ssb, released from ribosomes, antagonizes propagation of the [PSI+] prion.
Differences in the effects of Ssa and Ssb proteins on [PSI+] prions are primarily controlled by their substrate-binding domains (Allen et al. 2005). Importantly, both genetic and biochemical data show that cytosolic Ssb antagonizes binding of Ssa to prion polymers (Kiktev et al. 2015). Ssb itself is apparently capable of only a transient interaction with a prion in vivo (Allen et al. 2005; Bagriantsev et al. 2008; Kiktev et al. 2015) and inhibits the seeded Sup35NM amyloidogenesis in combination with Hsp104 in vitro (Shorter and Lindquist 2008). Thus, Ssb competition with Ssa impairs prion propagation (Fig. 2C).
Interestingly, a release of Ssb from ribosomes into cytosol can be induced not only by mutational disruption of RAC but also by certain physiological conditions (e.g., by growth in poor synthetic medium) (Kiktev et al. 2015). Propagation of a “weak” variant of the [PSI+] prion is destabilized under these conditions much more efficiently in the wild-type strain compared with the ssb1/2Δ strain. This implies the role of cytosolic (not bound to ribosomes) Ssb in [PSI+] destabilization and confirms the physiological importance of the Ssb-based prion-modulating circuit. Although the overall rate of protein synthesis decreases in unfavorable conditions, synthesis of some proteins must be maintained or increased (Verghese et al. 2012; Conrad et al. 2014). This may result in their misfolding and accumulation of prion-like self-perpetuating aggregates, either toxic or cytoprotective (e.g., via promoting formation of quality-control protein deposits discussed below or other protective structures such as stress granules) (Miller et al. 2015b). However, even persistent propagation of cytoprotective aggregates could become toxic, especially if conditions were to change. Cytosolic Ssb may prevent efficient aggregate propagation, thus compensating in part for the shortage of Ssb on ribosomes.
PRIONS AND PROTEIN QUALITY CONTROL
Notably, the same Hsp104/Hsp70-Ssa/Hsp40 chaperone machinery that controls prion propagation is also involved in protecting yeast cells against stress and promoting disaggregation of stress-damaged proteins (Glover and Lindquist 1998). Apparently, the effects of chaperones on aggregates of stress-damaged proteins and on amyloid polymers are based on the same molecular mechanisms but lead to different consequences because of different features of the aggregated proteins. Chaperones tend to solubilize monomeric proteins from aggregates of damaged proteins, resulting in aggregate disassembly. However, because of the highly ordered organization of prion polymers, chaperones fragment them into oligomeric propagons. In this way, prions hijack the cellular stress-defense machinery for the purpose of their own propagation. Essentially, it is the Hsp-mediated fragmentation that enables prions to be “replicated” and transmitted on cell division and therefore makes the amyloid a prion.
Clearance of prions and prionogenic proteins is still poorly understood, although some data point to the role of the ubiquitin-proteasome system (UPS) in prion formation. Increased levels of free ubiquitin stimulate de novo formation of the [PSI+] prion in the presence of excess Sup35 by an unknown mechanism (Chernova et al. 2003), whereas deletion of the gene coding for one of the major yeast ubiquitin-conjugating enzymes, Ubc4, increases spontaneous [PSI+] formation (Allen et al. 2007). Because no evidence of Sup35 ubiquitination has been uncovered despite numerous attempts (Peng et al. 2003; Allen et al. 2007; Kabani et al. 2014), it appears that the effect of ubc4Δ on [PSI+] formation is mediated by other proteins. Notably, increased [PSI+] formation in the ubc4Δ strain does not require the presence of other prions but is antagonized by the deletion of RNQ1, a gene coding for the prion protein Rnq1 (Allen et al. 2007). Several other UPS deficiencies increase [PSI+] formation similarly to ubc4Δ (Tyedmers et al. 2008), and some proteins with Q/N-rich prion-like domains that can stimulate formation of the [PSI+] prion on overproduction (Derkatch et al. 2001) are known to be ubiquitinated and degraded by the proteasome. One well-studied example is the Lsb2/Pin3 protein (Chernova et al. 2011), which is discussed in the following section. Ubc4Δ also antagonizes [PSI+] curing by excess Hsp104 (Allen et al. 2007). This effect does not depend on Rnq1 protein but rather is associated with increased binding of Ssa protein to Sup35 prion polymers. Overall, it appears that UPS exercises its effects on the [PSI+] prion primarily by modulating auxiliary factors involved in prion formation and propagation.
PRIONS AND QUALITY-CONTROL COMPARTMENTS
Yeast cells contain various protein quality-control deposits (for review, see Sontag et al. 2014; Miller et al. 2015b). Misfolded proteins, which cannot be degraded by UPS, are concentrated into such deposits, which possibly leads to their detoxification via preventing sequestration of other proteins and UPS components, their degradation via autophagy, and/or their clearance from daughter cells via asymmetric cell division (see below).
Amyloidogenic proteins accumulate in perivacuolar insoluble protein deposits (IPODs) in a highly ordered form (Kaganovich et al. 2008). Production of the polyglutamine-extended exon 1 of huntingtin (a protein associated with Huntington’s disease in humans) leads to the formation of the perinuclear and perivacuolar deposits, co-localized with the spindle body (the yeast analog of the centrosome) marker Spc42, and therefore resembling the mammalian aggresome (Wang et al. 2009). Notably, the P-rich region of huntingtin, located next to the polyQ-expanded region, is important for aggresome formation and polyQP detoxification in yeast.
Several lines of evidence indicate that yeast prions interact with the quality-control deposits of insoluble proteins, such as IPODs and aggresome, and modify their distribution and composition. Overproduction of Rnq1 protein in a strain containing [RNQ+] prions leads to the formation of a structure combining some features of IPODs and aggresome, because it is located on the cell periphery but is co-localized with Spc42 (Treusch and Lindquist 2012). This structure is cytotoxic, which is apparently a result of sequestration of the spindle body components. In contrast, the polyQP-induced aggresome does not lead to spindle body mislocalization and is normally not cytotoxic (Wang et al. 2009). These results indicate that formation of IPOD-like deposits of different proteins could be based on similar molecular mechanisms; however, their eventual localization and cytotoxicity patterns could be influenced by prion composition of the yeast cell. Consistent with this model is the observation that the normally cytoprotective polyQP aggresome becomes cytotoxic in cells containing the Sup35 prion, [PSI+] (Gong et al. 2012). In this case, cytotoxicity is a result of sequestration of the essential termination factor Sup45 (eRF1), a partner of Sup35 in translation termination. The prion form of Sup35, which interacts with polyQP aggregates, mediates sequestration of Sup45, whereas overproduction of Sup45 ameliorates cytotoxicity.
