Abstract
There are numerous heritable diseases associated with mutations in the LMNA gene. Most of these laminopathic diseases, including several muscular dystrophies, are autosomal dominant and have tissue-specific phenotypes. Our previous studies have shown that the globally expressed Emery-Dreifuss muscular dystrophy (EDMD)-linked lamin mutation, L535P, disrupts nuclear mechanical response specifically in muscle nuclei of C. elegans leading to atrophy of the body muscle cells and to reduced motility. Here we used RNA sequencing to analyze the global changes in gene expression caused by the L535P EDMD lamin mutation in order to gain better understanding of disease mechanisms and the correlation between transcription and phenotype. Our results show changes in key genes and biological pathways that can help explain the muscle specific phenotypes. In addition, the differential gene expression between wild-type and L535P mutant animals suggests that the pharynx function in the L535P mutant animals is affected by this lamin mutation. Moreover, these transcriptional changes were then correlated with reduced pharynx activity and abnormal pharynx muscle structure. Understanding disease mechanisms will potentially lead to new therapeutic approaches toward curing EDMD.
Keywords: laminopathies, motility, muscular dystrophy, nuclear lamina, pharynx, transcription
Introduction
Intermediate filaments (IFs) are a large group of proteins that play a key role in regulating signal transduction and providing cells with tensile strength.1–3 Most IFs are cytoplasmic and are differentially expressed in tissues. Lamins are similar in their structure to the cytoplasmic IFs; they contain an N-terminal head domain, a central α helical rod domain and a carboxy tail domain. Lamins are nuclear intermediate filaments, which are expressed in all cells. The tail domain of lamins contains an Ig fold, an unstructured region, a nuclear localization signal (NLS) and a CaaX motif.4,5 In mammals, lamins are encoded by 3 genes, which give rise to 4 major isoforms. Lamins A and C (A type lamins) are encoded by the LMNA gene through alternative splicing, and are mostly expressed in differentiated cells. Lamin B1 and lamin B2 (B type lamins) are encoded by LMNB1 and LMNB2 respectively and are expressed in all mammalian cells.6 Lamins, together with an assortment of lamin associated proteins, are the key components of the nuclear lamina, which lies directly underneath the inner nuclear membrane (INM). Lamins interacts with many INM trans-membrane proteins, as well as with chromatin, and contribute to transcription regulation and signal transduction.7,8 Coupling of the nucleus to the cytoskeleton is mediated by the nuclear lamina. This allows external forces to be transmitted to the nucleus and proper response to be initiated.9–11 The nuclear lamina is also required for cell cycle progression, DNA replication, providing structural support to the nucleus and is required for nuclei to maintain their shape.12–14
Mutations in the lamin genes lead to over 16 different diseases, collectively called laminopathies. To date, over 500 distinct missense mutations, which are spread throughout the LMNA gene, were identified. The resulting diseases are mostly autosomal dominant, appear postnatally and vary significantly in phenotypes. LMNA-based diseases can be roughly divided into 4 disease groups: Striated muscle diseases, peripheral neuropathies and metabolic diseases all affecting specific tissues, and accelerated aging diseases, which affect multiple tissues. The majority of lamin mutations cause striated muscle diseases, which include Emery-Dreifuss muscular dystrophy (EDMD), limb girdle muscular dystrophy type 1B (LGMD1B), LMNA-related congenital muscular dystrophy (L-CMD) and dilated cardiomyopathy with conduction disease (DCM-CD). These diseases range in severity and involve different muscle cells. All LMNA based muscle diseases have cardiac muscle defects.15–18
The muscle-specific degeneration observed in the case of muscle diseases can be explained by the mechanical hypothesis; certain lamin mutations cause nuclei to lose their ability to resist mechanical strain, which in turn leads to nuclear rupture, cell death and tissue deterioration. Mechanically stressed tissues are particularly susceptible as they are constantly under strain, and therefore damage is initially observed in these cells.19 Indeed, dominant expression of the L535P EDMD-linked lamin mutation in C. elegans caused the muscle nuclei to become more rigid, altering their mechanical response to strain. Restoring proper muscle nuclear response rescued disease phenotypes in these animals.20 However, this hypothesis does not fully explain how a globally expressed mutation can have such a tissue specific effect, as not all muscle diseases caused by lamin mutations affect the exact same muscle sub-types. Furthermore, expressing the Y59C EDMD linked mutation did not affect muscle nuclear mechanical response, but rather the transcription of muscle specific developmentally expressed genes.20,21 Also, the skin, which is under constant mechanical tension, has no apparent phenotypes in this group of diseases. The many nuclear roles attributed to lamins are most likely mediated through their interactions with a variety of inner nuclear membrane proteins, which are differentially expressed within tissues.22,23 Several lamin mutations can disrupt the interaction with a specific nuclear trans-membrane protein, resulting in a tissue specific phenotype. The prevailing opinion is that more than one mechanism underlies disease pathology.
Here, we take advantage of the previously established laminopathy model organism; C. elegans24 to study early global changes in transcription resulting from the expression of an EDMD disease linked lamin mutation. C. elegans has one lamin gene (lmn-1), with functions and properties of both A and B type lamins.25 Dominant expression of lamin disease-linked mutations causes phenotypes in the worm, which mimic the corresponding phenotypes in humans.24,26 In order to understand how genetic changes contribute to disease phenotypes, the well-characterized L535P EDMD-linked lamin mutation in C. elegans was chosen for in-depth analyses. Expression of the L535P EDMD mutation significantly down regulated genes involved in body wall- and pharyngeal-muscle organization and function. This down regulation correlates with the muscle sub-types affected by EDMD both in worms and in human patients.
