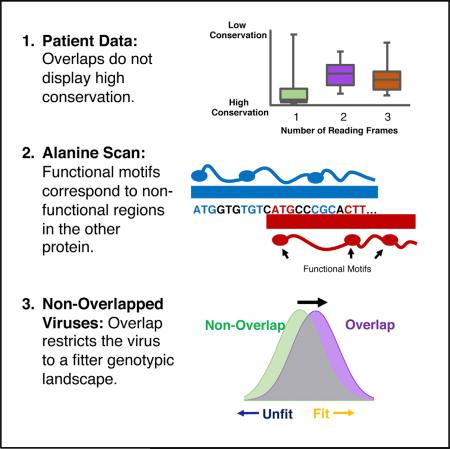
SUMMARY
Overlapping genes pose an evolutionary dilemma as one DNA sequence evolves under the selection pressures of multiple proteins. Here, we perform systematic statistical and mutational analyses of the overlapping HIV-1 genes tat and rev and engineer exhaustive libraries of non-overlapped viruses to perform deep mutational scanning of each gene independently. We find a “segregated” organization in which overlapped sites encode functional residues of one gene or the other, but never both. Furthermore, this organization eliminates unfit genotypes, providing a fitness advantage to the population. Our comprehensive analysis reveals the extraordinary manner in which HIV minimizes the constraint of overlapping genes and repurposes that constraint to its own advantage. Thus, overlaps are not just consequences of evolutionary constraints, but rather can provide population fitness advantages.
Graphical abstract
INTRODUCTION
The sequencing of the first complete DNA genome, ΦX174, revealed the startling discovery that genes can overlap with one another (Barrell et al., 1976; Sanger et al., 1977). Since this initial observation, overlapping reading frames have been observed in most viruses and across all domains of life (Belshaw et al., 2007; Makalowska et al., 2005; Rogozin et al., 2002). In viruses, these regions are traditionally thought to arise as consequences of error-prone polymerases and constraints on the size of viral capsid proteins (Belshaw et al., 2007; Chirico et al., 2010). For instance, high polymerase error rates favor short genomes thereby decreasing the probability of catastrophic mutations, while the viral capsid imposes a biophysical limit on genome size. Other models suggest that overlap formation is driven by selection pressures favoring evolutionary innovation (Brandes and Linial, 2016; Keese and Gibbs, 1992; Rancurel et al., 2009; Sabath et al., 2012), as overlaps are also found in large genomes. Regardless, once present in a genome, overlapping genes must balance nucleotide usage so that the functions of each reading frame are satisfied. Several studies have used computational methods to estimate gene-wide selective forces (Hein and Støvlbaek, 1995; Sabath et al., 2008; Wei and Zhang, 2014) but only a few have generated experimental data (Kawano et al., 2013). Computational analyses of protein structure have demonstrated that overlapped proteins in all viruses tend toward intrinsic disorder (Rancurel et al., 2009), but how structured and/or functional regions are divided at the amino acid level remains unknown. It is possible to envisage two extreme models for this simultaneous evolution: (1) a “segregated” model in which the amino acid/nucleotide preferences for one gene dominate and the other gene accommodates with no observable benefit to itself, or (2) a “shared” model in which both genes exert selective forces at the same site, enforcing strong conservation in both frames (Figure S1). It is unlikely that segregated or shared decisions are uniform over an entire overlap, so defining the selective forces on a per residue basis becomes critical to understanding how the functions of a pair of proteins can be properly balanced.
HIV-1 provides a compelling model as it contains eight distinct areas of coding overlap (Figure S2A) constituting ~8% of its entire genome, and extensive sequence information from many virus isolates is available (Foley et al., 2013) (https://www.hiv.lanl.gov/content/index). The tat and rev regulatory genes (Figure 1A) are a particularly interesting case as both are essential for virus replication and thus experience strong simultaneous selective pressure, both have well-established functions and assays, and both have partial structures available to help interpret the functional consequences of sequence variants (Figure S2B) (Daugherty et al., 2010; DiMattia et al., 2010; Tahirov et al., 2010). Tat activates transcriptional elongation at the HIV-1 promoter via its interactions with host transcription factors (most notably P-TEFb) and an RNA element at the 5′ end of viral transcripts known as trans-activation response element (TAR) (Ott et al., 2011). Rev facilitates the nuclear export of partially spliced and un-spliced viral RNAs that encode essential late-stage viral proteins and genomic RNA for packaging (Pollard and Malim, 1998). Rev binds as an oligomer to an RNA element present in viral introns known as the Rev response element (RRE) and guides the RNAs to the cytoplasm via interactions with the Crm1 nuclear export machinery.
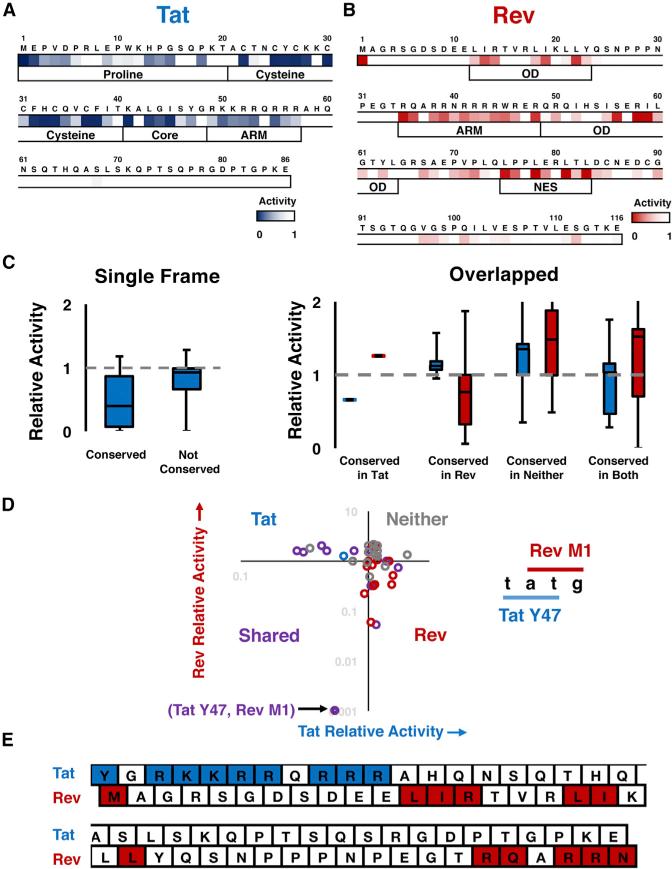
Figure 1. Organization and Conservation of HIV-1 Overlaps.
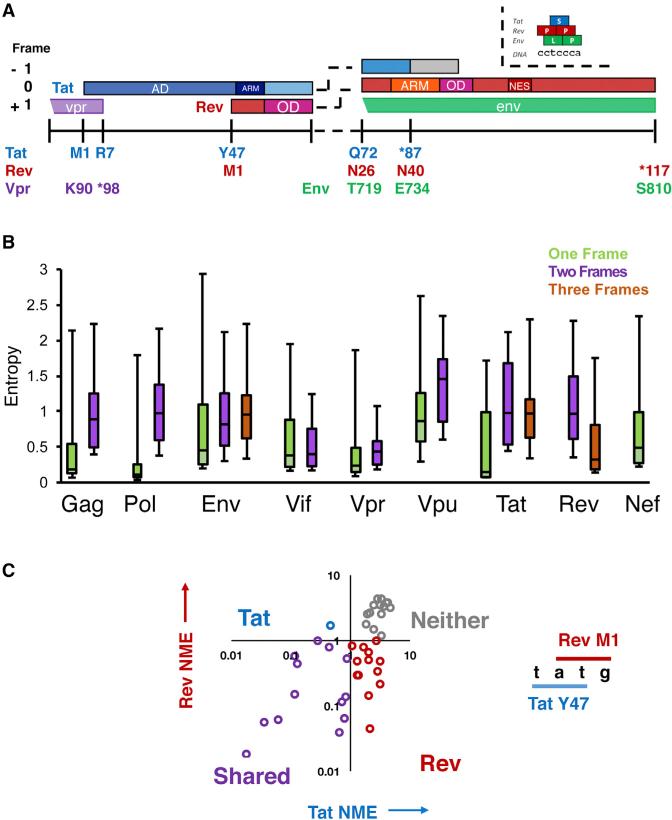
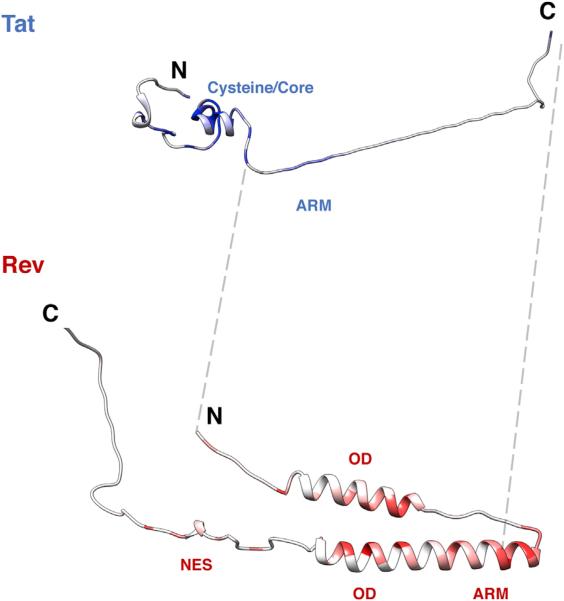
(A) Layout of the Genetic Organization of the tat/rev overlap in HIV-1. ARM, arginine rich motif/nuclear localization sequence; OD, oligomerization domain; NES, nuclear export sequence. In HIV-1 NL4-3 Tat is 86 residues, although many patient genes are 101 residues (gray box).