The [URE3] prion (Kryndushkin et al. 2008) and artificial prion [NPR1C+], which contains the PrD-like region of the yeast protein Nrp1 (Malinovska et al. 2012), are cured by overproduction of Btn2 or Cur1, yeast members of the Hook protein family whose metazoan counterparts are involved in microtubule-dependent centrosome attachment to the nucleus (Malone et al. 2003) and aggresome formation (Szebenyi et al. 2007). Normal levels of Btn2 and Cur1 proteins also counteract propagation of de novo induced [URE3] prions, as shown in experiments with deletion strains (Wickner et al. 2014). This implicates Btn2 and Cur1 as components of the antiprion system. Yeast proteins Btn2 and Cur1, which are stress-inducible, are involved in redistribution of misfolded proteins between IPOD and a juxtanuclear compartment (Malinovska et al. 2012), which investigators equate with JUNQ (Kaganovich et al. 2008) or InQ (Miller et al. 2015a), a deposit of soluble misfolded proteins apparently distinct from the aggresome. The effect of Cur1 on quality-control compartments could at least be partly attributable to targeting of the Hsp40 chaperone Sis1 from cytosol to the nucleus (Malinovska et al. 2012). Prion curing by Cur1 (and possibly Btn2) overexpression could be a result of Sis1 sequestration or relocation, as supported by the observation that curing is inhibited in the presence of high levels of Sis1 (Malinovska et al. 2012). It is more difficult to explain the effects of the normal levels of Btn2 or Cur1 in this way, because Sis1 is more abundant than Cur1 (Wickner et al. 2014).
THE CYTOSKELETON AS A SCAFFOLD FOR PRIONOGENESIS
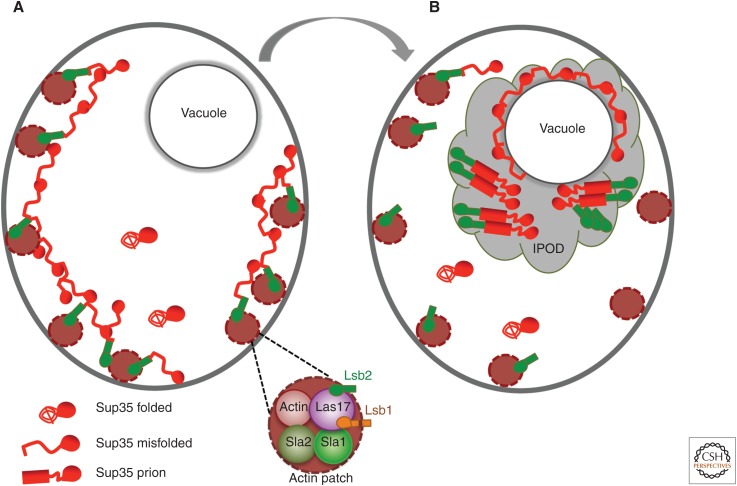
Large filamentous or ring-like structures are formed by overproduced green fluorescent protein (GFP)-tagged constructs containing Sup35 PrD during the process of de novo [PSI+] induction (Zhou et al. 2001; Ganusova et al. 2006). Although these structures are typically not seen in the cells containing mature [PSI+] prions, cells with filaments or rings preferentially produce [PSI+] colonies, implicating filamentous structures as intermediates in the prion-formation pathway. Peripheral filamentous structures overlap with actin patches, associated with the endocytic sites, whereas internal rings are assembled around the vacuole (Ganusova et al. 2006). Filamentous Sup35 structures accumulate actin assembly proteins involved in endocytosis (Ganusova et al. 2006), such as Sla1 and Sla2, a yeast homolog of mammalian huntingtin-interacting protein, Hip1 (Kalchman et al. 1997). Sup35 physically and genetically interacts with Sla1 (Bailleul et al. 1999), and deletions of SLA1 or genes coding for other actin assembly proteins, END3 or SLA2, impair de novo [PSI+] induction by excess Sup35 (Bailleul et al. 1999; Ganusova et al. 2006). Some ring-like Sup35 structures also co-localize with the autophagy markers characteristic of IPODs (Tyedmers et al. 2010). Overall, it appears that misfolded Sup35 is assembled into filamentous structures with the help of the peripheral actin cytoskeleton (Fig. 3A) and is then transported to the IPOD-like and/or autophagosome-like compartments attached to the vacuole (Fig. 3B). This process, apparently aimed at detoxifying and possibly eliminating the misfolded proteins, eventually promotes de novo prion formation via accumulating prionogenic molecules in these compartments.
Figure 3.
Model for the role of the actin cytoskeleton in the formation of the Sup35 prion. (A) Initial assembly of misfolded Sup35 in the peripheral cytoskeletal sites. (B) Cytoskeleton-mediated accumulation of Sup35 in the quality-control compartment, followed by conversion into a prion form. Sup35 regions and isoforms are shown as in Figure 1.
Some cytoskeletal assembly proteins (including Sla2) contain Q/N-rich prion-like domains (Michelitsch and Weissman 2000; Alberti et al. 2009). One such protein, termed Pin3, has been identified among proteins whose overproduction facilitates formation of the [PSI+] prion by excess Sup35 in cells lacking other preexisting prions (Derkatch et al. 2001). Pin3, more commonly called Lsb2, binds a major yeast actin-nucleating protein, Las17 (Madania et al. 1999). Although Lsb2 is not conserved in mammals, Las17 is a homolog of the mammalian actin nucleator, Wiscott–Aldrich syndrome protein (WASP) (Takenawa and Suetsugu 2007). [PSI+] induction by Lsb2 requires its interaction with Las17 (Chernova et al. 2011). Overproduced Lsb2 forms large structures that promote accumulation of misfolded Sup35 protein in adjacent cytoskeleton-associated locations, which serve as “prion induction” sites. The [PSI+]-inducing phenotype can persist in the cells for a certain number of cell generations after Lsb2 overproduction is turned off (Chernova et al. 2011), suggesting that Lsb2 can form a metastable prion-like state that facilitates formation of other prions. In addition to [PSI+], Lsb2 apparently promotes formation of the prion form of Rnq1 (Chernova et al. 2011). Notably, Lsb2 is a short-lived ubiquitinated protein whose levels are rapidly increased during environmental stress, such as heat shock (HS), which is followed by rapid degradation during adaptation to stress conditions (Chernova et al. 2011). Therefore, reversible variations of Lsb2 levels under physiological conditions may modulate some of the previously reported (e.g., Tyedmers et al. 2008) effects of stress-inducing agents on de novo prion formation.