Results
Bioinformatic analysis of transcriptional changes resulting from the expression of the L535P EDMD lamin mutation in living C. elegans
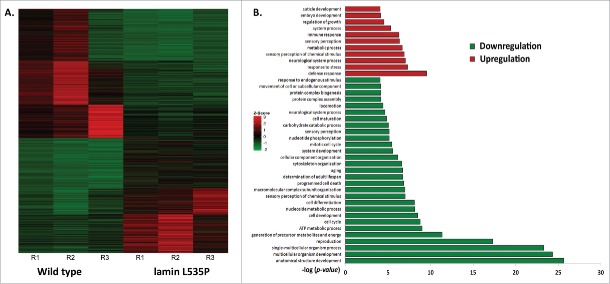
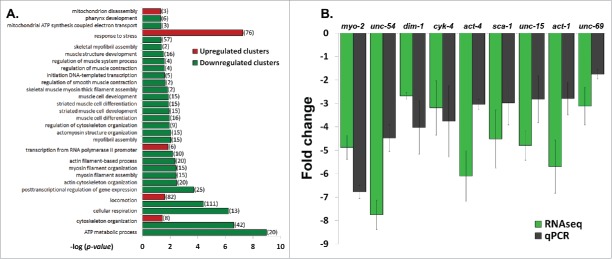
Previous studies demonstrated that C. elegans expressing the L535P EDMD-linked mutation at levels of 15-30% of the endogenous wild-type lamin show specific muscle- and motility-phenotypes, similar to expressing the homologous lamins A/C mutation in human cells.27 In order to gain better understanding of these EDMD disease-like phenotypes and to study changes in nuclear mechanical response, we analyzed the early transcription changes resulting from the dominant expression of the L535P EDMD mutation in C. elegans. mRNA of wild-type (N2)-, and L535P-animals at the fourth larval stage were extracted from roughly 3 hundred animals and these mRNA samples were used to prepare cDNA libraries that were subjected to deep sequencing analysis. These transcriptome experiments were independently repeated 3 times. In order to compare global expression patterns between the different samples, gene counts and FPKM (Fragments Per Kilobase of transcript per Million mapped reads) distributions were used. Differential expression was calculated using a count threshold of at least 15 for statistical significance testing (elaborated in the material and methods section). Significant differentially expressed genes were defined as ones with at least 1 FPKM level of expression in at least one of the conditions, a log2 fold change of more than 1 in each direction and a p-value less than 0.05. Based on these parameters 960 genes were significantly downregulated and 838 genes were significantly up regulated in L535P EDMD animals as compared to wild-type animals (Fig. 1A). In order to identify key biological processes these genes are involved in, we performed cluster and gene ontology (GO) analysis. The most significant upregulated clusters contained genes involved in the defense and stress response mechanisms. Clusters that were significantly down regulated pointed to biological processes involved in the proper anatomical development of the organism, reproduction and cellular energy production (Fig. 1B). Many of the altered clusters demonstrated both up and down regulation of their genetic components. For example, the cluster of animal locomotion contained 111 genes that were downregulated and 82 genes that were up regulated. The cluster involving C. elegans stress response included 57 genes that were down regulated and 76 genes that were upregulated. Other clusters such as cytoskeleton organization had 42 downregulated genes and 8 up regulated genes, transcription showed 35 down regulated genes and 6 upregulated genes and the cluster involving mitochondria function had 31 downregulated genes and 3 up regulated genes. Interestingly, genes involved in pharyngeal muscle development (6) as well as muscle structure (16) and function (4) were mostly down regulated as a result of the L535P EDMD lamin mutation, which correlate with the animal muscle phenotypes (Fig. 2A and Table 1). We concluded that the regulation of numerous genes involved in muscle organization and motility, cytoskeleton organization and stress response by lamin is abnormal by the L535P EDMD-linked mutation in a dominant way.
Figure 1.
Genetic changes caused by the L535P EDMD lamin mutation. (A) Heat map representing genes that were significantly (p-value ≤ 0.05) up regulated (red, n = 838) or down regulated (green, n = 960) in EDMD L535P animals as opposed to wild-type animals. R = repetition of the experiment (B) (–log) of the p-value score of the most significant upregulated (red) and downregulated (green) gene clusters following Gene Ontology (GO) Enrichment Analysis (http://geneontology.org/page/go-enrichment-analysis).
Figure 2.
Down regulation of muscle and cytoskeleton genes in L535P animals. (A) (–log) of the p-value score of up regulated (red) and down regulated (green) genes following Gene Ontology (GO) Enrichment Analysis. Displayed are gene clusters involved in biological processes of stress response, transcription, animal locomotion, cytoskeleton organization, mitochondria function and muscle related processes. (B) Fold change comparison between RNA sequencing (RNAseq) experiments and quantitative PCR (qPCR) experiments. Error bars represent SEM. P values of the fold change between qPCR and RNAseq are: 0.12, 0.06, 0.44, 0.8, 0.2, 0.43, 0.19, 0.26 and 0.32, respectively.
Table 1.
Representative clusters that were altered as a result of the L535P lamin mutation.