(B) Individual gene entropy analysis for overlapped and single-frame regions in the HIV-1 genome (see Figure S2A). Entropy values were computed at the protein level for each frame and Shannon entropy values for alignments of HIV-1 patient sequences are shown. Median, range, and interquartile range (IQR) are shown in the box and whiskers plot. A score of 0 indicates absolute conservation and a score of 3 indicates near-absolute degeneracy.
(C) Categorization of sites by normalized mean entropy (NME) in the tat/rev overlap. Residues are grouped into pairs that share two nucleotides, and their NME plotted accordingly (Tat NME, Rev NME). Quadrants are labeled to indicate which genes are conserved in that region.
See also Figure S1.
In order to understand the consequences of the tat/rev overlap for viral evolution, we compare sequence conservation in patient isolates to comprehensive residue-by-residue functional maps of Tat and Rev generated by alanine scanning. We further compare these datasets to replication experiments in viruses in which we removed the tat/rev overlap and subsequently measured the experimental fitness of every amino acid at each position in each protein independently. We find that HIV-1 has evolved in a segregated manner—to separate functionally important amino acids for Tat and Rev—and moreover, the arrangement decreases sampling of unfit genotypes, thereby turning an apparent genetic constraint into a fitness advantage. The combination of these orthogonal datasets provides the most complete picture of an overlap to date and demonstrates another way in which HIV-1 has efficiently utilized the coding capacity of its small genome to optimally arrange its core regulatory machinery.
RESULTS
Overlapped Regions in HIV-1 Are Not More Conserved Than Single-Frame Regions
In order to explore the constraints imposed in the tat/rev overlap, we first examined the conservation of each protein residue in the HIV-1 proteome using patient data from the Los Alamos Database (https://www.hiv.lanl.gov/content/index). These datasets comprise ~2,000 HIV-1 sequences in high-quality alignments. We then calculated the Shannon entropy for each residue of each HIV-1 protein. We used this relatively simple metric—in which a low value indicates high amino acid conservation—as it requires no assumptions about selection or substitution rates, which are challenging to approximate in overlapped regions. Furthermore, this metric has previously been used to quantify diversity of viral sequences, to identify interaction surfaces on proteins, and analyze overlapping reading frames (Pan and Deem, 2011; Zaaijer et al., 2007). We examined each residue in every HIV-1 gene and compared the single, double, and triple reading frame regions to one another (Figure S2C) and discovered that overlapped regions were, surprisingly, not more conserved than single-frame regions. This behavior held for almost every HIV-1 gene (Figure 1B), although this may reflect inherent biases in the structural features of these proteins as opposed to the presence of dual coding. Regardless, these results demonstrate that regions of overlap do not experience high conservation, providing evidence against a shared organization (with the caveat that positive selection and variations in mutational tolerance can confound this analysis). Genome-wide selection analyses using sophisticated evolutionary rate calculations have produced results consistent with this data, although the inability to accurately estimate neutral substitution (or determine selective forces on a per-residue basis) have prevented these studies from definitively interpreting these findings (Ngandu et al., 2008; Snoeck et al., 2011).
Classification of Overlapped Residues as Segregating or Sharing Based on Entropy
To narrow our focus and analyze specific residues within Tat and Rev, we defined a statistic, normalized-to-mean entropy (NME), in which the entropy of each residue was normalized to the average entropy of the one-frame regions of the viral genome. We reasoned that amino acid conservation within single-frame regions reflects a simple requirement for protein function without the confounding constraints of an overlapping gene. Thus a NME <1 would be suggestive of functional importance even within overlapped regions. We calculated the NME values of all residues from Tat and Rev that share two nucleotides within a codon—for example, Y47 of Tat (tAT) shares two nucleotides with M1 of Rev (ATg)—and plotted Rev NME versus Tat NME (Figure 1C). By separating the plot into quadrants, we can speculatively classify residues according to their conservation for one protein alone (Tat or Rev), for both (Shared), or for neither. Thus, we can tentatively hypothesize that residues highly conserved in only either the Tat or Rev reading frame are segregated, devoted to a single protein while residues conserved in both frames may share functionally important positions. However, this naive classification requires detailed experimental testing as it does not attempt to estimate a neutral rate of substitution or consider the extent to which the organization of the genetic code may constrain the codon possibilities at the overlapped positions.
Functionally Important Residues in Tat and Rev Are Segregated
To evaluate which amino acids in Tat and Rev are conserved for function versus those restricted by codon usage in the alternative frame, we generated complete sets of alanine point mutants for both proteins and tested their activities in separate cell-based reporter systems, effectively removing the coding constraint of the overlap. Tat activity was measured by its ability to enhance transcription of an integrated firefly luciferase gene under the control of the HIV-1 promoter (D'Orso and Frankel, 2010) (Figure S3A), and Rev activity was measured by co-transfecting each mutant with a CMV-driven, RRE-containing, gag-pol reporter that produces viral capsid (p24) in a Rev-dependent manner (Smith et al., 1990) (Figure S3B). Heatmaps (Figures 2A and 2B) and graphs (Figures S3C and S3D) show the relative activities of each mutant, normalized to a reference sequence (HXB2 and NL4-3 isolates), and highlight the functionally important residues, which correspond well to established motifs of these proteins. As expected, mutation of conserved residues among patient isolates in the single-frame region of Tat (Figure 1A) correlated well with loss of function (Figure 2C). The remainder of tat overlaps with rev, env, or vpr, whereas rev overlaps with tat or env over its entire length. Within the tat/rev overlapped region, mutation of residues conserved in either only Tat or only Rev correlated with their respective loss of function, whereas mutation of residues not conserved in either protein generally did not affect function (Figure 2C). Somewhat surprisingly, however, mutation of residues that show conservation in both frames generally correlated with loss of function of only one of the proteins (Figure 2C).
Figure 2. Functional Dissection of Tat and Rev.
(A and B) Alanine scanning of Tat (A) and Rev (B) in functional reporter assays.
(C) Relationship between entropy and function. Left: highly conserved (NME <1) residues in the Tat single-frame region generally result in a large loss of Tat activity when mutated to alanine, while residues that are not conserved (NME > 1) do not. A similar pattern is seen in the overlap (right) when only a single frame is conserved (blue, Tat activities; red, Rev activities). When neither gene is conserved there is generally no loss of function. When both frames are conserved the relationship between entropy and function is less clear.
(D) Categorization of residues in the tat/rev overlap by function. Residues are grouped into sites that share two nucleotides and colored as in Figure 1D. The only site that is required for the activity of both proteins is (Tat Y47, Rev M1) (labeled). Coloring of points represents classifications made by NME as in Figure 1D.
(E) Residues determined to be functionally important by alanine scan in the context of the overlap. Dark blue residues are important for Tat function (<50% activity) while dark red residues are important for Rev (<50%activity). Only Tat Y47 and Rev M1 share nucleotides.
See also Figure S3.
To examine more closely if residues in this overlapping region might be simultaneously important for both proteins, we plotted the activities of Tat and Rev mutants against one another and found, remarkably, that almost no sites contributed to both functions, with one exception (Figure 2D, “Shared” quadrant). This result stands in stark contrast to the naive expectation from the entropy analysis (Figure 1D versus Figure 2D). This functional segregation at the single amino acid level, despite the overlap, is readily apparent when the important functional amino acids are mapped onto their primary structures (Figure 2E). The lone exception is the overlap between Tat Y47 and the Rev initiation codon M1, where Y47 is important for Tat activity and M1 is important only for Rev protein synthesis, as mutating the Rev start codon in the context of an N-terminally tagged Rev has no effect on activity (Figure S3D). The striking segregation of functional residues suggests that Tat and Rev conform to a segregated model of evolution.
Deep Mutational Scanning and Selection of Viruses with Uncoupled tat and rev Genes
Our patient dataset contained several sites that were conserved in both the tat and rev frames. Our functional data suggests that this conservation is not a consequence of simultaneous selection for the function of both proteins, but rather reflects the requirements of only a single protein with subsequent restriction imposed by the genetic code in the alternative frame. If this is true, then we would expect the conserved but non-functional sites to mutate more freely absent the constraint of the overlap. To examine this, we created viruses (NL4-3 isolate) in which the tat and rev genes were engineered into different parts of the viral genome by first mutating either the tat or rev start codon and introducing downstream stop co-dons, synonymous in the alternative frame, to ablate the endogenous locus. We then generated exhaustive libraries of tat or rev variants in which every individual codon was randomized and cloned the resulting codon library into the non-essential nef region of the genome (Figure S4A). This resulted in 202 virus pools (116 for rev and 86 for tat), each composed of 64 different alleles. We performed independent, competitive replication experiments with each pool and determined allele frequencies, pre- and post-selection, by deep sequencing (Figure S4B).