Proteins containing PrD-like domains are also involved in generating other cytoprotective assemblies such as stress granules (Buchan 2014) and P-bodies (Gilks et al. 2004; Decker et al. 2007; Reijns et al. 2008; Kato et al. 2012; Newby and Lindquist 2013; Anderson et al. 2014). The PrD-like domain of the stress-granule assembly protein Tia1/Pub1 interacts with Sup35 PrD in yeast, and the tubulin-associated dimeric complex formed by this interaction is important for the integrity of tubulin cytoskeleton (Li et al. 2014).
It is likely that interactions of some PrDs with cytoskeletal components reflect the role of PrDs in forming intracellular structures, and accumulation of misfolded prionogenic proteins into certain cellular locations, mediated by cytoskeletal structures, may also serve a cytoprotective purpose (Chernoff 2007). Prion formation may arise as a by-product of such an adaptive process. It is also possible that in some cases, conversion of misfolded proteins into self-perpetuating prions may minimize cellular damage when other protective strategies are exhausted. Indeed, it is shown that the Sis1 chaperone suppresses the cytotoxicity of overproduced Rnq1 protein by promoting formation and/or propagation of its prion form (Douglas et al. 2008).
PRION MAINTENANCE DURING STRESS AND AGING
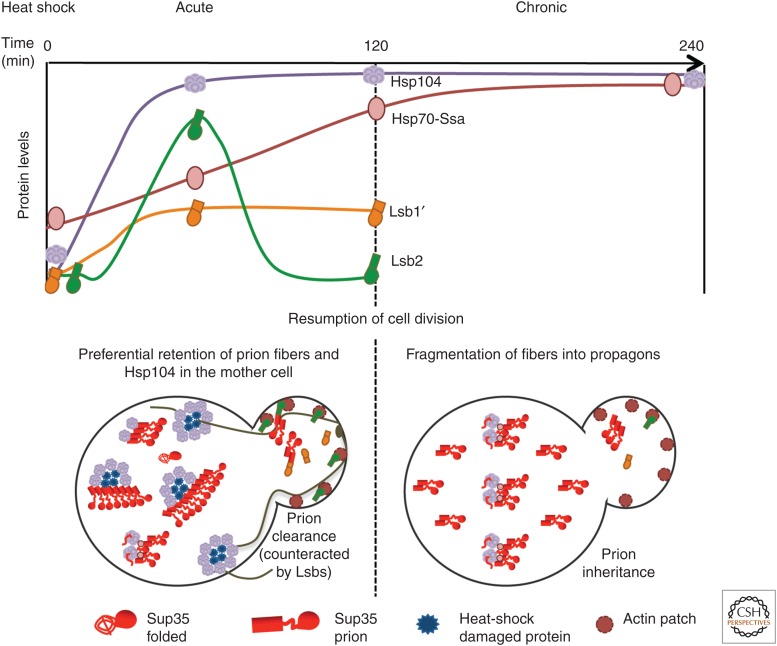
Short-term exposure to some stresses destabilizes propagation of certain prions (e.g., [PSI+]) (Cox et al. 1988). Indeed, short-term (15–60-min) exposure of an exponentially growing yeast culture to mild HS (39°C–42°C) destabilizes “weak” variants of [PSI+] (Newnam et al. 2011; Klaips et al. 2014). Most prion loss takes place during cell division, occurring after a return to normal temperature and resumption of growth. This generates sectored (“mosaic”) colonies. Notably, prion destabilization is not seen if cells are incubated at 39°C for a longer time (Newnam et al. 2011). Although Hsp104 (which has low background levels) is accumulated quickly during HS, the increase in levels of Ssa (starting from significantly higher background) proceeds more slowly. Thus, the “window” of maximal prion destabilization coincides with the period of maximal imbalance between the Hsp104 and Ssa proteins, although after several hours, Ssa levels catch up with the levels of Hsp104 (Fig. 4). Indeed, deletions of SSA genes increase prion destabilization by short-term HS and impair prion “recovery” during long-term HS (Newnam et al. 2011).
Figure 4.
Model for prion destabilization after HS. (Top) Dynamics of Hsp104, Hsp70-Ssa, actin assembly protein Lsb2, and cytosolic-processed isoform (Lsb1′) of its paralog Lsb1 during short-term (acute) and prolonged (chronic) temperature stress. (Bottom) Asymmetric segregation of prions, heat-shock damaged proteins, and chaperones in cell division after stress. Sup35 regions and isoforms are shown as in Figure 1.
HS-induced [PSI+] loss, at least in the first post-HS cell division, is clearly asymmetric, with daughter cells (new buds) losing prions more frequently than mother cells (Newnam et al. 2011; Ali et al. 2014). In this way, prion segregation after HS resembles segregation of oxidatively damaged proteins, which are preferentially accumulated in mother cells (Aguilaniu et al. 2003; Liu et al. 2011; Nystrom and Liu 2014). This mother-specific retention of protein aggregates requires Hsp104 (Erjavec et al. 2007; Tessarz et al. 2009; Newnam et al. 2011; Zhou et al. 2011) and has been explained by various, not necessarily mutually exclusive, mechanisms such as decreased passive diffusion through bud neck, active retention through association with organelles, quality-control compartments and/or cytoskeletal components, and retrograde transport mediated by the actin cytoskeleton (Erjavec et al. 2007; Tessarz et al. 2009; Liu et al. 2010; Zhou et al. 2011, 2014; Spokoini et al. 2012; Higuchi et al. 2013; Coelho et al. 2014; Song et al. 2014). Our data show that the actin cytoskeleton apparently plays a role in prion segregation, because [PSI+] destabilization is significantly increased in strains lacking the actin assembly protein Lsb2 or its paralog Lsb1; whereas in the strain lacking both proteins, [PSI+] remains destabilized even after prolonged HS (Chernova et al. 2011; Ali et al. 2014). As mentioned previously, Lsb2 synthesis is induced by HS (Chernova et al. 2011), whereas in the case of Lsb1, the proteolytic processing leading to accumulation of membrane nonbound (cytosolic) protein isoform is HS-inducible (Ali et al. 2014). It is possible that association with actin assembly proteins promotes delivery of prion seeds to daughter cells (Fig. 4).
In addition to asymmetric distribution of prion aggregates per se, chaperone distribution in cell divisions may also play a role in [PSI+] destabilization by HS. Although mother cells accumulate more protein aggregates, they also accumulate more aggregate-associated chaperones, specifically Hsp104. Indeed, when mother cells retain a prion in the first post-HS division, they produce colonies with an increased proportion of [psi−] cells generated in subsequent cell divisions compared with daughters (Klaips et al. 2014). One explanation for this could be an increased level of Hsp104 in mother cells.
