Abstract
Complex regulatory landscapes control the pleiotropic transcriptional activities of developmental genes. For most genes, the number, location, and dynamics of their associated regulatory elements are unknown. In this work, we characterized the three-dimensional chromatin microarchitecture and regulatory landscape of 446 limb-associated gene loci in mouse using Capture-C, ChIP-seq, and RNA-seq in forelimb, hindlimb at three developmental stages, and midbrain. The fine mapping of chromatin interactions revealed a strong preference for functional genomic regions such as repressed or active domains. By combining chromatin marks and interaction peaks, we annotated more than 1000 putative limb enhancers and their associated genes. Moreover, the analysis of chromatin interactions revealed two regimes of chromatin folding, one producing interactions stable across tissues and stages and another one associated with tissue and/or stage-specific interactions. Whereas stable interactions associate strongly with CTCF/RAD21 binding, the intensity of variable interactions correlates with changes in underlying chromatin modifications, specifically at the viewpoint and at the interaction site. In conclusion, this comprehensive data set provides a resource for the characterization of hundreds of limb-associated regulatory landscapes and a framework to interpret the chromatin folding dynamics observed during embryogenesis.
Gene regulation in higher eukaryotes involves complex interactions between regulatory sequences and their associated promoters, which can be separated by hundreds of kilobases from each other. The communication between these distal genomic regions is made possible through chromatin interactions (Lettice et al. 2003; Montavon et al. 2011; Andrey et al. 2013). The advent of Chromosome Conformation Capture-based technologies (4C, 5C, Hi-C), which combine next-generation sequencing and proximity ligation assays to probe chromatin interactions, transformed our understanding of the genomic 3D architecture (Dekker et al. 2002; de Laat and Duboule 2013). Studies using Hi-C that capture interactions genome-wide revealed topologically associated domains (TADs) as modules of chromatin organization with high, local interactions. TADs are proposed to be stable and isolated regulatory units, which are conserved across cell types and species (Dixon et al. 2012, 2015; Nora et al. 2012; Sexton et al. 2012). Although their overall structure is stable, cell-type–specific changes in intra-TADs interactions were observed in these studies and were predicted to associate with transcription (Giorgetti et al. 2014, 2016). In contrast to genome-wide Hi-C interaction maps, the 4C technology generates one-dimensional interaction profiles of a single viewpoint with the rest of the genome. 4C requires much less sequencing to achieve a high resolution but limits the analysis to a single viewpoint. Because of its high resolution and low production cost, 4C has been extensively used to explore the regulatory microarchitecture of specific loci and as a proxy to recapitulate and assess TADs structure and variability (Montavon et al. 2012; Andrey et al. 2013; Ghavi-Helm et al. 2014; Lupianez et al. 2015; Acemel et al. 2016).
Despite an abundant use of the 4C technology, little is known about the dynamics and properties underlying interactions in different cell types, tissues, or time points in development. Several authors have described the dynamics of tissue-specific enhancer-promoter interactions (Montavon et al. 2011; Phillips-Cremins et al. 2013; Ghavi-Helm et al. 2014; Lonfat et al. 2014). A study at 103 Drosophila loci revealed remarkably little tissue-specific looping, with only 6% of the interacting fragments showing changes between tissues (Ghavi-Helm et al. 2014). In contrast to Drosophila, a study at six loci during the differentiation of mouse ESCs into neural progenitor cells showed that almost half of the detected interactions are cell-type–specific rather than constitutive (Phillips-Cremins et al. 2013). At these loci, changes in looping were associated with changes in enhancer activity and with the presence of the architectural proteins Mediator, Cohesin, and CTCF. However, the 5C technology used in the latter work limited the number of studied loci to six. It is thus unclear if these extensive interaction changes are a general phenomenon or locus-specific.
In this work, we aim to understand the principles underlying chromatin interactions and their dynamics by characterizing a comprehensive set of regulatory landscapes in developing mouse tissues.
