ABSTRACT
The nucleolus forms as a consequence of ribosome biogenesis, but it is also implicated in other cell functions. The identification of nucleolus-associated chromatin domains (NADs) in animal and plant cells revealed the presence of DNA sequences other than rRNA genes in and around the nucleolus. NADs display repressive chromatin signatures and harbour repetitive DNA, but also tRNA genes and RNA polymerase II-transcribed genes. Furthermore, the identification of NADs revealed a specific function of the nucleolus and the protein Nucleolin 1 (NUC1) in telomere biology. Here, we discuss the significance of these data with regard to nucleolar structure and to the role of the nucleolus and NUC1 in global genome organization and stability.
KEYWORDS: genome stability, heterochromatin, nucleolus, nuclear architecture, nucleolus-associated chromatin domains
Introduction
In eukaryotic cells, a precise organization of chromatin is essential for proper gene regulation and genome integrity. Epigenetic changes such as DNA methylation and post-translational histone modifications participate in the establishment of different states of chromatin condensation.1 According to a classic textbook definition, heterochromatin is highly condensed and transcriptionally repressed, being mainly composed of repetitive DNA elements and transposons. Euchromatin is in a relaxed state permissive for transcription and is enriched in genes. However, deeper classifications of chromatin organization delineate several subcategories of chromatin.2 One major challenge in cell biology today is to accurately situate the epigenome in its nuclear context. Chromatin organization within the nucleus is governed by several aspects of cell organization. Among these are intra and inter-chromosomal interactions, nuclear periphery association and localization to specialized nuclear bodies. It is clear now that the nuclear distribution of heterochromatin is regulated by several factors. In mammals, the nuclear lamina was shown to anchor a large portion of heterochromatin at the nuclear periphery. Although the positioning of heterochromatin at the nuclear periphery is not solely dependent on the nuclear lamina, this process is crucial for genome integrity and its misregulation can provoke diseases in humans that include muscle dystrophies, neurological disorders or progeria syndromes.3
Heterochromatin was also shown to organize in and around the nucleolus, as demonstrated by the characterization of nucleolus-associated chromatin domains (NADs) in human cells.4,5 Several megabases of genomic DNA encompassing at least one genomic region from each of the 23 chromosomes physically associate with the nucleolus, demonstrating that NAD identity is not determined by genetic linkage to rRNA (rRNA) gene arrays located on Chr, 13, 14, 15, 21 and 22.
In plant cells, we recently reported the identification of NADs from Arabidopsis thaliana leaf cells6: DNA purified from isolated nucleoli was analyzed by high-throughput sequencing.7 In A. thaliana, like in human cells, NADs display heterochromatic signatures and encompass genomic domains from all 5 A. thaliana chromosomes. Moreover, NAD identification in a nucleolin 1 (nuc1) mutant, where the nucleolus structure is affected,8 demonstrated several important aspect of NAD biology. Indeed, our recent study demonstrated the importance of rRNA gene expression for NAD composition, and the role of NUC1 protein in nucleolar heterochromatin organization and telomere maintenance. Here, we discuss the emerging role of the nucleolus in the nuclear organization of the epigenome and the impact of its alteration in the nuc1 mutant.
NAD composition in A. thaliana
In A. thaliana leaf cells, NADs are mainly composed of sequences displaying 2 types of epigenetic features: regions enriched in dimethylated lysine 9 of Histone 3 (H3K9me2) or regions displaying trimethylated lysine 27 of Histone 3 (H3K27me3). H3K9me2 enriched regions correspond to genomic domains mainly composed of repeated elements such as transposons, while H3K27me3 enriched regions are mainly composed of silent genes or pseudogenes. In total, NADs represent around 4% of the genome, 3% of the genes and 11% of transposable elements in A. thaliana.6
This organization may differ depending on the cell type or stress condition analyzed, but we observed that 45S rRNA gene regulation is one important determinant of NAD composition. 45S rRNA genes are arranged in large tandem arrays called Nucleolus Organizer Regions (NORs). A change in NOR transcriptional activity modifies its subnuclear localization, as demonstrated in mutant backgrounds that show perturbations in rRNA gene transcription, such as nuc1 or histone deacetylase 6 (hda6).9-11 As a consequence, changes in NOR transcription affect the global organization of chromosomes in the nucleus. In wild-type A. thaliana (ecotype Col-0), only the short arm of chromosome 4 (Chr4S) associates with the nucleolus, due to NOR4-derived rRNA gene expression. However, in the nuc1 mutant where NOR2 rRNA genes also become transcriptionally active, the short arm of chromosome 2 (Chr2S) now associates with the nucleolus. This observation demonstrates the importance of rRNA gene regulation in the global organization of nuclear DNA.