| Upregulated genes | Downregulated genes | |
|---|---|---|
| Locomotion: | bli-2, nid-1, mig-2, acn-1, nmy-1, mrps-30, R02D5.3, bli-5, ptr-23, D2045.9, unc-68, mlt-9, dig-1, Y11D7A.9, Y37D8A.16, noah-1, mlt-8, T19B10.2, epi-1, blmp-1, Y47D3B.1, chs-2, tat-5, rem-8, nkb-1, prp-17, efn-2, T04F3.1, F10D11.6, E03H4.8, nhr-49, tag-297, dpy-7, ced-12, hog-1, sec-8, egal-1, grd-2, itr-1, B0511.12, nas-36, Y47G6A.15, ptr-4, lpr-4, bus-8, mboa-1, dpy-18, rol-6, wrt-6, wts-1, trr-1, sdz-24, aex-1, Y4C6B.2, cyp-42a1, C56E6.9, dpy-2, cul-4, pes-8, glf-1, osm-7, cuti-1, osr-1, ins-33, rnf-121, wrt-1, wrt-10, hmgr-1, C26B9.3, ZK688.2, sma-1, lrp-1, asns-1, lpr-5, bec-1, mus-101, pyr-1, unc-13, C32D5.12, qua-1, wrt-4, dpy-10 | kin-10, col-103, paa-1, emb-9, egl-30, col-98, hsp-1, attf-3, tbb-2, tba-2, dyn-1, K04H4.2, cct-7, col-166, Y69H2.3, sid-3, rsp-4, act-1, gsp-3, act-3, gsa-1, gbf-1, gsp-4, clec-1, col-139, ssq-2, F44E5.1, cmd-1, vab-10, xrn-2, F53F8.3, W03G9.3, act-4, let-49, set-16, lin-17, bir-1, unc-15, let-2, ahcy-1, lit-1, K09E4.1, alh-7, col-122, act-2, col-168, col-94, pccb-1, pfn-1, his-40, rab-7, ostd-1, tnt-2, col-170, cdc-37, rack-1, col-93, mig-6, grd-3, lin-66, gsk-3, mesp-1, pbs-1, alg-1, zyg-8, cav-1, osm-11, mcm-7, his-74, C38D4.1, cyk-4, pod-2, cri-3, atp-2, sams-1, spo-2, Y46G5A.4, C04B4.2, unc-69, fib-1, pdf-2, mec-12, mdt-17, col-92, htz-1, eft-3, tbb-1, sap-49, unc-50, K01G5.3, smd-1, max-2, dpy-4, snr-5, xpo-2, aldo-2, dcap-1, grd-13, mig-14, pdi-2, vha-13, stf-4, unc-51, dpy-30, F08D12.1, unc-54, rnp-5, dlst-1, col-167, T12A2.2, hsp-3 |
| Stress response: | abu-13, K09D9.1, mig-2, pek-1, M60.2, mrps-30, pme-4, atg-13, cyp-37b1, mtk-1, gei-17, drh-3, mom-4, F16A11.2, dct-5, F52E1.5, C49G7.10, F53C11.1, gst-6, ZK909.3, F55G11.8, epi-1, dct-17, irg-1, ikb-1, fmo-2, T19D12.4, clec-258, comt-2, efl-3, nhr-49, Y41C4A.8, gst-24, dod-17, C04F12.1, F53A9.8, F53B1.4, clec-67, col-54, cup-2, cyp35a5, F10G8.2, F35E12.9, trr-1, cdr-4, clec-66, C08E8.4, hsp-70, flr-4, glf-1, lys-3, osm-7, gad-3, abu-12, rnf-121, dod-22, spp-2, rtel-1, F56D2.5, H20E11.2, smc-5, clec-86, cnc-4, H20E11.3, B0024.4, bec-1, cnc-2, Y41C4A.11, F49E10.4, mus-101, pgp-5, cnx-1, gpx-1, nlp-29, C17H12.8, F08G2.5 | pqn-54, hsp-1, T06E4.8, sid-3, thn-1, abu-10, F41E6.11, vit-2, abu-6, ubc-26, vit-4, Y54G2A.17, cebp-1, abu-8, hsp-6, lys-7, pcn-1, col-94, irg-3, mev-1, vit-6, rack-1, col-93, grd-3, hlh-30, set-26, F22H10.2, sea-2, set-9, ser-1, osm-11, mpk-1, F35E12.5, zip-2, flp-21, K08F4.2, tut-1, F10A3.4, col-179, col-178, col-92, R03D7.2, abu-15, max-2, eel-1, pqn-74, dcap-1, pdi-2, F55G11.4, dss-1, aak-2, T28D6.4, vit-5, abu-7, sca-1, vit-3, hsp-3 |
| Transcription regulation: | mef-2, set-27, blmp-1, nhr-49, nhr-69, daf-6 | taf-11.2, ceh-49, ceh-21, B0336.13, mxl-3, rpb-11, zip-2, sna-1, ell-1, ama-1, kin-10, paa-1, K12H4.5, sid-3, mom-2, rpl-25.2, alg-2, F59A3.4, gld-1, xrn-2, set-16, gcn-1, tsn-1, puf-6, alg-1, T19B4.5, ncl-1, mex-3, rde-4, fib-1, htz-1, xpo-2, F54D11.2, puf-7, puf-5, tbp-1, ttb-1 |
| Cytoskeleton organization: | mig-2, pyk-2, Y65B4BL.6, ced-12, lim-9, inx-15, sma-1, hpo-28 | kin-10, col-103, paa-1, emb-9, par-2, tbb-2, wip-1, tba-2, mom-2, act-1, vab-19, air-1, act-3, cmd-1, vab-10, act-4, bir-1, unc-15, lit-1, cyb-3, F47G4.4, act-2, klp-7, tomm-7, npp-11, gsk-3, zyg-8, cyk-4, mdh-2, F52a8.1, mei-2, tbb-1, rga-4, car-1, max-2, dim-1, mig-14, ify-1, T27E9.2, ama-1, unc-54, sca-1 |
| Mitochondria: | atg-13, atg-1, F49E10.4 | pgk-1, F58F12.1, gpd-1, Y82E9BR.3, mev-1, H28O16.1, W09C5.8, asb-2, atp-2, enol-1, gpd-3, gpd-4, Y69A2AR.18, aldo-1, pfk-1, aldo-2, vha-13, gpd-2, T27E9.2, ogdh-1, tomm-7, F32B4.2, sdha-1, C05G5.4, cts-1, sdhb-1, F47B10.1, fum-1, mdh-2, mdh-1, dist-1 |
| Muscle contraction regulation: | msp-77, flp-16, tnt-2, msp-38 | |
| Muscle structure: | kin-10, col-103, emb-9, act-1, act-3, vab-10, unc-15, lit-1, act-2, tomm-7, mdh-2, F52A8.1, dim-1, T27E9.2, unc-54, sca-1 | |
| Pharyngeal development: | abu-6, abu-8, mex-3, spn-4, abu-15, abu-7 |
Quantitative PCR analysis (qPCR) was used to test the results of the RNA-sequencing (RNAseq) data. The results of the gene expression levels of selected genes that are involved in proper muscle function and animal locomotion were analyzed using qPCR and compared to the transcriptome results (Fig. 2B). In these group of genes, the changes in gene expression observed in the RNAseq data was always consistent with the qPCR analysis, thus verifying the transcriptome analysis (Fig. 2B).
Down regulation of pharyngeal muscle genes correlates with aberrant pharyngeal muscle organization and function
Previous studies show that C. elegans expressing dominant lamin mutations, homologous to the human EDMD mutations, display disease phenotypes which resemble those observed in human EDMD patients.27 Our transcriptional changes in the locomotion cluster (Table 1) correlate with the impaired motility observed in L535P animals.20,24 Many of the lamin based muscle diseases have impaired ability to transmit forces from the cytoplasm to the nucleus and vice versa. Indeed we observed many changes in genes expression involved in cytoskeleton organization and mechano-transduction (Fig. 2A and Table 1).