Asymmetric segregation of prion aggregates can also be detected in other conditions. When Hsp104 activity is chemically inhibited and propagons are diluted by subsequent cell divisions, mother cells are more likely to retain prions than daughters (Cox et al. 2003). Aged yeast [PSI+] cells accumulate larger-sized aggregates, indicative of the asymmetry of aggregate segregation (Derdowski et al. 2010). The ability of yeast cells to get rid of prion aggregates in cell divisions may explain why in mammalian systems amyloids and prions are most damaging to terminally differentiated cells like neurons.
IS THERE AN AMYLOID SENSOR?
Triggering a protective response in a cell that has accumulated amyloid deposits requires “amyloid sensor(s)” that can orchestrate cellular responses. One candidate is the small glutamine-rich tetratricopeptide co-chaperone Sgt2, a yeast counterpart of mammalian SGT, specifically SGTA (Cziepluch et al. 1998; Liu et al. 1999). Sgt2 is involved in the guided entry of tail-anchored (GET) proteins trafficking pathway (Schuldiner et al. 2008; Wang et al. 2010). It binds hydrophobic tails of newly synthesized membrane-bound proteins and mediates their association with cytosolic chaperones (including Ssa and Ydj1) and other components of the GET pathway (Get3, 4, and 5 proteins), thus preventing uncontrolled aggregation of tail-anchored proteins and facilitating their delivery to membranes (Liou et al. 2007; Jonikas et al. 2009).
Under conditions when major Hsps are not induced, Sgt2 is increased in abundance in response to the presence of a prion in the yeast cells (Kiktev et al. 2012). The animal ortholog of Sgt2 is also shown to interact with amyloids in some assays (Fonte et al. 2002), suggesting that the amyloid-binding properties of Sgt2/SGT could be conserved in evolution. Recent data show that Sgt2 also interacts with polyglutamine (Wang et al. 2007, 2009) and prion (Kiktev et al. 2012) aggregates and facilitates binding of Ssa to Sup35 prion fibers, thus promoting [PSI+] propagation even in the presence of high levels of Hsp104 (Kiktev et al. 2012). Formation of amyloid-like aggregates and quality-control deposits by expanded polyglutamines in yeast also promotes formation of cytosolic aggregates of TA and Get proteins whose formation depends on Sgt2 (Kiktev et al. 2012) and leads to the appearance of detergent-insoluble aggregates of Sgt2, which are similar to those formed by yeast prion proteins (Nizhnikov et al. 2014a). These data implicate Sgt2 as an “amyloid sensor,” connecting chaperones to amyloidogenic/prionogenic proteins and potentially capable of acquiring at least a transient amyloid-like form by itself. It remains to be determined whether the amyloid-like state of Sgt2 is transmissible; if so, this will be the first proven case of a chaperone or co-chaperone that possesses prion properties.
PARALLELS AND DIFFERENCES BETWEEN CHAPERONE EFFECTS ON AMYLOIDS IN LOWER AND HIGHER EUKARYOTES
Chaperone proteins of the Hsp70 and Hsp40 families, as well as their major co-chaperones and cofactors, are conserved from yeast to humans. The constitutive protein of the human Hsp70 family, Hsc70, can support yeast viability and [PSI+] prion propagation in the absence of yeast Ssa, albeit with reduced efficiency (Tutar et al. 2006). Mutations in human Hsp40 protein DNAJB6 that are associated with myopathy differentially impact yeast prions [PSI+] and [RNQ+] when introduced to the Sis1-DNAJB6 chimeric construct (Stein et al. 2014). Thus, it is likely that mammalian and human chaperones are involved in modulation of amyloid-like aggregation in their native environments. Indeed, the Hsp40 proteins from mammals and other animals have been reported to influence aggregation-related disorders such as polyglutamine diseases (Kazemi-Esfarjani and Benzer 2000; Auluck et al. 2002; Fayazi et al. 2006; Brehme et al. 2014; Paul and Mahanta 2014). Binding to the Hsp70/Hsp40 chaperones prevents heat shock–induced intranuclear aggregation of mammalian protein TDP-43 (Udan-Johns et al. 2014), which is associated with a variety of human neurodegenerative diseases. The animal ortholog of the Sgt2 co-chaperone is shown to interact with amyloids in some assays (Fonte et al. 2002), suggesting that the role of Sgt2/SGT in “amyloid sensing” could be conserved in evolution. The Hsp70-Ssb chaperone is fungi-specific (Peisker et al. 2010); however, the RAC complex is conserved (Jaiswal et al. 2011), and, therefore, it is likely that another member of the Hsp70 family works as a functional analog of Ssb in mammals.
In contrast to other members of the yeast prion-propagating machinery, orthologs of Hsp104 (except for its mitochondrial counterparts) are not found in multicellular animals, although they are present in plants, protists, and prokaryotes (Zolkiewski et al. 2012). Highly expressed or “potentiated” hyperactive Hsp104 constructs counteract some metazoan amyloids in yeast and animal models (Satyal et al. 2000; Jackrel et al. 2014; Jackrel and Shorter 2015); however, some other protein(s) should take the place of Hsp104 in native animal cells. It appears that such functional analogs of Hsp104 exist, as constructs based on yeast Sup35NM can propagate the prion state in cultured mammalian cells (Hofmann and Vorberg 2013; Hofmann et al. 2013). The Hsp110 protein (homologous to yeast Sse1) has been implicated in helping Hsp70/Hsp40 to disaggregate stress-damaged mammalian proteins (Shorter 2011; Torrente and Shorter 2013). The human Hsp110/Hsp70/Hsp40 complex, with the help of the small Hsp, HspB5, can promote depolymerization of α-synuclein (associated with Parkinson’s disease) in vitro (Duennwald et al. 2012). Some mammalian proteins of the AAA+ superfamily are also obvious candidates for the role of Hsp104. Of special interest are proteins RuvbL1 and RuvbL2, which were previously described as helicases but were recently shown to perform some chaperone functions and participate in the control of protein aggregation and aggregate detoxification (Zaarur et al. 2015). Notably, RuvbL can promote disassembly of amyloid β (Aβ) aggregates in vitro, and overexpressed yeast homologs of Ruvb1 or 2 (Rvb1 or 2, respectively) compensate for the viability defect during severe HS in the absence of Hsp104. It is likely that different chaperones may interact with mammalian amyloid- and prion-like proteins expressed in different compartments (e.g., intracellular and excreted extracellular). Recent data (Cohen et al. 2015) identify a BRICHOS domain, implicated in chaperone function and present in a variety of mammalian proteins as a factor binding Aβ polymers (associated with Alzheimer’s disease) and inhibiting formation of oligomeric seeds. Amelioration of Aβ toxicity by BRICHOS indicates that the progression of Alzheimer’s disease may depend on fragmentation and propagation of Aβ polymers in a manner similar to yeast prions.