Heterochromatin and the nucleolus
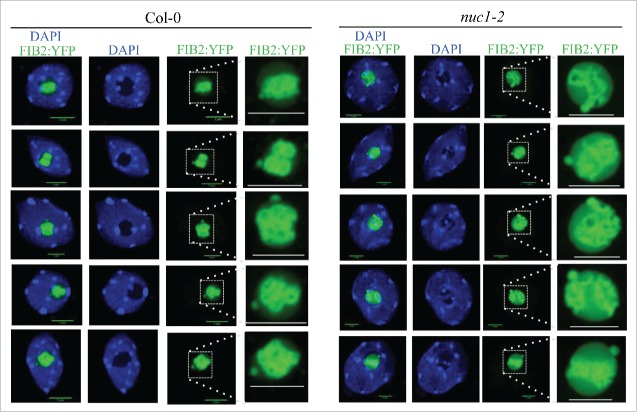
It has been known for decades that the nucleolus is surrounded by a layer of highly condensed late-replicating heterochromatin, often called the nucleolar shell.12 Whether this localization was an indirect consequence of the transcriptional activity of DNA recruited to the nucleolar shell or whether nucleolar proteins were directly implicated in nucleolar heterochromatin tethering remained an open question for several years. However, studies performed in Drosophila demonstrated a direct role for the respective protein homologs of Nucleolin (known as Modulo in Drosophila) and Nucleoplasmin.13 These two nucleolar proteins form a stable platform together with the CCCTC-binding factor to anchor centromeres at the periphery of the nucleolus in Drosophila.13 In our recent study in A. thaliana, analyses of NADs in the nuc1 mutant revealed a perturbed association of centromeres with the nucleolus: centromere association with the nucleolus was slightly increased in the nuc1 mutant. In addition, heterochromatin can be detected in a larger proportion of nucleoli in the nuc1 mutant as compared to wild-type nucleoli. Previous analyses of subnucleolar compartments in nuc1 by transmission electron microscopy revealed defects in their organization. All the 3 nucleolar compartments normally observed in eukaryotic cells—the granular component (GC), dense fibrillar component (DFC) and fibrillar center (FC)—are disrupted in nuc1.8 We know that rRNA genes transcription and processing is affected in these plants,10,14 but we wanted to see whether intra-nucleolar heterochromatin could be responsible for this disorganization. Fibrillarin is a nucleolar protein implicated in the snoRNA-guided methylation of rRNA precursors in the DFC.15,16 Therefore, Fibrillarin mainly localizes to the DFC, forming ring-like structures in the nucleolus (Fig. 1A). However, in the nuc1 mutant, A. thaliana homolog Fibrillarin 2 (FIB2) subnucleolar localization varies dramatically; instead of forming ring-like structures, it surrounds the intra nucleolar heterochromatin signals (Fig. 1B). These data suggest that the intra-nucleolar chromatin accumulating in nuc1 nucleoli affect the typical organization of nucleolar factors implicated in ribosome biogenesis. Additional analyses should reveal whether or not this observation is also true for other factors required for proper pre-rRNA processing.
Figure 1.
Chromatin and Fibrillarin distribution in WT and nuc1 nuclei Confocal images of 5 nuclei from WT (A) or nuc1 mutant (B) expressing the Fibrillarin fused to YFP (green). DNA is labeled by DAPI (blue) and scale bars represent 5 μm.