Interestingly, genes involved in maintaining proper muscle structure and function were negatively affected by the L535P EDMD lamin mutation; sca-1 (ortholog of human ATP2A1) is required for muscle development and function. Actin isoforms act-4 act-3 and act-1 (homolog of human cytoplasmic Actin 1) are needed for proper body wall muscle structure; dim-1 is required for muscle organization and proper anchoring of the myofilament lattice to the muscle cell membrane; vab-10 (homologous of human Microtubule - actin cross-linking factor 1), tomm-7 (Mitochondrial import receptor subunit TOM7 human homolog) and F52A8.1 are required for body wall myosin organization, as well as mechanical resilience, proper mitochondria structure and striated muscle thick myosin filament assembly, respectively; unc-54 (human myosin heavy chain ortholog) is the major myosin heavy chain expressed in C. elegans and unc-15 (homolog to human myosin-7) is a paramyosin which physically interacts with MHC-A and requires unc-54 in order to prevent its degradation (Fig. 2, Table 1). The down regulation of all these genes corresponds to the musculature disorganization and degeneration present in L535P animals.
In addition to skeletal muscle abnormalities, all forms of LMNA dependent muscular dystrophies, including EDMD, affect cardiac muscle structure and function. Cardiomyocytes of these patients demonstrate nuclear envelope abnormalities, changes in lamins expression and mitochondria miss-localization.28,29 C. elegans pharynx is often used as a model of the function and muscular composition of the human heart.30 In our analysis, we detected down regulation of several genes involved in the proper function and structure of the pharyngeal muscle; abu-6, abu-7, abu-8 and abu-15 (homologs of Keratin-associated protein 10-4) have a role in protecting against miss-folded protein stress, as well as proper pharyngeal grinder function; act-1 is required for pharyngeal muscle structures and myo-2 (human myosin heavy chain ortholog) encodes a muscle pharynx specific myosin heavy chain isoform. This prompted us to test pharyngeal muscle function of L535P EDMD animals. We analyzed the number of pharyngeal contractions per minute of both wild-type and L535P animals. The pharynx of L535P EDMD animals contracted significantly less than the pharynx of wild-type animals (Fig. 3; 166 and 226 contractions per minute, respectively; p value = 7.5 × 10−22). The aberrant function of both body wall muscles and of the pharyngeal muscles in our L535P EDMD animals can also be explained, in part, by changes in genes involved in proper mitochondria function. As in the case of genes involved in proper muscle function and structure, genes involved in cellular respiration and ATP metabolism were negatively affected by the L535P mutation; several genes encoding mitochondria subunits were downregulated; mev-1 (a subunit of mitochondrial respiratory chain complex II), H28O16.1 (ortholog of the α subunit of mitochondrial ATP synthase), W09C5.8 (ortholog of the cytochrome c oxidase subunit IV), atp-2 (β subunit of the catalytic F1 portion of ATP synthase) required for pharyngeal pumping, Y69A2AR.18 (ortholog of human ATP synthase of the mitochondrial F1 complex, gamma polypeptide 1) expressed in the pharynx and the body wall musculature. Also down regulated were; asb-2 - an ATP synthase B analog, T27E9.2 which is involved in striated muscle myosin thick filament assembly and predicted to have ubiquinol-cytochrome-c reductase activity and tomm-7 which is required for normal mitochondrial morphology. The few upregulated genes are involved in mitochondria disassembly (Fig. 2A). Analysis of pharyngeal muscle structure using thin section transmission electron microscopy revealed overall loss of proper pharyngeal structure in animals expressing the L535P lamin mutation (Compare Fig. 4A-D and Fig. 4E-F). Furthermore, the uniform organization of sarcomeres and muscle fibers was lost (Fig. 4C-D, G-J). In wild type animals, mitochondria are evenly spaced between the muscle sarcomeres (Fig. 4C). However, in L535P animals, this organization is lost, and mitochondria seem to localize within sarcomeres (Fig. 4J). When looking at the global transcriptional changes resulting from the dominant expression of the L535P EDMD-linked lamin mutation, we find a good correlation between down regulation in gene expression and disease phenotypes.
Figure 3.
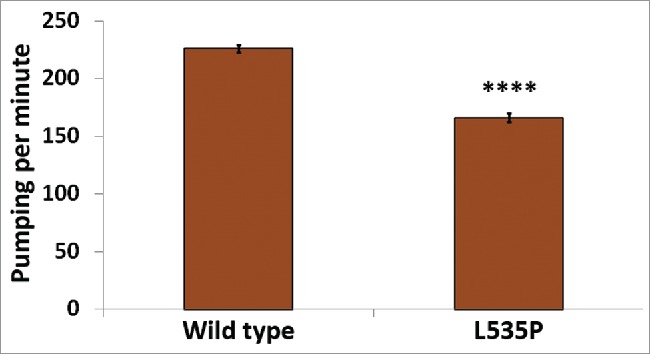
Aberrant pharyngeal muscle function in L535P animals. The number of pharyngeal contractions per minute was analyzed in both wild-type and L535P animals (226 and 166 contractions/min, respectively, n = 60 for each strain). Pumping was significantly reduced in L535P animals (p value = 7.5 × 10−22).
Figure 4.