CONCLUDING REMARKS
Overall, an overwhelming body of data supports the notion that the formation, propagation, and elimination of yeast prions are mediated by the protein quality-control machinery of the yeast cell. Protein networks that are responsible for proteostasis can both antagonize prions and be hijacked by prions for the purpose of their own proliferation. Therefore, modulation of proteostasis networks provides a most promising tool for controlling prions and amyloids, both in pathology and disease. Available evidence strongly shows relevance of yeast-derived results to mammalian prion and amyloid phenomena and underlines the importance of yeast as an experimental model leading to mechanistic understanding of their molecular and cellular foundations.
ACKNOWLEDGMENTS
This work is supported in part by grant GM093294 from the National Institutes of Health (K.D.W.), and by St. Petersburg University, Project 15.61.2218.2013 and Grant 14-50-00069 from the Russian Science Foundation (Y.O.C.).
Footnotes
Editor: Stanley B. Prusiner
Additional Perspectives on Prion Biology available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Aguilaniu H, Gustafsson L, Rigoulet M, Nyström T. 2003. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 299: 1751–1753. [DOI] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137: 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M, Chernova TA, Newnam GP, Yin L, Shanks J, Karpova TS, Lee A, Laur O, Subramanian S, Kim D, et al. 2014. Stress-dependent proteolytic processing of the actin assembly protein Lsb1 modulates a yeast prion. J Biol Chem 289: 27625–27639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen KD, Wegrzyn RD, Chernova TA, Müller S, Newnam GP, Winslett PA, Wittich KB, Wilkinson KD, Chernoff YO. 2005. Hsp70 chaperones as modulators of prion life cycle: Novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 169: 1227–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen KD, Chernova TA, Tennant EP, Wilkinson KD, Chernoff YO. 2007. Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J Biol Chem 282: 3004–3013. [DOI] [PubMed] [Google Scholar]
- Amor AJ, Castanzo DT, Delany SP, Selechnik DM, van Ooy A, Cameron DM. 2015. The ribosome-associated complex antagonizes prion formation in yeast. Prion 9: 144–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, Ivanov P. 2014. Stress granules, P-bodies and cancer. Biochim Biophys Acta 1849: 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM. 2002. Chaperone suppression of α-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science 295: 865–868. [DOI] [PubMed] [Google Scholar]
- Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW. 2008. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol Biol Cell 19: 2433–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailleul PA, Newnam GP, Steenbergen JN, Chernoff YO. 1999. Genetic study of interactions between the cytoskeletal assembly protein sla1 and prion-forming domain of the release factor Sup35 (eRF3) in Saccharomyces cerevisiae. Genetics 153: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchsenius AS, Wegrzyn RD, Newnam GP, Inge-Vechtomov SG, Chernoff YO. 2001. Yeast prion protein derivative defective in aggregate shearing and production of new “seeds.” EMBO J 20: 6683–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehme M, Voisine C, Rolland T, Wachi S, Soper JH, Zhu Y, Orton K, Villella A, Garza D, Vidal M, et al. 2014. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep 9: 1135–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JC, Lindquist S. 2009. A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Dev 23: 2320–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR. 2014. mRNP granules. Assembly, function, and connections with disease. RNA Biol 11: 1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Szczesniak B, Kochneva-Pervukhova NV, Kushnirov VV, Ter-Avanesyan MD, Boguta M. 2001. Ssb1 chaperone is a [PSI+] prion-curing factor. Curr Genet 39: 62–67. [DOI] [PubMed] [Google Scholar]
- Chernoff YO. 2007. Stress and prions: Lessons from the yeast model. FEBS Lett 581: 3695–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268: 880–884. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD. 1999. Evidence for a protein mutator in yeast: Role of the Hsp70-related chaperone Ssb in formation, stability, and toxicity of the [PSI] prion. Mol Cell Biol 19: 8103–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernova TA, Allen KD, Wesoloski LM, Shanks JR, Chernoff YO, Wilkinson KD. 2003. Pleiotropic effects of Ubp6 loss on drug sensitivities and yeast prion are due to depletion of the free ubiquitin pool. J Biol Chemy 278: 52102–52115. [DOI] [PubMed] [Google Scholar]
- Chernova TA, Romanyuk AV, Karpova TS, Shanks JR, Ali M, Moffatt N, Howie RL, O’Dell A, McNally JG, Liebman SW, et al. 2011. Prion induction by the short-lived, stress-induced protein Lsb2 is regulated by ubiquitination and association with the actin cytoskeleton. Mol Cell 43: 242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernova TA, Wilkinson KD, Chernoff YO. 2014. Physiological and environmental control of yeast prions. FEMS Microbiol Rev 38: 326–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho M, Lade SJ, Alberti S, Gross T, Tolić IM. 2014. Fusion of protein aggregates facilitates asymmetric damage segregation. PLoS Biol 12: e1001886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SI, Arosio P, Presto J, Kurudenkandy FR, Biverstål H, Dolfe L, Dunning C, Yang X, Frohm B, Vendruscolo M, et al. 2015. A molecular chaperone breaks the catalytic cycle that generates toxic Aβ oligomers. Nat Struct Mol Biol 22: 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM. 2014. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 38: 254–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BS, Tuite MF, McLaughlin CS. 1988. The psi factor of yeast: A problem in inheritance. Yeast 4: 159–178. [DOI] [PubMed] [Google Scholar]
- Cox B, Ness F, Tuite M. 2003. Analysis of the generation and segregation of propagons: Entities that propagate the [PSI+] prion in yeast. Genetics 165: 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr DM, Ramos CH. 2015. Specification of Hsp70 function by Type I and Type II Hsp40. Subcell Biochem 78: 91–102. [DOI] [PubMed] [Google Scholar]
- Cziepluch C, Kordes E, Poirey R, Grewenig A, Rommelaere J, Jauniaux JC. 1998. Identification of a novel cellular TPR-containing protein, SGT, that interacts with the nonstructural protein NS1 of parvovirus H-1. J Virol 72: 4149–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R. 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol 179: 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdowski A, Sindi SS, Klaips CL, DiSalvo S, Serio TR. 2010. A size threshold limits prion transmission and establishes phenotypic diversity. Science 330: 680–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Liebman SW. 2013. The story of stolen chaperones: How overexpression of Q/N proteins cures yeast prions. Prion 7: 294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. 2001. Prions affect the appearance of other prions: The story of [PIN+]. Cell 106: 171–182. [DOI] [PubMed] [Google Scholar]