Population Genetics Modeling Allows Inference of Significant Selection
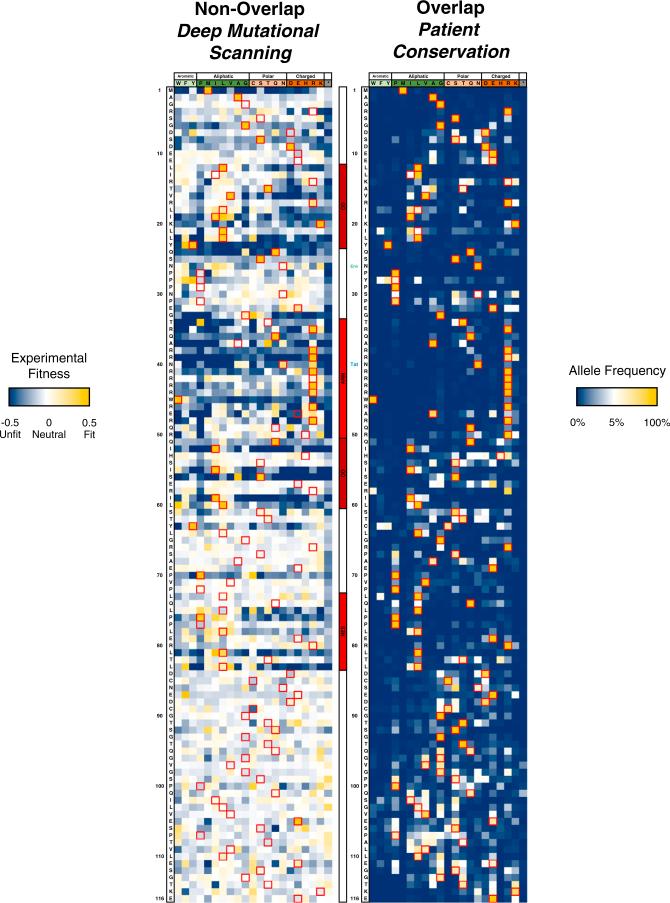
In order to infer selection from the changes in allele frequency, we developed a Wright-Fisher based model (Feder et al., 2014; Wright, 1931) using population numbers from our own replication data (Figure S4B). This model allowed us to estimate the experimental fitness for each allele and statistically test if the allele was significantly under selection (given experimental error and the possibility of neutral variation) (Figure S5A). This method has an overall high correspondence (Figure S5B) with logarithmic fitness values often calculated in deep mutational scanning experiments (McLaughlin et al., 2012) but has the advantage that statistical testing, rooted in population genetics principles, can be performed to assess confidence in selection values (Figure S5C). We used this model to calculate the mean selection coefficient (experimental fitness) for each amino acid allele (grouping synonymous codons) to produce non-overlapped mutational profiles of Tat and Rev (Figures 3 and 4). Amino acid experimental fitness profiles were clustered by chemical properties (Firnberg et al., 2014) to help visualize side chain preferences at every position, and the resulting landscapes were compared to the overlapped patient frequencies (Figures 3 and 4).
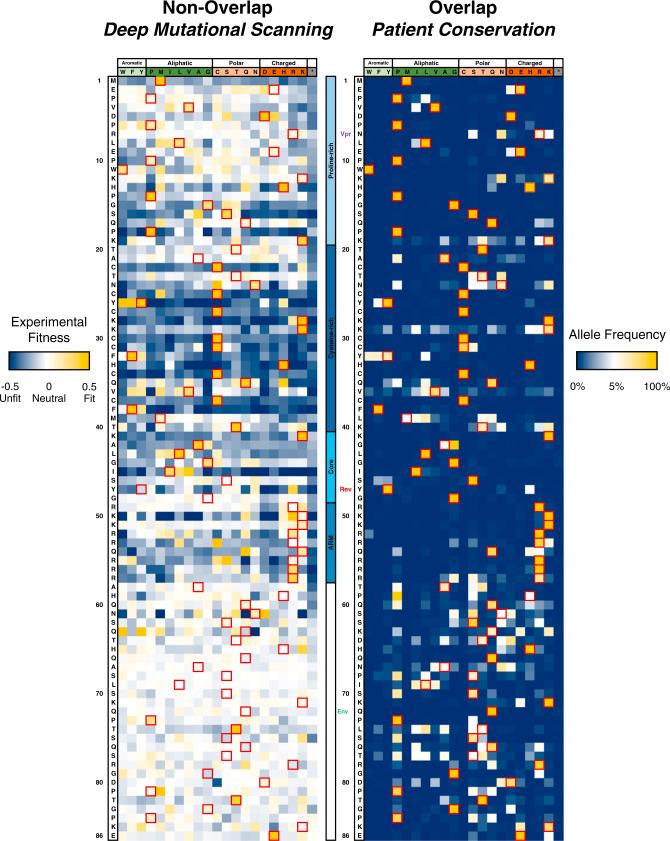
Figure 3. Mutational Profiling of HIV-1 Tat.
Left: experimental fitness of every residue (all synonymous codons grouped) of Tat in non-overlapped tat-in-nef viruses after approximately six generations of selection in SupT1 cells. Red boxes and row headers denote the NL4-3 sequence. Dark blue indicates negative selection, white indicates neutral, and gold indicates positive selection. Center: motif and overlap organization of Tat. Labels of other indicate stop (vpr) and start codons (rev, gp41). Right: overlapped/Patient conservation of Tat residues. The neutral expectation (white) is approximated as a residue being equally represented by all amino acids (1/20). Red boxes denote the NL4-3 sequence while row headers denote the consensus sequence.
See also Figures S4 and S5.
Figure 4. Mutational Profiling of HIV-1 Rev.
Left: experimental fitness of every residue (all synonymous codons grouped) of Rev in non-overlapped rev-in-nef viruses and (right) overlapped, patient conservation of Rev residues. Coloring and labeling are consistent with those shown in Figure 3. Tat stop codon and start of Env coding overlap are labeled.
See also Figures S4 and S5.
Additionally, we identified a subset of alleles from the selection data that displayed consistent and statistically significant selection (Figure S5D). The complete results of this analysis are provided (Table S1) as a resource for further study of individual alleles and comparison to existing deep mutational scanning datasets (Doud and Bloom, 2016).
Disordered Residues Mutate Freely in the Selection Dataset
Strikingly, the patient datasets show much stricter conservation than the selection datasets (Figures 3 and 4). This may in part reflect different selective forces in patients (i.e., immune system pressure, virus latency, or length of selection) but this phenomenon is commonly observed in deep mutational scanning experiments, where codon randomization is able to sample sequence space much more rapidly than natural evolution (McLaughlin et al., 2012; Thyagarajan and Bloom, 2014). Regardless, both proteins show strong selection within their known functional motifs (Figures 3 and 4; gold is positive selection and blue is negative) whereas the linker regions show generally neutral variation (white). In order to map the data to the protein structures, we used the median experimental fitness for each residue to approximate the strength of selection for each residue (Figure S6; low median value = high selection) and plotted these values onto the structures of Tat and Rev (Figure 5). Interaction and folding surfaces between Tat and P-TEFb (cysteine/core) and TAR (ARM) are readily visible as are surfaces between Rev, RRE (ARM), Crm1 (NES), and other Rev molecules (OD). In sharp contrast, the disordered regions experience a large amount of non-synonymous, but neutral substitution. Notably these regions do not express strong signals of positive selection even absent the constraint of the overlap, suggesting that the presence of an overlapped gene does not prevent the formation of additional, extended interaction surfaces. Interestingly, the neutral selection values for stop codon alleles at residue 67 of Tat and residue 86 of Rev indicate that the C termini of both proteins are dispensable for replication in this context. In some cases, we observed shifts from the NL4-3 reference sequence to the patient consensus (e.g., Rev P28Y), suggesting that NL4-3 sequences are not fully optimized for individual protein function. We also identified important alanine residues not captured by alanine mutagenesis, such as Tat A42, which showed strong conservation of small side chains (A/G) in both patient and selection datasets. Positions such as this, as well as the fact that experimental fitness of alanine variants does not always match the median experimental fitness, suggests that alanine scanning, while grounded in biophysical reasoning, may not always provide the best measure of functional importance of any particular site.
Figure 5. Structural Profiles of Tat and Rev.
Crystal structures of Tat (3MI9) and Rev (3LPH, 3NBZ) colored by median experimental fitness values. Dark blue/red coloring indicates strong selection for a particular amino acid at that site (white indicates median experimental fitness of 0). Unstructured regions in Tat (residues 50–86) and Rev (residues 1–9, 65–73, 86–116) were built in PyMOL and added to the structures for visualization purposes. The gray dashed lines flank the overlap in both proteins.
See also Figure S6.
All Three Datasets Exhibit Signatures of Segregated Evolution
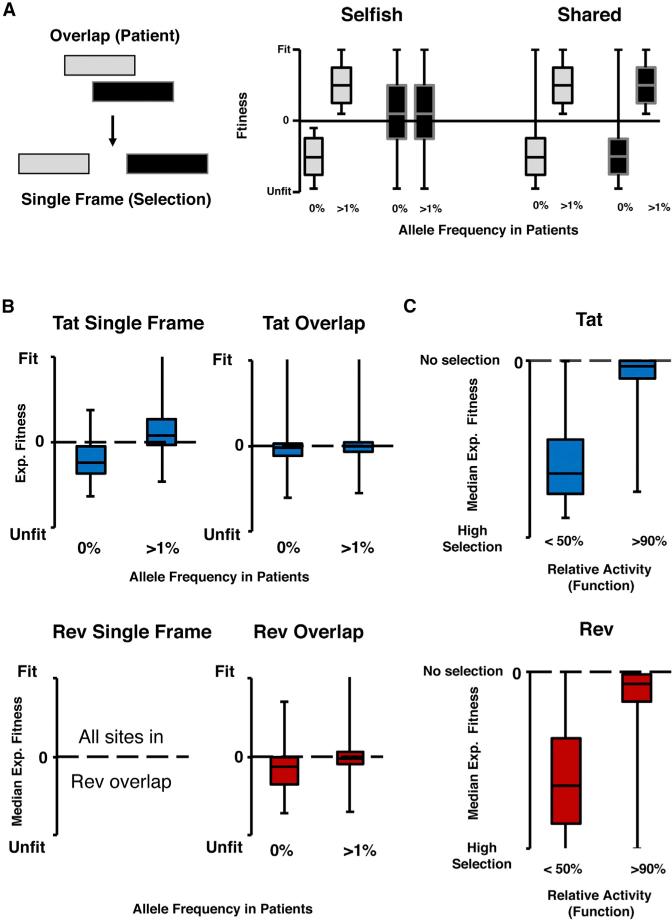
To assess the agreement between datasets, and compare them to our models of segregated and shared evolution, we grouped alleles into two categories: those present in the patient dataset (frequency >1%) and those absent (frequency = 0%). In a segregated model of evolution, a dominant gene (Figure 6A: gray gene), should act similarly regardless of whether it is overlapped or not, as its own requirements for protein function drive selection at these nucleotides. In a single-frame context, the dominant gene should continue to match the overlapped dataset with absent alleles remaining unfit and present alleles remaining fit. In contrast, the accommodating gene (black) should have many alleles that are absent in the patient population (due to functional requirements of the gray gene) but are actually fit in a single-frame context. In a shared model, the selective pressures for the functions of both proteins constrain the nucleotide sequence and thus removing the overlap provides only a minimal relaxation. At best, only a small number of chemically conservative amino acids, absent in the overlapped dataset due to the alternative frame, will be fit (Figure 6A: 0% long tail).