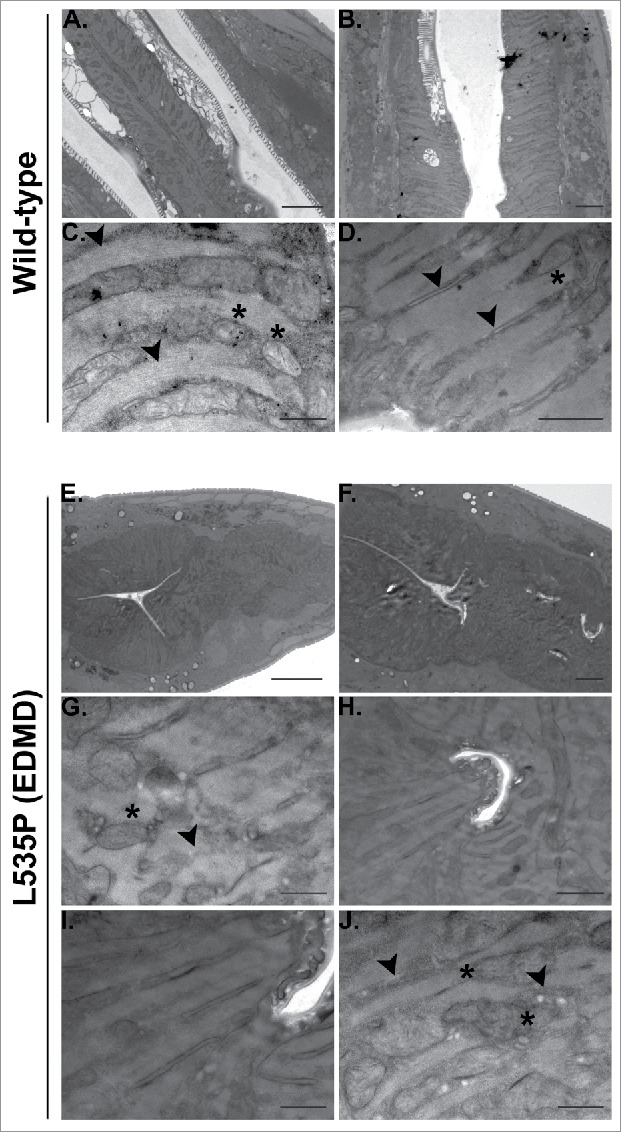
Aberrant pharyngeal muscle structure in L535P animals. Electron microscopy was used to analyze the effects of the L535P EDMD lamin mutation on pharyngeal muscle organization. (A-D) wild type animals. (E-J) L535P animals. In wild-type animals sarcomeres and muscle fibers were well aligned with mitochondria embedded between them (C-D; black arrowheads and Asterisk, respectively). In contrast, L535P animals had overall aberrant pharynx morphology (E-F), with loss of sarcomere, muscle fiber and mitochondria organization (G and J; black arrowheads and Asterisk, respectively).
Discussion
The diversity of LMNA-based diseases raises questions of how so many different missense mutations in a single gene cause a wide variety of tissue phenotypes. One of the major goals in the study of laminopathies is to gain a mechanistic understanding of the effects of different lamin mutations. A promising way to achieve this goal is to study transcriptional changes resulting from lamin disease mutations. In this study we utilized the well-characterized transgenic C. elegans strain dominantly expressing the L535P EDMD lamin mutation in order to analyze genetic changes resulting from this mutation. To date, gene expression analysis of lamin based diseases was performed on patient fibroblasts. This analysis of Hutchinson-Gilford progeria syndrome (HGPS) fibroblasts showed down regulation of a transcription factor involved in the negative regulation of mesodermal proliferation, which correlated with the tissues affected by this disease.31 However, since lamins play pivotal roles in many of the biological processes taking place in nuclei, we believe that in order to gain better understanding of disease mechanisms, transcriptome analysis of an entire living organism would provide further useful information. Indeed, our results show many genes which change expression as a result of the L535P EDMD lamin mutation. Many of the affected clusters include both up and downregulated genes. However, our analysis shows an overall down regulation of genes important for proper muscle organization and function. This correlates with the muscle disorganisation and impaired motility characteristic of L535P animals.20,24
A common phenotype in all forms of lamin-based muscle diseases is a disruption in cardiac muscle function. Indeed, our results show down regulation of genes which are important for the proper structure and function of the pharyngeal muscle. These expression changes again correlate with the significantly slowed pharyngeal muscle pumping of L535P animals. It is worth mentioning that the C. elegans pharynx shares functional and molecular similarities with cardiac muscles of other species30.
The unidirectional change in expression of genes involved in proper body wall and pharyngeal muscle structure and function can potentially explain the tissue specificity of the L535P EDMD mutation. In other biological pathways affected by this mutation, down regulation of certain genes could conceivably be compensated for by the up regulation of other genes involved in the relevant pathway and vice versa.
Material and methods
Strains
The Bristol strain N2 was used as a standard wild-type C. elegans strain. YG499 ([baf-1p::GFP::lmn-1 L535P unc119(+)]; unc-119(ed3)) was used as EDMD disease model. Both N2 and YG499 were maintained and manipulated under standard conditions as previously described.32 Mutant strains were out-crossed 3 times to ensure a clean background.
RNA isolation
Bleached N2 and YG499 embryos were placed on NGM plates at 20°C, and late L4 progeny was collected after 48 hr for analysis. Worms were washed 3 times using M9 buffer (3g KH2PO4, 6g Na2HPO4, 5g NaCl, 1ml 1M MgSO4, H2O to 1 liter. Sterilize by autoclaving) then transferred to an RNase DNase-free eppendorf and additionally washed with 1ml DEPC treated DDW. Samples were centrifuged at 2000 rpm for 1 min and a ratio of 2 parts BIO TRI RNA (Bio-Lab ltd., Cat. #90102331) to 1 part worm pellet was added and tubes were gently mixed. Samples were kept overnight at (−80°C). Samples were subjected to 3 rounds of crushing using an eppendorf mortar and pestle, thawing at (42°C) and then freezing in liquid nitrogen, with an additional 2 cycles of thawing at (42°C) and then freezing in liquid nitrogen. Samples were kept for 10 minutes at room temperature (∼22°C) than centrifuged at 12,000 × g for 15 minutes at (4°C). Supernatant was transferred to a new tube, and a ratio of 1 part chloroform to 5 parts supernatant was added and shaken vigorously. The samples were incubated for 5 min in RT and centrifuged at 12,000 × g for 30 min in (4°C). The aqueous transparent phase was transferred to a new tube and a ratio of 1 part isopropanol to 2 parts starting volume (worm pellet + Tri-reagent) and 0.7 μl glycogen was added. The samples were then shaken and incubated overnight at (−20°C). Samples were centrifuged at 12,000 × g for 30 min in (4°C) and 1ml of 75% ethanol was added to the pellet. Samples were briefly vortexed than centrifuged at 7500 × g for 15 min in (4°C). All samples were placed on ice, the ethanol was discarded and the pellets were air-dried for ∼20 min until fully dried. RNA pellet was eluted using 30 μl of PCR grade DDW and incubated at 55°C for 10 min. RNA concentration was measured using a BioAnalyzer machine (Agilent Technologies, Inc., serial #DE54700884, firmware C.01.069, type G2939A) and frozen at (−80°C) until use. Three separate isolations were performed for each strain.
cDNA (cDNA) preparation
2 μg RNA was treated with RQ1 RNase-Free DNase (Promega Corporation, cat. #M6101). Protocol performed according to manufacturer's instructions. First-strand cDNA synthesis was performed using M-MLV RT kit (Promega Corporation, cat. #M170A). Protocol performed according to manufacturer's instructions. Samples were stored at (−80°C) until use.