- DiSalvo S, Derdowski A, Pezza JA, Serio TR. 2011. Dominant prion mutants induce curing through pathways that promote chaperone-mediated disaggregation. Nat Struct Mol Biol 18: 486–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas PM, Treusch S, Ren HY, Halfmann R, Duennwald ML, Lindquist S, Cyr DM. 2008. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc Natl Acad Sci 105: 7206–7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Park KW, Yu H, Fan Q, Li L. 2008. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet 40: 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald ML, Echeverria A, Shorter J. 2012. Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol 10: e1001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erjavec N, Larsson L, Grantham J, Nyström T. 2007. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev 21: 2410–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q, Park KW, Du Z, Morano KA, Li L. 2007. The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics 177: 1583–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayazi Z, Ghosh S, Marion S, Bao X, Shero M, Kazemi-Esfarjani P. 2006. A Drosophila ortholog of the human MRJ modulates polyglutamine toxicity and aggregation. Neurobiol Dis 24: 226–244. [DOI] [PubMed] [Google Scholar]
- Fonte V, Kapulkin WJ, Taft A, Fluet A, Friedman D, Link CD. 2002. Interaction of intracellular β amyloid peptide with chaperone proteins. Proc Natl Acad Sci 99: 9439–9444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganusova EE, Ozolins LN, Bhagat S, Newnam GP, Wegrzyn RD, Sherman MY, Chernoff YO. 2006. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol Cell Biol 26: 617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M, Lilie H, Fünfschilling U, Mun A, Ross S, Lithgow T, Rücknagel P, Rospert S. 2001. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc Natl Acad Sci 98: 3762–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15: 5383–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. 1998. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82. [DOI] [PubMed] [Google Scholar]
- Glover JR, Lum R. 2009. Remodeling of protein aggregates by Hsp104. Protein Pept Lett 16: 587–597. [DOI] [PubMed] [Google Scholar]
- Gokhale KC, Newnam GP, Sherman MY, Chernoff YO. 2005. Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the yeast model. J Biol Chem 280: 22809–22818. [DOI] [PubMed] [Google Scholar]
- Gong H, Romanova NV, Allen KD, Chandramowlishwaran P, Gokhale K, Newnam GP, Mieczkowski P, Sherman MY, Chernoff YO. 2012. Polyglutamine toxicity is controlled by prion composition and gene dosage in yeast. PLoS Genet 8: e1002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsen CW, Glover JR. 2012a. Insight into molecular basis of curing of [PSI+] prion by overexpression of 104-kDa heat shock protein (Hsp104). J Biol Chem 287: 542–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsen CW, Glover JR. 2012b. A new perspective on Hsp104-mediated propagation and curing of the yeast prion [PSI+]. Prion 6: 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R, Vevea JD, Swayne TC, Chojnowski R, Hill V, Boldogh IR, Pon LA. 2013. Actin dynamics affect mitochondrial quality control and aging in budding yeast. Curr Biol 23: 2417–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higurashi T, Hines JK, Sahi C, Aron R, Craig EA. 2008. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc Natl Acad Sci 105: 16596–16601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines JK, Li X, Du Z, Higurashi T, Li L, Craig EA. 2011. [SWI], the prion formed by the chromatin remodeling factor Swi1, is highly sensitive to alterations in Hsp70 chaperone system activity. PLoS Genet 7: e1001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J, Vorberg I. 2013. Life cycle of cytosolic prions. Prion 7: 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann JP, Denner P, Nussbaum-Krammer C, Kuhn PH, Suhre MH, Scheibel T, Lichtenthaler SF, Schätzl HM, Bano D, Vorberg IM. 2013. Cell-to-cell propagation of infectious cytosolic protein aggregates. Proc Natl Acad Sci 110: 5951–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Gautschi M, Walter W, Rospert S, Craig EA. 2005. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat Struct Mol Biol 12: 497–504. [DOI] [PubMed] [Google Scholar]
- Hung GC, Masison DC. 2006. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics 173: 611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackrel ME, Shorter J. 2015. Engineering enhanced protein disaggregases for neurodegenerative disease. Prion 9: 90–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackrel ME, DeSantis ME, Martinez BA, Castellano LM, Stewart RM, Caldwell KA, Caldwell GA, Shorter J. 2014. Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell 156: 170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal H, Conz C, Otto H, Wölfle T, Fitzke E, Mayer MP, Rospert S. 2011. The chaperone network connected to human ribosome-associated complex. Mol Cell Biol 31: 1160–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Pfund C, Craig EA. 1997. Functional specificity among Hsp70 molecular chaperones. Science 275: 387–389. [DOI] [PubMed] [Google Scholar]
- Jarosz DF, Lancaster AK, Brown JC, Lindquist S. 2014. An evolutionarily conserved prion-like element converts wild fungi from metabolic specialists to generalists. Cell 158: 1072–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G, Song Y, Chung S, Masison DC. 2004. Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol Cell Biol 24: 3928–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, et al. 2009. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323: 1693–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M, Redeker V, Melki R. 2014. A role for the proteasome in the turnover of Sup35p and in [PSI+] prion propagation. Mol Microbiol 92: 507–528. [DOI] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. 2008. Misfolded proteins partition between two distinct quality control compartments. Nature 454: 1088–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalchman MA, Koide HB, McCutcheon K, Graham RK, Nichol K, Nishiyama K, Kazemi-Esfarjani P, Lynn FC, Wellington C, Metzler M, et al. 1997. HIP1, a human homologue of S. cerevisiae Sla2p, interacts with membrane-associated huntingtin in the brain. Nat Genet 16: 44–53. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. 2012. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 149: 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemi-Esfarjani P, Benzer S. 2000. Genetic suppression of polyglutamine toxicity in Drosophila. Science 287: 1837–1840. [DOI] [PubMed] [Google Scholar]
- Kiktev DA, Patterson JC, Müller S, Bariar B, Pan T, Chernoff YO. 2012. Regulation of chaperone effects on a yeast prion by cochaperone Sgt2. Mol Cell Biol 32: 4960–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiktev DA, Melomed MM, Lu CD, Newnam GP, Chernoff YO. 2015. Feedback control of prion formation and propagation by the ribosome-associated chaperone complex. Mol Microbiol 96: 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland PA, Reidy M, Masison DC. 2011. Functions of yeast Hsp40 chaperone Sis1p dispensable for prion propagation but important for prion curing and protection from prion toxicity. Genetics 188: 565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaips CL, Hochstrasser ML, Langlois CR, Serio TR. 2014. Spatial quality control bypasses cell-based limitations on proteostasis to promote prion curing. eLife 3: e04288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D, Wickner RB. 2007. Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Mol Biol Cell 18: 2149–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. 2003. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem 278: 49636–49643. [DOI] [PubMed] [Google Scholar]