Figure 6. Comparisons between Datasets.
(A) Model for how overlapped proteins might behave in a single-frame context. For a dominant gene (gray) in a segregated organization, alleles that are absent (0% frequency) remain unfit while those that are present (>1%) remain fit. For an accommodating gene (black) many absent alleles are fit. In a shared organization, removal of the overlap does not change the fact that both proteins experience strong selection at that site.
(B) The single-frame region of Tat acts in a segregated manner, with high correspondence between the patient and selection data. In the overlapped regions of Tat and Rev many absent alleles are fit in the single-frame context, with Rev as the dominant gene (on average).
(C) Functional data matches selection data with functional residues (activity <50%) correlating well with selection (low median experimental fitness) while non-functional residues (activity >90%) have a median experimental fitness near 0 (neutral).
See also Figure S6.
The single-frame region of Tat provides a good test for this behavior as Tat, the only gene present, should act in a segregated manner in both patients and our randomized selection dataset. As expected, there is a strong correlation between patient allele frequency and selection (Figure 6B) with absent alleles unfit and present alleles fit. The correlation is much weaker in the multiple frame regions largely due to a subset of alleles that produce fit viruses but are absent in patients, presumably due to the constraint of the overlap (Figure 6A). These results are consistent with our segregated model, with Rev acting as the dominant gene over the majority of sites in the overlap (Figure 6B, lower panel). However, the degree to which each gene dominates varies over the region of overlap.
To compare the selection dataset to the functional dataset, we once again used the median experimental fitness for each residue to approximate overall strength of selection at each position in each protein (Figure 6C). We then segregated sites into those identified as functionally important by the alanine scan (<50% relative activity) and those identified as unimportant (>90%). As expected, those sites identified as important for both Tat and Rev, have strong signals of selection, while those identified by the alanine scan as unimportant for function have median experimental fitness near zero, suggesting no preference for any particular amino acid (Figure 6C).
Comparison of Single-Frame and Overlapped Viral Mutational Profiles
Although the segregation of function for Tat and Rev helps minimize the evolutionary penalty of the overlap, we wondered whether there might be measurable benefits to overlapping genes as opposed to encoding each gene in a single frame. Simulation studies have hypothesized that simply decreasing the length of a genome by introducing overlaps can increase viral fitness by decreasing the probability of detrimental mutations introduced by polymerase (Belshaw et al., 2007). In addition, the actual coding requirements of each protein can provide additional buffering by decreasing the number of detrimental allelic combinations. Specifically, an overlapped virus has, by definition, a limited number of possible genotypes compared to its single-frame equivalent. This restriction can result in an overlapped viral fitness landscape that is, proportionally, more, less, or equivalently fit when compared to a virus with both genes in single frames (Figure 7A). The manner in which the overlap restricts the landscape depends on each gene's functional requirements and the genetic code: detrimental restriction re sults when fit alleles in one gene encode unfit alleles in the alternative frame thereby preventing the fit allele from existing in the population. Conversely, gain of fitness restriction occurs when fit alleles are unable to encode unfit alleles in the alternative frame. This arrangement can effectively purge unfit viral genotypes, in which the fit allele is sabotaged by an unfit allele in the other gene, from the genotypic landscape of the virus.
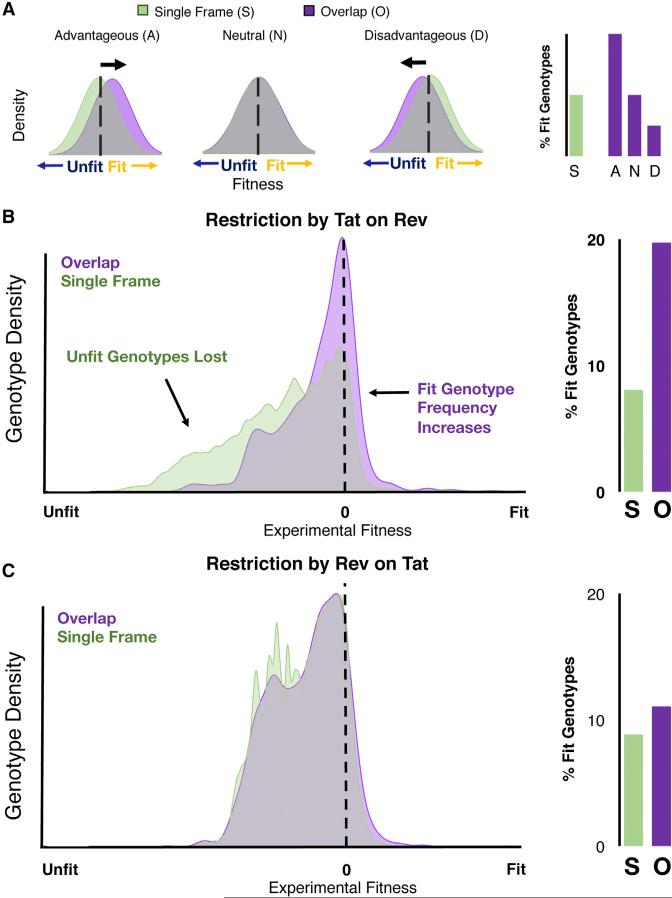
Figure 7. Advantageous Restriction by the Overlap.
(A) The fitness distribution of an overlapped virus (purple) is restricted compared to an equivalent single-frame virus (S, green) restriction can be advantageous (A), neutral (N), or disadvantageous (D).
(B) Approximated fitness distribution of viral point mutations in a single-frame virus compared to an overlapped virus in which Rev alleles are calculated for every possible Tat allele (right). The resulting overlapped landscape (O) has twice the percentage of fit genotypes (fitness >0) as the single-frame (S) virus.
(C) Equivalent comparison between a single-frame virus and overlapped one, in which Tat alleles are calculated for all Rev alleles (right). The resulting overlapped landscape is approximately equivalent to the single frame with a slight increase in the percentage of fit genotypes.
See also Figure S7.
To examine how HIV-1 Tat and Rev affect each other, we used the selection data from the non-overlapped viruses to approximate the differences in the fitness distributions of both overlapped and non-overlapped viruses. For example, if both genes were encoded in a single-frame context, viruses with the Tat Y47 allele would not affect the composition of the Rev allele at position 1, resulting in 21 possible Rev alleles (including stop codons). In the overlapped context, however, Tat Y47 restricts Rev codons to M1, I1, and T1, resulting in the loss of 18 viral genotypes (e.g., Tat Y47, Rev G1 cannot exist in an overlapped context). Importantly, the codon restrictions are not the same in both directions, as fixing the Rev M1 allele constrains Tat alleles (Y, H, D, and R) at position 47 in a different manner. Therefore, we approximated the effect of each gene's constraint on the other in both directions and compared the unconstrained single-frame context to the overlapped case (Figures 7B and 7C). Because most sites in the overlap actually restrict two adjacent codons in the alternative frame (for example, Tat G48 influences both Rev M1 and A2), we made a simplifying assumption that the experimental fitness of the two amino acids were additive and approximated the experimental fitness of the overlapped protein (Figure S7). Finally, as both Rev and Tat are required for viral replication, we approximated the overall viral fitness as the lower value of either the Rev or Tat allele.
Tat Restricts Rev to Increase the Fitness of the Overlapped Viral Landscape
Examining the restriction Tat imposes on Rev (Figure 7B) reveals a large population of potential genotypes in the single-frame context, with only a small fraction of fit alleles. By overlapping the genes, the total number of possible phenotypes is drastically reduced; however a disproportionate number of unfit combinations are purged. Explicit calculation of the genotypic proportions shows that this restriction results in a doubling of the proportion of fit genotypes (Figure 7B). In essence, the overlap takes advantage of subtle preferences in the alternative frame to reduce the number of unfit genotypes. For example, the preference for R53 in Tat biases the overlapping but relatively tolerant residue 9 of Rev toward either D or A, but away from the highly unfit R, K, and stop codons. In contrast, Rev's constraint on Tat does not significantly alter the overall percentage of fit viruses (Figure 7C), most likely because the majority of the functional motifs in Rev overlap with the readily mutable C terminus of Tat. Thus, the genomic arrangement and functional amino acid requirements of Tat and Rev appear to confer a distinct selective advantage to the virus.
DISCUSSION
Overlapping genes were originally considered unfortunate consequences of other selection pressures such as high polymerase error rates and capsid size (Belshaw et al., 2007; Chirico et al., 2010). However, more recently, this dogma has been questioned (Brandes and Linial, 2016) with gene delivery experiments demonstrating that HIV-1 is capable of packaging genomes of considerably larger sizes (Kumar et al., 2001) and with polymerase error appearing to insufficiently explain overlap proportion (Chirico et al., 2010). Indeed, the creation of fit, uncoupled viruses in this work and others (Chan et al., 2005; Neuveut and Jeang, 1996) makes explanations of overlaps as simply negative consequences of pressures on genome size highly unsatisfactory.