Role of NUC1 in the phase separation of sub-nucleolar domains
The anchoring of centromeres and heterochromatin at the nucleolar periphery probably helps keep heterochromatin away from the nucleoplasm, where most RNA Polymerase II transcription of genes occurs. Conversely, this heterochromatin tethering would also protect the nucleolus from being invaded by heterochromatin. In human cells, recent work demonstrated how 2 nucleolar proteins, Fibrillarin and Nucleophosmin, can phase separate into droplets similar to subnucleolar compartments in vitro and in vivo.17 This is partially due to the physical properties of the Arginine/Glycine (R/G) rich disordered domain of Fibrillarin. The combination of the R/G-rich domain and at least one RNA Recognition Motif (RRM) in the Fibrillarin protein is required for proper subnucleolar compartment formation and maintenance.17
In A. thaliana there is no obvious homolog of Nucleophosmin, but NUC1, one of the most abundant nucleolar proteins, also combines 2 RRM domains with a disordered R/G rich domain in its C-terminal region (Fig. 2). Knock-out mutations of nuc1 provoke nucleolar disorganisation and, as mentioned before, heterochromatin entering the nucleolus (Fig. 1). The A. thaliana genome encodes a second Nucleolin homolog, NUC2, which is not expressed in the leaf cell. However, in a nuc1 mutant background, NUC2 becomes expressed and appears to partially complement NUC1 deficiency because the nuc1 nuc2 double mutant is lethal.8,18 Why NUC2 complementation of NUC1 is only partial remains an open question, but sequence comparison reveals the presence of less (R/G) residues in the C-terminal part of NUC2 (Fig. 2). 45 R/G residues out of 60 are found in NUC1, while only 26 out 59 are present in NUC2, affecting its disordered domain as analyzed using the Protein DisOrder prediction System PrDOS19 (Fig. 2). We propose that the subnucleolar disorganisation observed in nuc1 mutants is due to the reduction of this R/G domain in the remaining Nucleolin homolog, NUC2, preventing it from establishing proper subnucleolar compartments. Whether the intranucleolar localization of heterochromatin is a cause or a consequence of subnucleolar disorganisation observed in nuc1 mutants is unknown, but a systematic analysis of mutants with disrupted heterochromatin organization or altered nucleolus function might help answering this question.
Figure 2.
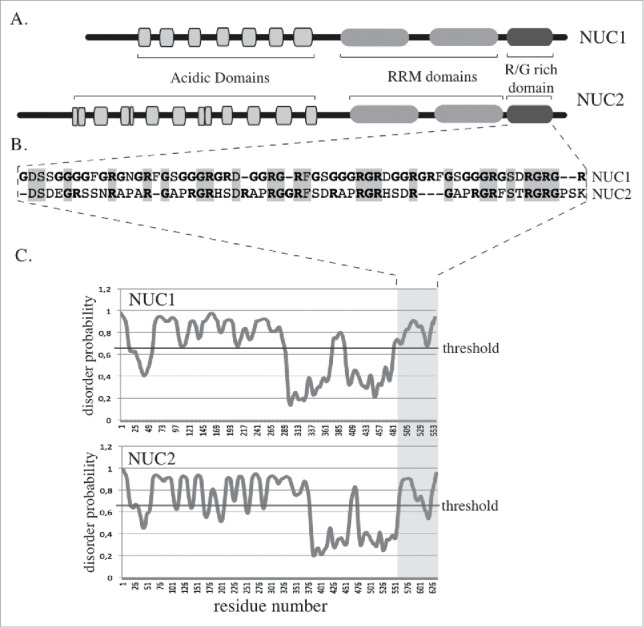
NUC1 and NUC2 proteins organization A. Schematic representation of NUC1 (AT1G48920) and NUC2 (AT3G18610) proteins from A. thaliana. B. Amino acid sequence alignment of the R/G rich domains of NUC1 and NUC2. C. Disorder profile plots of NUC1 and NUC2 amino acid sequences, obtained with the prediction software PrDOS. The black lines correspond to the threshold for a 2% prediction false rate.
NUC1 is required for global genome organization and stability
In A. thaliana, telomeric and subtelomeric regions cluster at the nucleolus.20-22 NAD identification in A. thaliana confirmed the association of telomeres with the nucleolus in wild-type plants, but also revealed decreased telomere association with the nucleolus in the nuc1 mutant.6 Further investigation demonstrated that NUC1 co-immunoprecipitates with a telomerase. In addition, telomeres are shortened in the nuc1 mutant, including telomeres not adjacent to NORs.6 These observations indicate that NUC1 itself may play a direct role in telomere biology. Human Nucleolin interacts with a double-stranded telomeric probe in vitro.23 But defects in telomere length could also be an indirect consequence of decreased telomere clustering at the nucleolus. The nucleolus may offer an environment that facilitates telomere protection and/or maintenance. In fact, the nucleolus has been described in animal and plant cells as a site where factors implicated in telomerase elongation are at least transiently detected (reviewed in24,25).