- Kryndushkin DS, Shewmaker F, Wickner RB. 2008. Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J 27: 2725–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV, Kryndushkin DS, Boguta M, Smirnov VN, Ter-Avanesyan MD. 2000. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr Biol 10: 1443–1446. [DOI] [PubMed] [Google Scholar]
- Li X, Rayman JB, Kandel ER, Derkatch IL. 2014. Functional role of Tia1/Pub1 and Sup35 prion domains: Directing protein synthesis machinery to the tubulin cytoskeleton. Mol Cell 55: 305–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman SW, Chernoff YO. 2012. Prions in yeast. Genetics 191: 1041–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou ST, Cheng MY, Wang C. 2007. SGT2 and MDY2 interact with molecular chaperone YDJ1 in Saccharomyces cerevisiae. Cell Stress Chaperones 12: 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu FH, Wu SJ, Hu SM, Hsiao CD, Wang C. 1999. Specific interaction of the 70-kDa heat shock cognate protein with the tetratricopeptide repeats. J Biol Chem 274: 34425–34432. [DOI] [PubMed] [Google Scholar]
- Liu B, Larsson L, Caballero A, Hao X, Öling D, Grantham J, Nyström T. 2010. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell 140: 257–267. [DOI] [PubMed] [Google Scholar]
- Liu B, Larsson L, Franssens V, Hao X, Hill SM, Andersson V, Höglund D, Song J, Yang X, Öling D, et al. 2011. Segregation of protein aggregates involves actin and the polarity machinery. Cell 147: 959–961. [DOI] [PubMed] [Google Scholar]
- Madania A, Dumoulin P, Grava S, Kitamoto H, Schärer-Brodbeck C, Soulard A, Moreau V, Winsor B. 1999. The Saccharomyces cerevisiae homologue of human Wiskott–Aldrich syndrome protein Las17p interacts with the Arp2/3 complex. Mol Biol Cell 10: 3521–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S. 2012. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol Biol Cell 23: 3041–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. 2003. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115: 825–836. [DOI] [PubMed] [Google Scholar]
- Michelitsch MD, Weissman JS. 2000. A census of glutamine/asparagine-rich regions: Implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci 97: 11910–11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SB, Ho CT, Winkler J, Khokhrina M, Neuner A, Mohamed MY, Guilbride DL, Richter K, Lisby M, Schiebel E, et al. 2015a. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J 34: 778–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SB, Mogk A, Bukau B. 2015b. Spatially organized aggregation of misfolded proteins as cellular stress defense strategy. J Mol Biol 427: 1564–1574. [DOI] [PubMed] [Google Scholar]
- Miot M, Reidy M, Doyle SM, Hoskins JR, Johnston DM, Genest O, Vitery MC, Masison DC, Wickner S. 2011. Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc Natl Acad Sci 108: 6915–6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosavi B, Wongwigkarn J, Tuite MF. 2010. Hsp70/Hsp90 co-chaperones are required for efficient Hsp104-mediated elimination of the yeast [PSI+] prion but not for prion propagation. Yeast 27: 167–179. [DOI] [PubMed] [Google Scholar]
- Moran C, Kinsella GK, Zhang ZR, Perrett S, Jones GW. 2013. Mutational analysis of Sse1 (Hsp110) suggests an integral role for this chaperone in yeast prion propagation in vivo. G3 (Bethesda) 3: 1409–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama H, Edskes HK, Wickner RB. 2000. [URE3] prion propagation in Saccharomyces cerevisiae: Requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol 20: 8916–8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. 1992. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71: 97–105. [DOI] [PubMed] [Google Scholar]
- Newby GA, Lindquist S. 2013. Blessings in disguise: Biological benefits of prion-like mechanisms. Trends Cell Biol 23: 251–259. [DOI] [PubMed] [Google Scholar]
- Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. 1999. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol 19: 1325–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newnam GP, Birchmore JL, Chernoff YO. 2011. Destabilization and recovery of a yeast prion after mild heat shock. J Mol Biol 408: 432–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizhnikov AA, Alexandrov AI, Ryzhova TA, Mitkevich OV, Dergalev AA, Ter-Avanesyan MD, Galkin AP. 2014a. Proteomic screening for amyloid proteins. PloS ONE 9: e116003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizhnikov AA, Antonets KS, Inge-Vechtomov SG, Derkatch IL. 2014b. Modulation of efficiency of translation termination in Saccharomyces cerevisiae. Prion 8: 247–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyström T, Liu B. 2014. The mystery of aging and rejuvenation—A budding topic. Curr Opin Microbiol 18C: 61–67. [DOI] [PubMed] [Google Scholar]
- Osherovich LZ, Cox BS, Tuite MF, Weissman JS. 2004. Dissection and design of yeast prions. PLoS Biol 2: E86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YN, Zhao X, Yim YI, Todor H, Ellerbrock R, Reidy M, Eisenberg E, Masison DC, Greene LE. 2014. Hsp104 overexpression cures Saccharomyces cerevisiae [PSI+] by causing dissolution of the prion seeds. Eukaryot Cell 13: 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel BK, Gavin-Smyth J, Liebman SW. 2009. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol 11: 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Mahanta S. 2014. Association of heat-shock proteins in various neurodegenerative disorders: Is it a master key to open the therapeutic door? Mol Cell Biochem 386: 45–61. [DOI] [PubMed] [Google Scholar]
- Peisker K, Chiabudini M, Rospert S. 2010. The ribosome-bound Hsp70 homolog Ssb of Saccharomyces cerevisiae. Biochim Biophys Acta 1803: 662–672. [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. 2003. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 21: 921–926. [DOI] [PubMed] [Google Scholar]
- Reidy M, Masison DC. 2011. Modulation and elimination of yeast prions by protein chaperones and co-chaperones. Prion 5: 245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy M, Sharma R, Shastry S, Roberts BL, Albino-Flores I, Wickner S, Masison DC. 2014. Hsp40s specify functions of Hsp104 and Hsp90 protein chaperone machines. PLoS Genet 10: e1004720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, Alexander RD, Spiller MP, Beggs JD. 2008. A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci 121: 2463–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikhvanov EG, Romanova NV, Chernoff YO. 2007. Chaperone effects on prion and nonprion aggregates. Prion 1: 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogoza T, Goginashvili A, Rodionova S, Ivanov M, Viktorovskaya O, Rubel A, Volkov K, Mironova L. 2010. Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc Natl Acad Sci 107: 10573–10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyal SH, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer JM, Morimoto RI. 2000. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc Natl Acad Sci 97: 5750–5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS. 2008. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134: 634–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwimmer C, Masison DC. 2002. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol Cell Biol 22: 3590–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Masison DC. 2008. Functionally redundant isoforms of a yeast Hsp70 chaperone subfamily have different antiprion effects. Genetics 179: 1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Masison DC. 2011. Single methyl group determines prion propagation and protein degradation activities of yeast heat shock protein (Hsp)-70 chaperones Ssa1p and Ssa2p. Proc Natl Acad Sci 108: 13665–13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Stanley RF, Masison DC. 2009. Curing of yeast [URE3] prion by the Hsp40 cochaperone Ydj1p is mediated by Hsp70. Genetics 181: 129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J. 2011. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PloS ONE 6: e26319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondheimer N, Lopez N, Craig EA, Lindquist S. 2001. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J 20: 2435–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Yang Q, Yang J, Larsson L, Hao X, Zhu X, Malmgren-Hill S, Cvijovic M, Fernandez-Rodriguez J, Grantham J, et al. 2014. Essential genetic interactors of SIR2 required for spatial sequestration and asymmetrical inheritance of protein aggregates. PLoS Genet 10: e1004539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag EM, Vonk WI, Frydman J. 2014. Sorting out the trash: The spatial nature of eukaryotic protein quality control. Curr Opin Cell Biol 26: 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spokoini R, Moldavski O, Nahmias Y, England JL, Schuldiner M, Kaganovich D. 2012. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep 2: 738–747. [DOI] [PubMed] [Google Scholar]
- Stein KC, Bengoechea R, Harms MB, Weihl CC, True HL. 2014. Myopathy-causing mutations in an HSP40 chaperone disrupt processing of specific client conformers. J Biol Chem 289: 21120–21130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers DW, Douglas PM, Cyr DM. 2009. Prion propagation by Hsp40 molecular chaperones. Prion 3: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki G, Shimazu N, Tanaka M. 2012. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science 336: 355–359. [DOI] [PubMed] [Google Scholar]
- Sweeny EA, Jackrel ME, Go MS, Sochor MA, Razzo BM, DeSantis ME, Gupta K, Shorter J. 2015. The hsp104 N-terminal domain enables disaggregase plasticity and potentiation. Mol Cell 57: 836–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebenyi G, Wigley WC, Hall B, Didier A, Yu M, Thomas P, Krämer H. 2007. Hook2 contributes to aggresome formation. BMC Cell Biol 8: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S. 2007. The WASP–WAVE protein network: Connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol 8: 37–48. [DOI] [PubMed] [Google Scholar]
- Tessarz P, Schwarz M, Mogk A, Bukau B. 2009. The yeast AAA+ chaperone Hsp104 is part of a network that links the actin cytoskeleton with the inheritance of damaged proteins. Mol Cell Biol 29: 3738–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrente MP, Shorter J. 2013. The metazoan protein disaggregase and amyloid depolymerase system: Hsp110, Hsp70, Hsp40, and small heat shock proteins. Prion 7: 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch S, Lindquist S. 2012. An intrinsically disordered yeast prion arrests the cell cycle by sequestering a spindle pole body component. J Cell Biol 197: 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutar Y, Song Y, Masison DC. 2006. Primate chaperones Hsc70 (constitutive) and Hsp70 (induced) differ functionally in supporting growth and prion propagation in Saccharomyces cerevisiae. Genetics 172: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Madariaga ML, Lindquist S. 2008. Prion switching in response to environmental stress. PLoS Biol 6: e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Treusch S, Dong J, McCaffery JM, Bevis B, Lindquist S. 2010. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc Natl Acad Sci 107: 8633–8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan-Johns M, Bengoechea R, Bell S, Shao J, Diamond MI, True HL, Weihl CC, Baloh RH. 2014. Prion-like nuclear aggregation of TDP-43 during heat shock is regulated by HSP40/70 chaperones. Hum Mol Genet 23: 157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese J, Abrams J, Wang Y, Morano KA. 2012. Biology of the heat shock response and protein chaperones: Budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol Mol Biol Rev 76: 115–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Meriin AB, Costello CE, Sherman MY. 2007. Characterization of proteins associated with polyglutamine aggregates: A novel approach towards isolation of aggregates from protein conformation disorders. Prion 1: 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Meriin AB, Zaarur N, Romanova NV, Chernoff YO, Costello CE, Sherman MY. 2009. Abnormal proteins can form aggresome in yeast: Aggresome-targeting signals and components of the machinery. FASEB J 23: 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Brown EC, Mak G, Zhuang J, Denic V. 2010. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell 40: 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, et al. 2004. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119: 653–665. [DOI] [PubMed] [Google Scholar]
- *.Wickner RB. 2016. Yeast and fungal prions. Cold Spring Harb Perspect Biol 8: a023531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Bezsonov E, Bateman DA. 2014. Normal levels of the antiprion proteins Btn2 and Cur1 cure most newly formed [URE3] prion variants. Proc Natl Acad Sci 111: E2711–E2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmund F, del Alamo M, Pechmann S, Chen T, Albanese V, Dammer EB, Peng J, Frydman J. 2013. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell 152: 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J, Tyedmers J, Bukau B, Mogk A. 2012a. Chaperone networks in protein disaggregation and prion propagation. J Struct Biol 179: 152–160. [DOI] [PubMed] [Google Scholar]
- Winkler J, Tyedmers J, Bukau B, Mogk A. 2012b. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J Cell Biol 198: 387–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KJ, Ren HY, Trepte P, Cyr DM. 2013. The Hsp70/90 cochaperone, Sti1, suppresses proteotoxicity by regulating spatial quality control of amyloid-like proteins. Mol Biol Cell 24: 3588–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Hong JY, Derkatch IL, Liebman SW. 2013. Heterologous gln/asn-rich proteins impede the propagation of yeast prions by altering chaperone availability. PLoS Genet 9: e1003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaarur N, Xu X, Lestienne P, Meriin AB, McComb M, Costello CE, Newnam GP, Ganti R, Romanova NV, Shanmugasundaram M, et al. 2015. RuvbL1 and RuvbL2 enhance aggresome formation and disaggregate amyloid fibrils. EMBO J 34: 2363–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Derkatch IL, Liebman SW. 2001. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI+] and [PIN+]. Mol Microbiol 39: 37–46. [DOI] [PubMed] [Google Scholar]
- Zhou C, Slaughter BD, Unruh JR, Eldakak A, Rubinstein B, Li R. 2011. Motility and segregation of Hsp104-associated protein aggregates in budding yeast. Cell 147: 1186–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Slaughter BD, Unruh JR, Guo F, Yu Z, Mickey K, Narkar A, Ross RT, McClain M, Li R. 2014. Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell 159: 530–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolkiewski M, Zhang T, Nagy M. 2012. Aggregate reactivation mediated by the Hsp100 chaperones. Arch Biochem Biophys 520: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]