In our investigation of this problem, we initially hypothesized two simple models for how overlapped genes might constrain one another: a segregated model in which one protein drives evolution with little regard for the other, or a sharing model in which common nucleotides encode important amino acids in each protein. To test these models, we analyzed three datasets: (1) a patient dataset of HIV-1 isolates in which we examined frame-specific conservation on the amino acid level, (2) a functional dataset in which we tested the functional contribution of each side chain in Tat and Rev, and (3) a virus selection dataset in which we created non-overlapped Tat and Rev viruses and determined experimental fitness values for every single amino acid allele. Analysis of these datasets led to three key findings, all consistent with a segregated organization of the overlap: (1) overlapped regions in HIV-1 are not more conserved than non-overlapped ones, (2) functional motifs of Tat and Rev are segregated from each other at the level of individual nucleotides and amino acids, and (3) the tat/rev overlap reduces the probability of generating unfit combinations of these genes.
The relatively low sequence conservation observed within the overlapped regions of Tat and Rev can be rationalized in light of a segregated organization. Both proteins consist of flexible linkers and short motifs that acquire their proper three-dimensional structures using host protein and viral RNA surfaces as templates (Ott et al., 2011; Pollard and Malim, 1998). By organizing their functional motifs such that critical functional residues in one protein overlap with the highly mutable linker regions of the other protein, HIV-1 can experience a high rate of non-synonymous substitutions that remain functionally neutral.
This segregation also allows the overlap to increase the fitness of the viral population by taking advantage of the drastic reduction in the number of possible allelic combinations of tat and rev. While initial models of overlaps suggested that such a reduction might reduce overall fitness because fit alleles would be linked with unfit alleles (Miyata and Yasunaga, 1978), this line of reasoning does not take into account that the equivalent single-frame virus can harbor every allelic combination, including all possibilities where just one of Tat or Rev is unfit and thus are replication incompetent. Whether an overlap is advantageous or disadvantageous to the viral population depends on which genotypes are lost when genes are overlapped: if most purged genotypes are fit, the overlapped viruses have a lower probability of producing fit progeny whereas if more purged viruses are unfit, the fitness of the population increases.
Segregation predisposes HIV-1 to exhibit beneficial purging of genotypes in a non-intuitive manner. Even though Tat and Rev are organized such that functionally important residues tend to overlap with flexible linker regions of the opposing protein, there is often selection against disruptive alleles (such as a proline or stop codon). Thus, even in these regions, if a functionally important allele in one frame prevents a disruptive and unfit allele in the alternative frame, the overlapped virus can have a selective advantage over the single-frame virus. In contrast, a shared (rather than segregated) organization is very likely to reduce the fit sequence space of the overlapped gene because synonymous (or chemically conservative) fit alleles of one protein would almost always be detrimental in the alternative frame. In a segregated organization, such changes are likely to be neutral in the alternative frame.
Taken together, these data support a simple model for how an overlap might arise and evolve. An ancestral ORF can encode a new ORF in an alternative frame via alternative splicing (Zhao et al., 2014) or atypical translation initiation (Touriol et al., 2003), which can gradually evolve its own function. Functional motifs in the new ORF are more likely to arise in regions that correspond to highly mutable regions of the ancestral ORF to minimize disruptions to the ancestral function. As the new gene function becomes refined, mutations that reduce fitness of the ancestral gene continue to be avoided and are therefore most likely to encode unfit or neutral alleles in the younger frame.
Such a model is consistent with our data as well as previous studies suggesting that Tat-like proteins are older than Rev-like proteins (Pavesi et al., 2013). In the context of the tat/rev overlap, the functionally important Y47 residue in Tat could have led to a start codon for Rev in the alternative reading frame. The nascent Rev ORF would then evolve only in the context of fit Tat alleles, avoiding simultaneously unfit mutations of both reading frames in the process. Most functional motifs of Rev overlap with the mutationally tolerant C terminus of Tat and thus mutations of these positions would not confer any particular advantage or disadvantage to Tat.
Despite the appeal of this speculative model, several caveats should be noted. First, two ancient endogenous lentiviruses have no known tat/rev overlap (Katzourakis et al., 2007; Keckesova et al., 2009), although this might result from gene loss events, independent evolutionary origins, or poor splicing annotation. The tat/rev overlap also may be more recent than the formation of the individual genes, with rev acquiring its first exon and tat acquiring its second exon at later times. Second, our analyses have made rough estimations of viral fitness and examined only single point mutations in one viral background in an artificial genomic locus, ignoring the potential for epistatic effects at multiple positions. However, positive epistasis is rare while negative epistasis is common (Bank et al., 2015), suggesting that our simplification is conservative and likely underestimates the severity of purged deleterious genotypes (Boucher et al., 2016), while missing only a few restorative double mutations. Regardless, further studies of overlaps in HIV-1 and other systems may shed more light on these aspects. Indeed, recent analyses of polyomaviruses have traced the clade-specific birth of an overlapped gene (Carter et al., 2013), and detailed functional studies in this system may provide good tests of the model.
It seems clear that overlaps are not necessarily constraints driven by other evolutionary forces, but may, in fact, reflect a common evolutionary strategy especially prevalent in small viruses. In large viruses, evolution of genomic novelty in response to a selection pressure can occur via gene duplication, adaptation and genome compression (Elde et al., 2012). An alternative mechanism—possibly preferentially deployed in viruses with small genomes and high recombination rates where gene duplication may not be feasible—is de novo gene creation via an overlapped reading frame. Indeed, there is ample evidence that the creation of genes via overlapped reading frames leads to a large degree of genetic novelty (Belshaw et al., 2007; Brandes and Linial, 2016; Keese and Gibbs, 1992; Rancurel et al., 2009) in viral proteomes and across the tree of life (Makalowska et al., 2005).
The unilateral division of genetic information between Tat and Rev likely reflects a common and clever evolutionary outcome that satisfies a mix of positive and negative pressures and may therefore occur in other systems. Computational studies suggest that other overlapped proteins also segregate (Hughes et al., 2001; Maman et al., 2011; Zaaijer et al., 2007), although detailed experimental validation is needed to determine the extent of segregated versus shared behavior on a per residue basis. At the same time, while functional segregation appears to be a favorable outcome, it is not an inevitable one. The intrinsically disordered nature of Tat and Rev allows considerable flexibility in the placement and ordering of functional motifs. Although overlapped proteins tend to be intrinsically disordered (Kovacs et al., 2010; Rancurel et al., 2009), other viral proteins, such as the HIV-1 capsid, are extremely genetically fragile (Rihn et al., 2013) and would likely be unable to completely segregate from another gene in an alternative frame. Even the ability of the tat/rev overlap to preferentially purge unfit genotypes is only advantageous given a fixed set of selection pressures. Indeed, the constraint of an overlap prevents the exploration of large regions of sequence space and suboptimal alleles, and thus limits the extent of adaptation (Stiffler et al., 2015). In the case of Tat and Rev, the need to preserve their essential regulatory functions may simply outweigh any loss of adaptation (Maman et al., 2011). Such a tradeoff may be detrimental for viral proteins that evolve rapidly to antagonize host defenses. Furthermore, selective advantages can emerge from shared organizations, as has been suggested for the co-evolution of tRNA synthetases encoded on opposite strands (Rodin and Ohno, 1995).
Finally, it is interesting to consider that adding positive selection to an overlapped region via a targeted therapeutic could prove challenging for viral escape. Studies of cytotoxic T cells in rhesus monkeys have demonstrated CTL targeting of a nonfunctional portion of SIV tat in the vpr/tat overlap, and viral escape from these pressures is limited to entirely predictable synonymous mutations (Hughes et al., 2001). Combinations of epitope targeting in overlapped regions could create a viral “Achilles’ heel,” in which combinations of selection pressures from multiple frames drives the virus to unescapable fitness minima. Given that our data suggests that the virus normally evolves to avoid such situations, the design of these kinds of therapeutic interventions will require systematic approaches such as the work presented here to reveal HIV's true mutational weak spots.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
- EXPERIMENTAL MODEL AND SUBJECT DETAILS
- ○ Cell Lines
- METHOD DETAILS
- ○ Patient Analysis
- ○ Tat and Rev Reporter assays
- ○ Creation of Uncoupled Viruses
- ○ Creation of Proviral Libraries
- ○ Virus Generation and Spread
- ○ Viral Competition and Selection
- ○ Amplicon Generation
- ○ Data Processing
- ○ Approximation of Overlap Effect on Viral Fitness
- ○ Experimental Replication
- QUANTIFICATION AND STATISTICAL ANALYSIS
- ○ Entropy Comparisons between Overlapped and Non-Overlapped Regions in Patient Data
- ○ Alanine Scanning
- ○ Estimations of Experimental Error
- ○ Estimating Population Growth
- ○ Neutral Simulations
- ○ Simulations with Selection
- ○ Point Estimate of Selection Coefficient
- ○ Calculations of Overlap Restriction
DATA AND SOFTWARE AVAILABILITY
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| Raw and processed deep mutational scanning data | This paper | ArrayExpress: E-MTAB-5154 |
| Patient Sequence Alignments | HIV LANL (https://www.hiv.lanl.gov/content/index) | 2014 alignments |
| Experimental Models: Cell Lines | ||
| Tat Reporter Cell Line | D'Orso and Frankel, 2010 | HeLa HIV-1 LTR |
| 293 Cell Line | American Type Cell Culture Collection | ATCC CRL-1573 |
| 293 FT Cell Line | ThermoFisher | R700-7 |
| Sup-T1 Cell Line | American Type Cell Culture Collection | ATCC CRL-1942 |
| Recombinant DNA | ||
| Tat Alanine Mutants | This paper | pcDNA4 Tat-Strep M1A, etc. |
| Rev Alanine Mutants | This paper | pcDNA4 NStrep-Rev M1A, etc. |
| Rev Reporter Plasmid | Smith et al., 1990 | pCMV gag-pol-RRE |
| Viruses for Rev deep mutational scanning (rev-in-nef) | This paper | pNL4-3 TxR M1X, etc |
| Viruses for Tat deep mutational scanning (tat-in-nef) | This paper | pNL4-3 xRT M1X, etc |
| Gene and Primer Sequences are listed in Table S2 | N/A | |
| Software and Algorithms | ||
| MiSeq Reporter | Illumina, Inc. | MiSeq Reporter v2.6 |
| Population Genetics Modeling | This paper | allele_frequency_trajectory_sim |
| Other | ||
| Code Resource Website for this work | This paper | https://github.com/nbstrauli/allele_frequency_trajectory_sim |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to Lead Contact Alan Frankel (frankel@cgl.ucsf.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines
The HeLa HIV-1 LTR FFL line (D'Orso and Frankel, 2010) was used for the Tat alanine scan. Cells were cultured under standard conditions (DMEM supplemented w/ 10% FBS, 1% Penn-Strep). Reporter assays were carried out in a 48-well format described below. 293 (ATCC CRL-1573) lines were used for the Rev reporter assay (described below) in standard culture conditions (DMEM supplemented w/ 10% FBS, 1% Penn-Strep) in a 24-well format. 293FT cells were used to generate virus (described below) under standard culture conditions (DMEM supplemented w/ 10% FBS, 1% Penn-Strep). SupT1 (ATCC CRL-1942) human T cell lines were used for HIV-1 spreading and competition assays in standard culturing (RPMI-1640 supplemented w/ 10% FBS, 1% Penn-Strep).