RNA sequencing
Sequencing of the cDNA libraries was performed using the NextSeq 500 High Output kit (75 cycles) #FC-404-2005 Illumina by the Center of Genomic Technology.
Bioinformatics analysis
Trimming and filtering of raw reads: The NextSeq base-calls files were converted to fastq files using the bcl2fastq (v2.15.0.4) program with default parameters. A fastq file was created for each sample separately. Raw reads (fastq files) were inspected for quality issues with FastQC (v0.11.2, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). According to the FastQC report, reads were quality-trimmed at both ends, using in-house Perl scripts, with a quality threshold of 32. In short, the scripts use a sliding window of 5 bases from the read's end and trim one base at a time until the average quality of the window passes the given threshold. Following quality-trimming, the SL1 and SL2 sequences were removed from the 5′ end by sequential runs of cutadapt (version 1.7.1, http://cutadapt.readthedocs.org/en/stable/), with a minimal overlap of 1 (-O parameter) and allowing for read wildcards. The adapter sequence was then similarly trimmed from the 3′ end. Reads that became shorter than 15 nucleotides were filtered out (-m parameter). The remaining reads were further filtered to remove very low quality reads, using the fastq_quality_filter program of the FASTX package (version 0.0.14, http://hannonlab.cshl.edu/fastx_toolkit/), with a quality threshold of 20 at 90 percent or more of the read's positions.
Mapping and differential expression analysis: The processed fastq files were mapped to the C. elegans transcriptome and genome using TopHat (v2.0.13). The genome version was WBcel235, with annotations from Ensembl release 82 (based on October 2014 annotations). Mapping allowed up to 5 mismatches per read, a maximum gap of 5 bases, and a total edit distance of 10. The short nature of the C. elegans introns was accounted for by setting the –i parameter to 20 (full command: tophat -G genes.gtf -N 5 –read-gap-length 5 –read-edit-dist 10 –segment-length 20 –read-realign-edit-dist 4 -i 20 –microexon-search –min-segment-intron 20 –coverage-search –min-coverage-intron 20 –max-coverage-intron 10000 genome processed.fastq). Quantification, normalization and differential expression were done with the Cufflinks package (v2.2.1). Quantification was done with cuffquant, using the genome bias correction (-b parameter), multi-mapped reads assignment algorithm (-u parameter) and masking for genes of type rRNA, tRNA, miRNA, snRNA, snoRNA, piRNA and ncRNA (-M parameter). Normalization was done with cuffnorm (using output format of Cuffdiff) and results were visualized in R, using the cummeRbund package (version 2.8.2) and in-house R scripts. Counts and FPKM distributions were used for comparing global expression between samples and for background expression level estimation. Differential expression was calculated with cuffdiff, using a count threshold (-c parameter) of at least 15 for statistical significance testing. Samples were assigned a condition (-L parameter) and all pair wise comparisons were performed. Gene-level cuffdiff output was combined with gene details (such as symbol, Entrez accession, etc.) taken from the results of a BioMart query (Ensembl, release 82). Significantly differentially expressed genes were defined as ones with at least 1 FPKM level of expression in at least one of the conditions and a q-value less than 0.05. Differential expression results were visualized in R with in-house scripts based on the cummeRbund code. Additional Cuffdiff run was performed with the same parameters.
Cluster analysis
All significant genes (log2 fold change of below -1 or above 1, with a p-value of 0.05 and below) were clustered using DAVID Bioinformatics Resources 6.7 (https://david.ncifcrf.gov/tools.jsp)33 and GO Enrichment Analysis | Gene Ontology Consortium annotation tool (http://geneontology.org/page/go-enrichment-analysis). Clusters of interest were analyzed with a Python program (see Supp. Fig 1; can be downloaded from https://www.dropbox.com/s/l1ctktlj3lyxaa3/WormbaseFetcher.zip?dl=0). Whereby, information about each gene was displayed in a graphic user interface (GUI). The information was fetched from the WormBase (http://www.wormbase.org) through the genes WormBase gene ID. The interface handled the visual representation of the information for one gene at the time (see Supp. Fig 1).
Quantitative PCR analysis
Quantitative amplification was performed on 2 separate RNA isolations from both wild-type and L535P animals with KAPA SYBR® FAST qPCR Kit Master Mix (2X) Universal (Roche, cat. #07959397001). Protocol was performed according to manufacturer's instructions. Analysis was performed using the Rotor-Gene™ machine (Corbett Life Science, Sydney, Australia, Model RG-6000 #R 120726) with software version 1.7 (Build 87). The threshold cycle (CT) for each gene was determined during primer calibration. The C. elegans housekeeping gene pmp-3 (F- AATTGGATTCGAGCTGGAAATGGC, R-CCACTGCGACCCTTGACTCTA) was used as a housekeeping control. Primers were designed using ApE (v2.0.3.7, Copyright © 2003-2009 by M. Wayne Davis) according to qPCR standards34; unc-54 F-CATGCCGACCTCGATGAGACC, R-CCTTCTTTCCTCCCTTAAGAGCAGC; unc-15 F-ACACGAGCTCGAGGATGCT, R-CCGTGACGAAAATCTTGGAGGATG; act-1 F-GGCCCAATCCAAGAGAGGTATCC, R-GGGGCAACACGAAGCTCATTG; act-4 F-GGCTCTCTTCCAGCCATCCT, R-TCTTCATGGTGGATGGGGCA; unc-69 F-GATGATCTCTCACAACGCGTCGA, R-TGTACTGGCCGAGCACTTGAT; D2063.1 F-AGACTCAAAGATTCTCGTCGACACA, R-AACTCTAATGGGACATTGACGGC; inx-15 F-GCATTGTTCTTCCTTACCGTCTGG, R-CGATTGTAGTCGTTCGAGTAAGTGC; dim-1 F-CAATCAAGGAGCTTGCTGATGCTG, R-GGAAGAGAATCCGGTAAGCTTCAGG; sca-1 F-TCTTGCTGTCGGAACCTACGTC, R-GCGCAGTCAAGATCGGCAAAG; cyk-4 F-GCAGGAATCAAAGCGATGCGAAAA, R-TTGGCTCCACGTAATGCGGAT and myo-2 F-AGGAAGAGCATCAGTTGACCGA, R-GCTCTTTCAGCATCATCTGTGCC.