In Drosophila and yeast, the number of rRNA genes influences heterochromatin formation, global gene expression and genome stability.26-28 However, shorter telomeres are a sign of genome instability in nuc1. Further analyses of the nuc1 mutant should reveal if telomeric regions defects is solely due to the absence of NUC1, or if it is also an indirect effect of changes in the global genome organization.
Concluding remarks
Our recent work demonstrated the importance of the nucleolus in global genome organization, as well as its role with the NUC1 protein in ensuring genome stability.6 This study extends previous investigations of NADs in human cells4,5 and addresses novel aspects of genome organization in Arabidopsis. However, several new questions have been raised by our own studies and those of other researchers. Is the nucleolar association of NAD-genes required for their transcriptional regulation? Does the pool of NAD loci depend on cell type? Are NADs affected by stress conditions? The identification of NADs in different cell types and growth conditions together with transcriptome analyses should allow us to determine whether the pool of NADs is dynamic, and to explore its potential role in gene regulation. Finally, our data suggest a direct link between ribosome biogenesis and genome organization within the nucleolus and further analyses should help clarify how these 2 fundamental aspects of the nucleus biology are interconnected.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgements
The authors thank Todd Blevins for English corrections and the Imaging facility of Perpignan University Via Domitia.
Funding
This work was supported by CNRS and ANR JCJC NucleoReg (ANR-15-CE12-0013-01) to F.P.
Notes on contributors
CP and FP performed the experiments. FP designed the experiments, analyzed the data, and wrote the manuscript.
References
- [1].Kouzarides T. Chromatin modifications and their function. Cell 2007; 128:693-705; PMID:17320507; http://dx.doi.org/ 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- [2].Sequeira-Mendes J, Araguez I, Peiro R, Mendez-Giraldez R, Zhang X, Jacobsen SE, Bastolla U, Gutierrez C. The functional topography of the arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell 2014; 26:2351-66; PMID:24934173; http://dx.doi.org/ 10.1105/tpc.114.124578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dobrzynska A, Gonzalo S, Shanahan C, Askjaer P. The nuclear lamina in health and disease. Nucl Austin Tex 2016; 7:233-48; PMID:27158763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nemeth A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Peterfia B, Solovei I, Cremer T, Dopazo J, Längst G. Initial genomics of the human nucleolus. PLoS Genet 2010; 6:e1000889; PMID:20361057; http://dx.doi.org/ 10.1371/journal.pgen.1000889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell 2010; 21:3735-48; PMID:20826608; http://dx.doi.org/ 10.1091/mbc.E10-06-0508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pontvianne F, Carpentier MC, Durut N, Pavlištová V, Jaške K, Schořová Š, et al. Identification of Nucleolus-Associated Chromatin Domains Reveals a Role for the Nucleolus in 3D Organization of the A. thaliana Genome. Cell Rep. 2016; 16(6):1574–87. http://dx.doi.org/ 10.1016/j.celrep.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pontvianne F, Boyer-Clavel M, Saez-Vasquez J. Fluorescence-Activated Nucleolus Sorting in Arabidopsis. Methods Mol Biol Clifton NJ 2016; 1455:203-11; PMID:27576720; http://dx.doi.org/ 10.1007/978-1-4939-3792-9_15 [DOI] [PubMed] [Google Scholar]
- [8].Pontvianne F, Matia I, Douet J, Tourmente S, Medina FJ, Echeverria M, Sáez-Vásquez J. Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol Biol Cell 2007; 18:369-79; PMID:17108323; http://dx.doi.org/ 10.1091/mbc.E06-08-0751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pontvianne F, Blevins T, Chandrasekhara C, Mozgova I, Hassel C, Pontes OMF, Tucker S, Mokros P, Muchová V, Fajkus J, et al.. Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev 2013; 27:1545-50; PMID:23873938; http://dx.doi.org/ 10.1101/gad.221648.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pontvianne F, Abou-Ellail M, Douet J, Comella P, Matia I, Chandrasekhara C, Debures A, Blevins T, Cooke R, Medina FJ, et al.. Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet 2010; 6:e1001225; PMID:21124873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Earley KW, Pontvianne F, Wierzbicki AT, Blevins T, Tucker S, Costa-Nunes P, Pontes O, Pikaard CS. Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev 2010; 24:1119-32; PMID:20516197; http://dx.doi.org/ 10.1101/gad.1914110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ferreira J, Paolella G, Ramos C, Lamond AI. Spatial organization of large-scale chromatin domains in the nucleus: a magnified view of single chromosome territories. J Cell Biol 1997; 139:1597-610; PMID:9412456; http://dx.doi.org/ 10.1083/jcb.139.7.1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Padeken J, Mendiburo MJ, Chlamydas S, Schwarz H-J, Kremmer E, Heun P. The nucleoplasmin homolog NLP mediates centromere clustering and anchoring to the nucleolus. Mol Cell 2013; 50:236-49; PMID:23562326; http://dx.doi.org/ 10.1016/j.molcel.2013.03.002 [DOI] [PubMed] [Google Scholar]
- [14].Kojima H, Suzuki T, Kato T, Enomoto K, Sato S, Kato T, Tabata S, Sáez-Vasquez J, Echeverría M, Nakagawa T, et al.. Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J Cell Mol Biol 2007; 49:1053-63; PMID:17286797; http://dx.doi.org/ 10.1111/j.1365-313X.2006.03016.x [DOI] [PubMed] [Google Scholar]
- [15].Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt EC. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J 1991; 10:573-83; PMID:1825809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gerbi SA, Borovjagin A. U3 snoRNA may recycle through different compartments of the nucleolus. Chromosoma 1997; 105:401-6; PMID:9211967; http://dx.doi.org/ 10.1007/BF02510476 [DOI] [PubMed] [Google Scholar]
- [17].Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. Coexisting liquid phases underlie nucleolar subcompartments. Cell 2016; 165:1686-97; PMID:27212236; http://dx.doi.org/ 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Durut N, Abou-Ellail M, Pontvianne F, Das S, Kojima H, Ukai S, de Bures A, Comella P, Nidelet S, Rialle S, et al.. A duplicated NUCLEOLIN gene with antagonistic activity is required for chromatin organization of silent 45S rDNA in Arabidopsis. Plant Cell 2014; 26:1330-44; PMID:24668745; http://dx.doi.org/ 10.1105/tpc.114.123893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ishida T, Kinoshita K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res 2007; 35:W460-4; PMID:17567614; http://dx.doi.org/ 10.1093/nar/gkm363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Armstrong SJ, Franklin FC, Jones GH. Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J. Cell Sci 2001; 114:4207-17; PMID:11739653 [DOI] [PubMed] [Google Scholar]
- [21].Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I. Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc Natl Acad Sci U S A 2002; 99:14584-9; PMID:12384572; http://dx.doi.org/ 10.1073/pnas.212325299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Schubert V, Rudnik R, Schubert I. Chromatin associations in Arabidopsis interphase nuclei. Front Genet 2014; 5:389; PMID:25431580; http://dx.doi.org/ 10.3389/fgene.2014.00389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pollice A, Zibella MP, Bilaud T, Laroche T, Pulitzer JF, Gilson E. In vitro binding of nucleolin to double-stranded telomeric DNA. Biochem Biophys Res Commun 2000; 268:909-15; PMID:10679304; http://dx.doi.org/ 10.1006/bbrc.2000.2237 [DOI] [PubMed] [Google Scholar]
- [24].Dvorackova M, Fojtova M, Fajkus J. Chromatin dynamics of plant telomeres and ribosomal genes. Plant J Cell Mol Biol 2015; 83:18-37; PMID:25752316; http://dx.doi.org/ 10.1111/tpj.12822 [DOI] [PubMed] [Google Scholar]
- [25].Pederson T. The nucleolus. Cold Spring Harb Perspect Biol. 2011 Mar 1;3(3). pii: a000638. http://dx.doi.org/ 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Paredes S, Branco AT, Hartl DL, Maggert KA, Lemos B. Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS Genet 2011; 7:e1001376; PMID:21533076; http://dx.doi.org/ 10.1371/journal.pgen.1001376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Paredes S, Maggert KA. Ribosomal DNA contributes to global chromatin regulation. Proc Natl Acad Sci U S A 2009; 106:17829-34; PMID:19822756; http://dx.doi.org/ 10.1073/pnas.0906811106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ide S, Miyazaki T, Maki H, Kobayashi T. Abundance of ribosomal RNA gene copies maintains genome integrity. Science 2010; 327:693-6; PMID:20133573; http://dx.doi.org/ 10.1126/science.1179044 [DOI] [PubMed] [Google Scholar]