METHOD DETAILS
Patient Analysis
Curated web alignments of protein sequences were downloaded from the Los Alamos HIV-1 Sequence using the protein alignments from 2014 for each HIV-1 protein. Each region of each protein was then divided into areas of one, two, or three coding overlaps and entropy statistics generated for each region and gene. Only positions present in the HXB2 reference sequence were considered in this analysis.
Tat and Rev Reporter assays
All 202 alanine mutants were created in independent site directed mutagenesis reactions in mammalian expression vectors. Reference alanines were not mutated. For the Tat reporter assays, 1ng of plasmid encoding wild-type HXB2 or mutant Tat proteins was cotransfected with 1ng CMV-Renilla control into a previously described HeLa cell line harboring an integrated copy of firefly luciferase under control of the HIV-1 LTR (D'Orso and Frankel, 2010). HeLa cells were plated and transfected in a 48 well plate with PolyJet (SignaGen) lipofection reagent for 48 hr. After 48 hr, cells were lysed with 1x Promega passive lysis buffer and firefly and Renilla luciferase values were measured using a luminometer. Tat-dependent firefly luciferase values were normalized to Renilla luciferase values. The assay was performed in biological quintuplet. For the Rev reporter assays, 250 ng of pCMVgagpolRRE was cotransfected with 25 ng of a pcDNA4 NL4-3 Nstrep-Rev mutant into 293 cells in a 24 well format (Smith et al., 1990) using PolyJet (SignaGen) lipofection reagent. Cells were then lysed and intracellular p24 levels measured by ELISA. The assay was performed in biological triplicate. For all mutants, activity was normalized to the unmutated reference activity.
Creation of Uncoupled Viruses
To create viruses with tat and rev in the nef locus, we introduced mutations into the start codon of the nef locus of pNL4-3 as well as sets of mutations into either Tat or Rev that ablated the start codon and introduced two downstream stops. The ablated frame was then reintroduced into nef. To create tat-in-nef and rev-in-nef viruses we introduced Sac II/Xba I sites upstream of the nef coding region and ablated the nef start codon. We then mutated either the endogenous start codon for tat and rev and introduced two downstream stops to prevent reversion. The ablated gene was then reintroduced into the engineered Sac II/Xba I sites. The reintroduced gene was synonymously substituted throughout the gene body to create codon-swapped versions (tat-cs and rev-cs, Table S2) in order to prevent recombination during viral replication.
Creation of Proviral Libraries
202 pools of 33 nt primers were synthesized containing NNN in the middle of the primer (see Table S2 for reference sequences). These primers were used in overlap extension PCRs then cloned into the nef locus of the appropriate knockout molecular clone using Gibson Assembly Master Mix (NEB) after digesting the backbone with Sac II and Xba I. A minimum of 1000 colonies was used to generate a single proviral library representing randomization of a single residue. The resulting colonies were scraped together and spin-column (EconoSpin) plasmid DNA purification was performed. Each randomized proviral library (202 total) was deep sequenced to ensure high codon diversity.
Virus Generation and Spread
Each proviral plasmid library was raised separately and complemented with reference rev and tat in trans to generate virus in 293T cells. Briefly, 60,000 293FT cells were transiently transfected with 200 ng of provial plasmid, and 2 ng each of pcDNA mammalian expression vectors containing tat or rev using PolyJet (SignaGen) lipofection reagent at a 1:5 ratio of the DNA:Polyjet. Virus-laden supernatant was collected 48 hr following transfection, cleared of cells via low speed centrifugation at 500 × g for 5 min, and stored at −80°C. Virus titers were determined by p24 ELISA.
Viral Competition and Selection
Selection assays were initiated via centrifugal inoculation of 106 SupT1 T cells with 5ng p24 and 8 ug/mL polybrene for 2 hr at 1200 × g at 32°C in 96-well microtiter plates. Following inoculation, cells were washed twice with PBS to remove input virus and 250 uL media (RPMI-HEPES + 10% fetal calf serum) replaced per well. The infections were then monitored by immunofluorescence assay for cellular HIV antigen synthesis and by supernatant p24 ELISA to quantify progeny virion release. At peak infection for reference or defective viral pools 150 uL of virus-laden supernatant was collected, cleared of cells, and the virion-associated RNA content extracted via ZR-96 Viral RNA extraction kits (Zymo) and eluted in 15 ul water. Once all RNA samples were collected, cDNA was generated from 5uL of extracted RNA using the iScript Advanced (BioRad) cDNA synthesis. Selection experiments for each residue (202 total + reference wells) were performed in biological duplicate.
Amplicon Generation
Amplicon specific primers were used to extract 175-nt fragments containing the randomized codon and add Illumina adaptor sequence (3 total primer sets for Tat and 5 for Rev; Table S2). Each amplicon specific primer consisted of a pool of 1-4 random nucleotides appended immediately 5′ of the gene-specific region in order to increase basecall diversity during the NGS runs. A second step PCR added barcode sequences (courtesy of J. DeRisi) and the remainder of the adaptor. DNA from each selection experiment was quantified using qPCR (Kapa Biosciences) and then pooled in equal amounts and gel extracted to create the NGS library. Library quantification was then performed using qPCR and sequenced using PE150s on a MiSeq (Illumina). Samples which produced low read counts, were either re-pooled in a smaller mixture or repeated starting at the initial infection stage.
Data Processing
Samples were de-multiplexed using MiSeqReporter to produce fastq files, each representing a single competition experiment. The resulting paired-end fastq files were then aligned to one another and the reference codon-swapped sequence (Table S2) and the frequencies of every codon were calculated. The randomized position was then extracted and a post-and pre-selection allele frequencies calculated. Synonymous codons were pooled as correlations between amino acids across biological replicates was much stronger than individual correlations of codons (data not shown).
Approximation of Overlap Effect on Viral Fitness
In order to calculate the manner in which one gene restricts the other we took the endogenous NL4-3 sequence and fixed one allele in one gene. For instance, to examine the effect of Tat Y47 on Rev Position 1, we substituted both Y codons (TAT, TAC) into the rev sequence (ATg (M) or ACg (T)) and estimated the resulting fitnesses of the viral genotypes (i.e., Tat Y47, Rev M1) as the smaller of the two alleles (i.e., either Tat Y47 or Rev M1). The comparable single-frame virus has 21 viral genotypes with Tat Y47 (i.e., Rev A1.Rev Y1) and the fitness of these genotypes can be approximated in a similar manner (Figure S7). The resulting single-frame landscape of these 21 viruses can be compared to the restricted landscape of the two overlapped genotypes. This procedure was applied for every possible amino acid at every position in the overlap, first holding Tat fixed and calculating the resulting Rev fitness (and subsequently vice versa). As holding most positions fixed influences a dipeptide in the alternative frame, we added the selection coefficients of the resulting alleles in the alternative frame. The resulting single frame and overlapped distributions were then plotted in R using ggplot2′s density function.
Experimental Replication
Alanine scanning was performed in biological quintuplet (Tat) or triplicate (Rev). For the deep mutational scanning dataset, each competition experiment (86 for Tat, 116 for Rev) was performed in biological duplicate.
QUANTIFICATION AND STATISTICAL ANALYSIS
Entropy Comparisons between Overlapped and Non-Overlapped Regions in Patient Data
To test for statistically significant differences between regions of overlap (Figure S2B), we used two approaches: 1) We used a Wilcoxon test to compare the empirical distributions of entropy values within a bin to the distribution of entropy values in another bin, and 2) we used a permutation-based approach where we compared the mean entropy value from the positions that correspond to a given bin class to randomly sampled similarly sized, contiguous segments of the HIV exome. We then determined if the observed mean entropy value was significantly outside of the distribution of entropy values from these randomly sampled segments. The latter method is best illustrated by example: Consider two regions in the exome that code for 3 genes, where each of these hypothetical regions are 5 amino acids long. First, we pool the entropy values that correspond to these regions, and calculate their mean entropy level. We then randomly sample (with replacement) two contiguous segments of the HIV exome that are each 5 amino acids long, and calculate the mean entropy level for these sampled segments. We then repeat this process 10,000 times to arrive at an empirical null distribution of mean entropy values. We then compare our observed mean entropy value for regions of 3 coding reading frames to this empirical null distribution to see if it is significantly outside of what one would expect from chance.
Alanine Scanning
Tat reporter assay values are the mean of five independent transfections with error bars representing SD of these replicates. Rev reporter assays represent the mean of three independent transfections (error bars are the SD).