Pharyngeal pumping
Bleached N2 and YG499 embryos were placed on NGM plates at 20°C, and late L4 progeny was collected after 48 h for analysis. Pumping rates of individual worms were measured as the number of pharyngeal contractions during a 1 min interval under a binocular microscope. Each experiment contained ∼30 worms.
Transmission electron microscopy
Late L4 worms were collected from NGM plates and prepared for imaging with an electron microscope (Philips Technai 12) equipped with a MegaView II CCD camera as described in.20
Supplementary Material
Abbreviations
- DCM
Dilated cardiomyopathy;
- EDMD
Emery-Dreifuss muscular dystrophy;
- FPKM
Fragments Per Kilobase of transcript per Million mapped reads;
- IFs
Intermediate filaments;
- NLS
Nuclear localization signal;
- qPCR
Quantitative PCR
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Alon Zaslaver the Gruenbaum lab members for critical reading of the manuscript. We also thank Dr. Yuval Nevo (Info-CORE, Bioinformatics Unit of the I-CORE Computation Center at the Hebrew University and Hadassah) for performing the bioinformatics analysis and Dr. Yael Friedmann and Hava Glickstein for helping with the EM analysis.
Funding
This work was supported by grants from The United States–Israel Binational Science Foundation (grant 2007215), the Muscular Dystrophy Association (MDA233701), the Niedersachsen-Israeli Research Cooperation and Israel Science Foundation (785/15).
References
- [1]. Wang N, Stamenovic D. Contribution of intermediate filaments to cell stiffness, stiffening, and growth. Am J Physiol Cell Physiol 2000; 279:C188-94; PMID:10898730 [DOI] [PubMed] [Google Scholar]
- [2]. Harada T, Swift J, Irianto J, Shin JW, Spinler KR, Athirasala A, Diegmiller R, Dingal PCDP, Ivanovska IL, Discher DE. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J Cell Biol 2014; 204:669-82; PMID:24567359; http://dx.doi.org/ 10.1083/jcb.201308029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PCDP, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. . Nuclear Lamin-A Scales with Tissue Stiffness and Enhances Matrix-Directed Differentiation. Science 2013; 341:1240104; PMID:23990565; http://dx.doi.org/ 10.1126/science.1240104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. Stuurman N, Heins S, Aebi U. Nuclear Lamins: Their Structure, Assembly, and Interactions. J Struct Biol 1998; 122:42-66; PMID:9724605; http://dx.doi.org/ 10.1006/jsbi.1998.3987 [DOI] [PubMed] [Google Scholar]
- [5]. Machowska M, Piekarowicz K, Rzepecki R. Regulation of lamin properties and functions: does phosphorylation do it all? Open Biol 2015; 5:150094; PMID:26581574; http://dx.doi.org/ 10.1098/rsob.150094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6]. Lehner CF. Differential expression of nuclear lamin proteins during chicken development. J Cell Biol 1987; 105:577-87; PMID:3301871; http://dx.doi.org/ 10.1083/jcb.105.1.577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Gruenbaum Y, Foisner R. Lamins: Nuclear Intermediate Filament Proteins with Fundamental Functions in Nuclear Mechanics and Genome Regulation. Annu Rev 2015; 84:131-64 [DOI] [PubMed] [Google Scholar]
- [8]. Gay S, Foiani M. Chapter Six – Nuclear Envelope and Chromatin, Lock and Key of Genome Integrity. Int Rev Cell Mol Biol 2015; 317:267-330; PMID:26008788; http://dx.doi.org/ 10.1016/bs.ircmb.2015.03.001 [DOI] [PubMed] [Google Scholar]
- [9]. Bertrand AT, Chikhaoui K, Yaou R, Ben, Bonne G. Clinical and genetic heterogeneity in laminopathies. Biochem Soc Trans 2011; 39:1687-92; PMID:22103508; http://dx.doi.org/ 10.1042/BST20110670 [DOI] [PubMed] [Google Scholar]
- [10]. Belaadi N, Aureille J, Guilluy C. Under Pressure: Mechanical Stress Management in the Nucleus. Cells 2016; 5:27; http://dx.doi.org/ 10.3390/cells5020027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11]. Osmanagic-Myers S, Dechat T, Foisner R. Lamins at the crossroads of mechanosignaling. Genes Dev 2015; 29:225-37; PMID:25644599; http://dx.doi.org/ 10.1101/gad.255968.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Prokocimer M, Davidovich M, Nissim-Rafinia M, Wiesel-Motiuk N, Bar DZ, Barken R, Meshorer E, Gruenbaum Y. Nuclear lamins: key regulators of nuclear structure and activities. J Cell Mol Med 2009; 13:1059-85; PMID: 19210577; http://dx.doi.org/ 10.1111/j.1582-4934.2008.00676.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13]. Dittmer TA, Misteli T. The lamin protein family. Genome Biol 2011; 12:1-14; http://dx.doi.org/ 10.1186/gb-2011-12-5-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Patricia MD, Lammerding J. Broken nuclei – lamins, nuclear mechanics, and disease. Trends Cell Biol 2014; 24:247-56; PMID:24309562; http://dx.doi.org/ 10.1016/j.tcb.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. Worman HJ. Nuclear lamins and laminopathies. J Pathol 2012; 226:316-25; PMID:21953297; http://dx.doi.org/ 10.1002/path.2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Schreiber KH, Kennedy BK. When Lamins Go Bad: Nuclear Structure and Disease. Cell 2013; 152:1365-75; PMID:23498943; http://dx.doi.org/ 10.1016/j.cell.2013.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]. Bonne G, Quijano-Roy S. Emery–Dreifuss muscular dystrophy, laminopathies, and other nuclear envelopathies. Handb Clin Neurol 2013; 113:1367-76; PMID:23622360; http://dx.doi.org/ 10.1016/B978-0-444-59565-2.00007-1 [DOI] [PubMed] [Google Scholar]