Estimations of Experimental Error
Because wild-type (non-randomized) positions were also sequenced in our data, we were able to use this information to estimate the overall error rate for each amino acid type. For wild-type positions, the expectation is that all sequenced reads will have the reference allele, and any reads that deviate from this expectation are the result of some type of error during the course of our protocol. These errors include random mutations during the experiment, PCR errors, or sequencing errors. We estimate the combined ‘experimental error’ for each amino acid type by using the non-randomized positions to count the number of instances that we observed a mutation ‘away’ from a given amino acid, and the number of times we observe a mutation ‘to’ that amino acid. We then normalized these counts by the total number of reads observed for the amino acid to determine mutation rates. Thus, each amino acid has an estimated ‘away’ mutation rate and an estimated ‘to’ mutation rate associated with it. We represent the rate at which amino acid Y mutates to any other amino acid as RY,·, and the rate at which any other amino acid mutates to Y as R·,Y.
Estimating Population Growth
We used data on the growth of a wild-type population of HIV under the same experimental protocol as in our evolution experiments, to estimate the overall population growth in each experiment. Specifically, we measured the p24 concentration (in pg/uL) for several time points during the course of the experiment in triplicate. We set the overall population size of HIV to be equal to the mean [p24] value at each time-point. This is a good estimate of the HIV population size, as we used this value to empirically determine our MOI and thus this is a reasonable estimate of the infectious population. We then fit a logistic curve to these observed values to get a population growth function over the time-course of the experiments. The result of this fit produced the following equation:
where Nt is the population size at generation t.
Neutral Simulations
We sought to simulate, in silico, our evolution experiments in order estimate the range that a neutral allele's frequency could feasibly change during the experiments. This simulated range served as our null distribution, and we generated a unique null distribution for each allele in our observed data. The neutral simulations had five parameters: the overall population growth function, the number of generations, the starting allele frequency, the ending read depth for the experiment, and the amino acid identity of the allele. Each of these will be explained below.
A set of 100,000 neutral simulations were run for each allele in the data, and each simulation was run as follows. The starting neutral allele frequency was set to begin at the same starting frequency of the observed allele. We then ran a Wright-Fisher simulation of neutral drift over 6 generations (2007). We model the neutral allele in question as allele A, with frequency p and collapsed all the other alleles in the population to be allele a, with frequency q = 1 – p. If the population size and frequency for allele A at generation t is Nt and pt, respectively, then the count of A is Pt = Ntpt, and the count of a is Qt = Nt – Pt. The probability of an allele count in the next generation (Pt + 1) follows the binomial distribution,
Thus, to get a random value for Pt + 1 in the simulation we simply randomly sample from the above distribution. Once the Wright-Fisher simulation is complete, we again use the binomial distribution to simulate sub-sampling of the population. We do this because the true population size of HIV at the end of the experiment is quite large, and when we sequence this population, we are only observing a subset. The size of this subsample is equal to the number of reads that were sequenced at the end of a given evolution experiment (one per randomized position). If N6 is the final population size, let the subsample size be . Similarly, if P6 is the count of A at the final generation, then is the count of A resulting from subsampling. We again use the binomial to model the probability of ,
and randomly sample from this distribution to simulate a value for . In order to simulate random additions to and subtractions from allele A that occur due mutations from the combined experimental error, we again use a binomial sampling approach. If the amino acid identity of A is Y then the rate at which Y is mutated to anything else is RY·, and the rate at which anything else mutates to Y is R·,Y. Let the number of units that is added to due to experimental error be represented as X·,Y, and the number of units subtracted be XY,·. The probability of these two values are again found using the binomial,
We randomly sample from these two distributions to get discrete values for X·,Y and XY,·. Finally, Let the count of allele A, at generation 6, after subsampling, and after introducing random experimental error be . is then given by,
is the final allele count that we use in the simulation. This process is then repeated 100,000 times to arrive at a simulated distribution for . This distribution represents the range of ending allele frequencies that one could reasonably expect for a neutral allele, if this allele had the same starting frequency, read depth, amino acid identity as a given observed allele in our data.
Simulations with Selection
We ran the same simulations of our evolution experiments as described above, but included a selection parameter in order to test the accuracy of our fitness estimates of observed alleles. The only component of the framework described above that changed in this case was the Wright-Fisher simulations. Here, the expected frequency of allele A with a fitness of s at generation t + 1 is given by,
and the probability of Pt + 1 with selection is given by,
Point Estimate of Selection Coefficient
We used population genetics theory to estimate the fitness, or selection coefficient (s) of an allele when given the starting frequency, ending frequency, and number of generations in-between (Felsenstein, 2016). The equation for s is given by,
Where i indexes the starting generation and t is the number of generations that have elapsed.
Our simulation framework with selection (described above) allowed us to test the accuracy of our fitness estimates. Because selection can be incorporated into our simulations, we can simulate an allele with a pre-specified ‘true’ fitness value. We can then compare this value with our estimate of fitness that is based only upon the results of our stochastic simulation (agnostic to the ‘true’ fitness value). To ensure that the parameters for these simulations with selection were relevant to our study, we gathered the parameters that pertain to each of the observed alleles in our data. These parameters are: starting allele frequency (to give the starting point of the simulation), the read depth at the end of the evolution experiment (to give the subsample size), and the amino acid identity of the allele (to give the experimental error rate). We then ran a single simulation pertaining to each of these alleles’ parameter sets, and importantly, included a fitness value with each of these simulations. To select a fitness value we randomly sampled from a uniform distribution between –2 and 2. This gives the true fitness. We then find the estimated fitness by incorporating the results of the stochastic simulation into the equation for the selection coefficient (given above). We found that the estimated fitness values track very well with the true fitness (Figure S5C), and thus our point estimate of fitness is accurate. However, we found that if the starting allele frequency is low (which by design is almost always the case for our data), one does not have the ability to infer extreme negative fitness values. This is because a moderately negatively selected allele will fall to a frequency of 0 just the same as an extremely negatively selected allele, if the starting frequency is low. This conceptual limit to one's ability to infer negative selection with low starting frequency is well illustrated by the plateauing of points on the left-hand side of Figure S5C.
Calculations of Overlap Restriction
In order to calculate the manner in which one gene restricts the other we took the endogenous NL4-3 sequence and fixed one allele in one gene. For instance, to examine the effect of Tat Y47 on Rev Position 1, we substituted both Y codons (TAT, TAC) into the rev sequence (ATg (M) or ACg (T)) and estimated the resulting fitnesses of the viral genotypes (i.e., Tat Y47, Rev M1) as the smaller of the two alleles (i.e., either Tat Y47 or Rev M1). The comparable single-frame virus has 21 viral genotypes with Tat Y47 (i.e., Rev A1.Rev Y1) and the fitness of these genotypes can be approximated in a similar manner. The resulting single-frame landscape of these 21 viruses can be compared to the restricted landscape of the two overlapped genotypes. This procedure was applied for every possible amino acid at every position in the overlap, first holding Tat fixed and calculating the resulting Rev fitness (and subsequently vice versa). As holding most positions fixed influences a dipeptide in the alternative frame, we added the selection coefficients of the resulting alleles in the alternative frame. The resulting single frame and overlapped distributions were then plotted in R using ggplot2′s density function.
Supplementary Material
Highlights.
Overlapped regions in HIV-1 are not more conserved than non-overlapped regions
Tat and Rev segregate motifs: functional regions overlap non-functional regions
When not overlapped, non-functional regions vary even more
Tat constrains Rev, increasing the fitness of the viral genotypic landscape
ACKNOWLEDGMENTS
We thank J. DeRisi and M. Stenglein for barcode sequences and advice for NGS experimental design, and R. Andino and A. Acevedo for use of equipment and advice for performing selection experiments. We also thank P. Babbitt, J. Gross, J. Fraser, R. Andino, M. Daugherty, L. Gitlin and members of the A.D.F. and Andino labs for helpful conversations and review of the manuscript. J.D.F. was supported in part by NIH training grant T32GM007175 and an Amgen Research Excellence Fellowship. This work was supported by NIH grant P50GM088250 to A.D.F.
Footnotes
AUTHOR CONTRIBUTIONS
J.D.F. and A.D.F. designed the experiments with input from T.B.F., D.C.C., and R.L.N. J.D.F. and A.D.F. wrote the manuscript with input from all authors. J.D.F. and N.B.S. analyzed the patient data. J.D.F. and T.B.F. performed the alanine scans. J.D.F., C.S., and D.C.C. performed viral selection and sequencing experiments. N.B.S. and R.D.H. developed the population genetics model and aided J.D.F. in analysis of the selection dataset.
DATA AND SOFTWARE AVAILABILITY
The accession number for the deep mutational scanning data reported in this paper is ArrayExpress: E-MTAB-5154.
Population Genetics Modeling and other code resources are available at https://github.com/nbstrauli/allele_frequency_trajectory_sim.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2016.11.031.