- [18]. Maggi L, Carboni N, Bernasconi P. Skeletal Muscle Laminopathies: A Review of Clinical and Molecular Features. Cells 2016; 5:33; http://dx.doi.org/ 10.3390/cells5030033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Zwerger M, Yee Ho C, Lammerding J. Nuclear Mechanics in Disease. Annu Rev 2011; 13:397-428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Zuela N, Zwerger M, Levin T, Medalia O, Gruenbaum Y. Impaired mechanical response of an EDMD mutation leads to motility phenotypes that are repaired by loss of prenylation. J Cell Sci 2016; 9:1781-91; http://dx.doi.org/ 10.1242/jcs.184309 [DOI] [PubMed] [Google Scholar]
- [21]. Mattout A, Pike BL, Towbin BD, Bank EM, Gonzalez-Sandoval A, Stadler MB, Meister P, Gruenbaum Y, Gasser SM. An EDMD Mutation in C. elegans Lamin Blocks Muscle-Specific Gene Relocation and Compromises Muscle Integrity. Curr Biol 2011; 21:1603-14; PMID:21962710; http://dx.doi.org/ 10.1016/j.cub.2011.08.030 [DOI] [PubMed] [Google Scholar]
- [22]. Korfali N, Wilkie GS, Swanson SK, Srsen V, de las Heras J, Batrakou DG, Malik P, Zuleger N, Kerr ARW, Florens L, et al. . The nuclear envelope proteome differs notably between tissues. Nucleus 2012; 3:552-64; PMID:22990521; http://dx.doi.org/ 10.4161/nucl.22257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Worman HJ, Schirmer EC. Nuclear membrane diversity: underlying tissue-specific pathologies in disease? Curr Opin Cell Biol 2015; 34:101-12; PMID:26115475; http://dx.doi.org/ 10.1016/j.ceb.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Bank EM, Gruenbaum Y. Caenorhabditis elegans as a model system for studying the nuclear lamina and laminopathic diseases. Nucleus 2011; 2:350-7; PMID:21970988; http://dx.doi.org/ 10.4161/nucl.2.5.17838 [DOI] [PubMed] [Google Scholar]
- [25]. Liu J, Ben-Shahar TR, Riemer D, Treinin M, Spann P, Weber K, Fire A, Gruenbaum Y. Essential Roles for Caenorhabditis elegans Lamin Gene in Nuclear Organization, Cell Cycle Progression, and Spatial Organization of Nuclear Pore Complexes. Mol Biol Cell 2000; 11:3937-47; PMID:11071918; http://dx.doi.org/ 10.1091/mbc.11.11.3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Wiesel N, Mattout A, Melcer S, Melamed-Book N, Herrmann H, Medalia O, Aebi U, Gruenbaum Y. Laminopathic mutations interfere with the assembly, localization, and dynamics of nuclear lamins. Proc Natl Acad Sci USA 2008; 105:180-5; PMID:18162544; http://dx.doi.org/ 10.1073/pnas.0708974105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Bank EM, Ben-Harush K, Feinstein N, Medalia O, Gruenbaum Y. Structural and physiological phenotypes of disease-linked lamin mutations in C. elegans. J Struct Biol 2012; 177:106-12; PMID:22079399; http://dx.doi.org/ 10.1016/j.jsb.2011.10.009 [DOI] [PubMed] [Google Scholar]
- [28]. Fidziańska A, Walczak E, Glinka Z, Religa G. Nuclear architecture remodelling in cardiomyocytes with. Folia Neuropathol 2008; 46:196-203; PMID:18825595 [PubMed] [Google Scholar]
- [29]. Gupta P, Bilinska ZT, Sylvius N, Boudreau E, Veinot JP, Labib S, Bolongo PM, Hamza A, Jackson T, Ploski R, et al. . Genetic and ultrastructural studies in dilated cardiomyopathy patients: a large deletion in the lamin A/C gene is associated with cardiomyocyte nuclear envelope disruption. Basic Res Cardiol 2010; 105:365-77; PMID:20127487; http://dx.doi.org/ 10.1007/s00395-010-0085-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Haun C, Alexander J, Stainier DY, Okkema PG. Rescue of Caenorhabditis elegans pharyngeal development by a vertebrate heart specification gene. Proc Natl Acad Sci 1998; 95:5072-5; PMID:9560230; http://dx.doi.org/ 10.1073/pnas.95.9.5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Csoka AB, English SB, Simkevich CP, Ginzinger DG, Butte AJ, Schatten GP, Rothman FG, Sedivy JM. Genome-scale expression profiling of Hutchinson–Gilford progeria syndrome reveals widespread transcriptional misregulation leading to mesodermal/mesenchymal defects and accelerated atherosclerosis. Aging Cell 2004; 3:235-43; PMID:15268757; http://dx.doi.org/ 10.1111/j.1474-9728.2004.00105.x [DOI] [PubMed] [Google Scholar]
- [32]. Brenner S. The genetics of Caenorhabditis elegans. Genetics 1974; 77:71-94; PMID:4366476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33]. Huang DW, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA. Extracting Biological Meaning from Large Gene Lists with DAVID. Curr Protoc Bioinforma Curr. Protoc. Bioinform 2009; 27:13.11.1-13.11.13; PMID: 19728287 [DOI] [PubMed] [Google Scholar]
- [34]. Udvardi MK, Czechowski T, Scheible W-R. Eleven Golden Rules of Quantitative RT-PCR. Plant Cell 2008; 20:1736-7; PMID:18664613; http://dx.doi.org/ 10.1105/tpc.108.061143 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.