REFERENCES
- Bank C, Hietpas RT, Jensen JD, Bolon DN. A systematic survey of an intragenic epistatic landscape. Mol. Biol. Evol. 2015;32:229–238. doi: 10.1093/molbev/msu301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell BG, Air GM, Hutchison CA., 3rd Overlapping genes in bacteriophage phiX174. Nature. 1976;264:34–41. doi: 10.1038/264034a0. [DOI] [PubMed] [Google Scholar]
- Belshaw R, Pybus OG, Rambaut A. The evolution of genome compression and genomic novelty in RNA viruses. Genome Res. 2007;17:1496–1504. doi: 10.1101/gr.6305707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher JI, Bolon DNA, Tawfik DS. Quantifying and understanding the fitness effects of protein mutations: Laboratory versus nature. Protein Sci. 2016;25:1219–1226. doi: 10.1002/pro.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes N, Linial M. Gene overlapping and size constraints in the viral world. Biol. Direct. 2016;11:26. doi: 10.1186/s13062-016-0128-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter JJ, Daugherty MD, Qi X, Bheda-Malge A, Wipf GC, Robinson K, Roman A, Malik HS, Galloway DA. Identification of an overprinting gene in Merkel cell polyomavirus provides evolutionary insight into the birth of viral genes. Proc. Natl. Acad. Sci. USA. 2013;110:12744–12749. doi: 10.1073/pnas.1303526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan LY, Kosuri S, Endy D. Refactoring bacteriophage T7. Mol. Syst. Biol. 2005;1:2005.0018. doi: 10.1038/msb4100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirico N, Vianelli A, Belshaw R. Why genes overlap in viruses. Proc. Biol. Sci. 2010;277:3809–3817. doi: 10.1098/rspb.2010.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Orso I, Frankel AD. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat. Struct. Mol. Biol. 2010;17:815–821. doi: 10.1038/nsmb.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty MD, Liu B, Frankel AD. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat. Struct. Mol. Biol. 2010;17:1337–1342. doi: 10.1038/nsmb.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMattia MA, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, Steven AC, Grimes JM. Implications of the HIV-1 Rev dimer structure at 3.2 A resolution for multimeric binding to the Rev response element. Proc. Natl. Acad. Sci. USA. 2010;107:5810–5814. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doud MB, Bloom JD. Accurate measurement of the effects of all amino-acid mutations on influenza hemagglutinin. Viruses. 2016;8:155. doi: 10.3390/v8060155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde NC, Child SJ, Eickbush MT, Kitzman JO, Rogers KS, Shendure J, Geballe AP, Malik HS. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell. 2012;150:831–841. doi: 10.1016/j.cell.2012.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder AF, Kryazhimskiy S, Plotkin JB. Identifying signatures of selection in genetic time series. Genetics. 2014;196:509–522. doi: 10.1534/genetics.113.158220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Theoretical Evolutionary Genetics. 2016 http://evolution.genetics.washington.edu/pgbook/pgbook.html.
- Firnberg E, Labonte JW, Gray JJ, Ostermeier M. A comprehensive, high-resolution map of a gene's fitness landscape. Mol. Biol. Evol. 2014;31:1581–1592. doi: 10.1093/molbev/msu081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley B, Leitner T, Apetrei C, Hahn B, Mizrachi I, Mullins J, Rambaut A, Wolinsky S, Korber B. HIV Sequence Compendium 2013. Theoretical Biology and Biophysics Group; Los Alamos National Laboratory, NM: 2013. [Google Scholar]
- Hein J, Støvlbaek J. A maximum-likelihood approach to analyzing nonoverlapping and overlapping reading frames. J. Mol. Evol. 1995;40:181–189. doi: 10.1007/BF00167112. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Westover K, da Silva J, O'Connor DH, Watkins DI. Simultaneous positive and purifying selection on overlapping reading frames of the tat and vpr genes of simian immunodeficiency virus. J. Virol. 2001;75:7966–7972. doi: 10.1128/JVI.75.17.7966-7972.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A, Tristem M, Pybus OG, Gifford RJ. Discovery and analysis of the first endogenous lentivirus. Proc. Natl. Acad. Sci. USA. 2007;104:6261–6265. doi: 10.1073/pnas.0700471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y, Neeley S, Adachi K, Nakai H. An experimental and computational evolution-based method to study a mode of co-evolution of overlapping open reading frames in the AAV2 viral genome. PLoS ONE. 2013;8:e66211. doi: 10.1371/journal.pone.0066211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keckesova Z, Ylinen LMJ, Towers GJ, Gifford RJ, Katzourakis A. Identification of a RELIK orthologue in the European hare (Lepus europaeus) reveals a minimum age of 12 million years for the lagomorph lentiviruses. Virology. 2009;384:7–11. doi: 10.1016/j.virol.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keese PK, Gibbs A. Origins of genes: “big bang” or continuous creation? Proc. Natl. Acad. Sci. USA. 1992;89:9489–9493. doi: 10.1073/pnas.89.20.9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs E, Tompa P, Liliom K, Kalmar L. Dual coding in alternative reading frames correlates with intrinsic protein disorder. Proc. Natl. Acad. Sci. USA. 2010;107:5429–5434. doi: 10.1073/pnas.0907841107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Keller B, Makalou N, Sutton RE. Systematic determination of the packaging limit of lentiviral vectors. Hum. Gene Ther. 2001;12:1893–1905. doi: 10.1089/104303401753153947. [DOI] [PubMed] [Google Scholar]
- Makalowska I, Lin C-F, Makalowski W. Overlapping genes in vertebrate genomes. Comput. Biol. Chem. 2005;29:1–12. doi: 10.1016/j.compbiolchem.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Maman Y, Blancher A, Benichou J, Yablonka A, Efroni S, Louzoun Y. Immune-induced evolutionary selection focused on a single reading frame in overlapping hepatitis B virus proteins. J. Virol. 2011;85:4558–4566. doi: 10.1128/JVI.02142-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin RN, Jr., Poelwijk FJ, Raman A, Gosal WS, Rangana-than R. The spatial architecture of protein function and adaptation. Nature. 2012;491:138–142. doi: 10.1038/nature11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Yasunaga T. Evolution of overlapping genes. Nature. 1978;272:532–535. doi: 10.1038/272532a0. [DOI] [PubMed] [Google Scholar]
- Neuveut C, Jeang KT. Recombinant human immunodeficiency virus type 1 genomes with tat unconstrained by overlapping reading frames reveal residues in Tat important for replication in tissue culture. J. Virol. 1996;70:5572–5581. doi: 10.1128/jvi.70.8.5572-5581.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngandu NK, Scheffler K, Moore P, Woodman Z, Martin D, Seoighe C. Extensive purifying selection acting on synonymous sites in HIV-1 Group M sequences. Virol. J. 2008;5:160. doi: 10.1186/1743-422X-5-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Geyer M, Zhou Q. The control of HIV transcription: keeping RNA polymerase II on track. Cell Host Microbe. 2011;10:426–435. doi: 10.1016/j.chom.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan K, Deem MW. Quantifying selection and diversity in viruses by entropy methods, with application to the haemagglutinin of H3N2 influenza. J. R. Soc. Interface. 2011;8:1644–1653. doi: 10.1098/rsif.2011.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A, Magiorkinis G, Karlin DG. Viral proteins originated de novo by overprinting can be identified by codon usage: application to the “gene nursery” of Deltaretroviruses. PLoS Comput. Biol. 2013;9:e1003162. doi: 10.1371/journal.pcbi.1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard VW, Malim MH. The HIV-1 Rev protein. Annu. Rev. Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- Rancurel C, Khosravi M, Dunker AK, Romero PR, Karlin D. Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J. Virol. 2009;83:10719–10736. doi: 10.1128/JVI.00595-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihn SJ, Wilson SJ, Loman NJ, Alim M, Bakker SE, Bhella D, Gifford RJ, Rixon FJ, Bieniasz PD. Extreme genetic fragility of the HIV-1 capsid. PLoS Pathog. 2013;9:e1003461. doi: 10.1371/journal.ppat.1003461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin SN, Ohno S. Two types of aminoacyl-tRNA synthetases could be originally encoded by complementary strands of the same nucleic acid. Orig. Life Evol. Biosph. 1995;25:565–589. doi: 10.1007/BF01582025. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Spiridonov AN, Sorokin AV, Wolf YI, Jordan IK, Tatusov RL, Koonin EV. Purifying and directional selection in overlapping prokaryotic genes. Trends Genet. 2002;18:228–232. doi: 10.1016/s0168-9525(02)02649-5. [DOI] [PubMed] [Google Scholar]
- Sabath N, Landan G, Graur D. A method for the simultaneous estimation of selection intensities in overlapping genes. PLoS ONE. 2008;3:e3996. doi: 10.1371/journal.pone.0003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath N, Wagner A, Karlin D. Evolution of viral proteins originated de novo by overprinting. Mol. Biol. Evol. 2012;29:3767–3780. doi: 10.1093/molbev/mss179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977;265:687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Smith AJ, Cho MI, Hammarskjöld ML, Rekosh D. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J. Virol. 1990;64:2743–2750. doi: 10.1128/jvi.64.6.2743-2750.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoeck J, Fellay J, Bartha I, Douek DC, Telenti A. Mapping of positive selection sites in the HIV-1 genome in the context of RNA and protein structural constraints. Retrovirology. 2011;8:87. doi: 10.1186/1742-4690-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiffler MA, Hekstra DR, Ranganathan R. Evolvability as a function of purifying selection in TEM-1 β-lactamase. Cell. 2015;160:882–892. doi: 10.1016/j.cell.2015.01.035. [DOI] [PubMed] [Google Scholar]
- Tahirov TH, Babayeva ND, Varzavand K, Cooper JJ, Sedore SC, Price DH. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature. 2010;465:747–751. doi: 10.1038/nature09131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan B, Bloom JD. The inherent mutational tolerance and antigenic evolvability of influenza hemagglutinin. eLife. 2014;3:e03300. doi: 10.7554/eLife.03300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touriol C, Bornes S, Bonnal S, Audigier S, Prats H, Prats A-C, Vagner S. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol. Cell. 2003;95:169–178. doi: 10.1016/s0248-4900(03)00033-9. [DOI] [PubMed] [Google Scholar]
- Wei X, Zhang J. A simple method for estimating the strength of natural selection on overlapping genes. Genome Biol. Evol. 2014;7:381–390. doi: 10.1093/gbe/evu294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaaijer HL, van Hemert FJ, Koppelman MH, Lukashov VV. Independent evolution of overlapping polymerase and surface protein genes of hepatitis B virus. J. Gen. Virol. 2007;88:2137–2143. doi: 10.1099/vir.0.82906-0. [DOI] [PubMed] [Google Scholar]
- Zhao L, Saelao P, Jones CD, Begun DJ. Origin and spread of de novo genes in Drosophila melanogaster populations. Science. 2014;343:769–772. doi: 10.1126/science.1248286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.