Results
Probing the interaction profiles of 446 loci using Capture-C
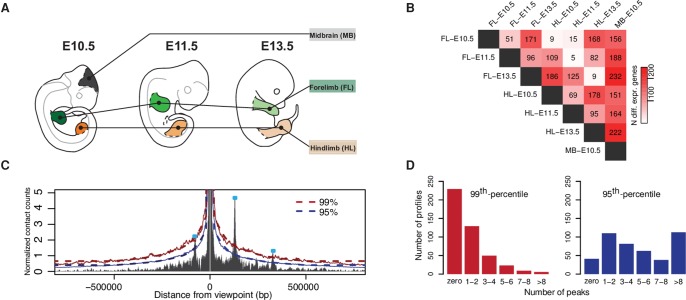
The developing limb is a dynamic system in which regulatory changes occur in a time- and tissue-specific manner. Despite many similarities between fore- and hindlimb development, the gradual changes in gene expression and minute differences in temporal patterning lead to different morphologies. Thus, developing limbs constitute a convenient model to survey the regulatory dynamic of chromatin architecture. In this project, we chose seven combinations of tissue and time points, i.e., forelimb and hindlimb, each at E10.5, E11.5, and E13.5, as well as midbrain at E10.5 (Fig. 1A). We probed the chromatin architecture of 446 viewpoints, including 434 main promoters and 12 alternative promoters. The 446 loci were selected based on their involvement in limb development (patterning, growth, and tissue differentiation), as well as their association with congenital limb malformations. These loci involve major developmental limb transcription factor families such as Tbx, Sox, Hox, and Runx; genes involved in signaling pathways such as FGF, WNT, RA, and BMP; as well as differentiation markers such as Sox9, Col1a1, and Myod1 (Supplemental Table S1). To evaluate the transcriptional variation among the tissues, we performed RNA-seq and identified genes which are expressed differentially between tissues or developmental stages (Fig. 1B). As expected, we observed a high similarity among the limb samples and major differences between limb and midbrain. Forelimb and hindlimb of the same stage were very similar to each other with only a few differentially expressed genes at all three time points. However, between developmental stages, transcriptional changes were much more pronounced.
Figure 1.
Experimental setup and calling of interaction peaks. (A) Seven tissues were selected to perform CC, RNA-seq, and ChIP-seq from: fore- (FL) and hindlimb (HL) at E10.5, E11.5, and E13.5, as well as midbrain (MB) at E10.5. (B) Number of differentially expressed genes from pairwise comparisons of RNA-seq data among the different tissues (threshold: abs[log2 fold change] > 1, adjusted P-value < 0.01). (C) Calling of interaction peaks using an empirical background model. The red and blue solid lines represent the 99th and 95th percentile, respectively. Dashed lines indicate smoothed threshold lines, values set fixed at 0.5 Mb from the viewpoint. (D) Number of peaks per viewpoint averaged over tissues and time points for two cutoff values: 99th and 95th percentile.
Using Capture-C (CC), we obtained high-resolution interaction profiles for all surveyed promoters in biological replicates (Supplemental Fig. S1; Hughes et al. 2014). In order to assess the intensity of chromatin interactions, we combined the 446 profiles across all time points, tissues, and replicates to generate a simple, empirical background. This model allows us to determine the interaction intensity of any region with respect to the distribution obtained from the whole data set at a given distance from its viewpoint (Fig. 1C). We defined interaction peaks as regions displaying intensity above the 99th or 95th percentile, thus as regions being among the top 1% or 5%, respectively, regarding their interaction intensity. Across all tissues and time points, we annotated, on average, 600 strong interactions when applying the 99th percentile and 3000 when lowering the threshold to the 95th percentile (Fig. 1D). For most analysis presented in this work, we applied the more conservative 99th percentile as a threshold to reduce the risk of false positive interactions. Finally, we tested whether these interactions could also be identified with an alternative approach, implemented in the software package CHiCAGO (Cairns et al. 2016), and found an overlap for 79%–89% of our called interactions (Supplemental Fig. S2).
3D-microarchitecture is associated with functional chromatin domains
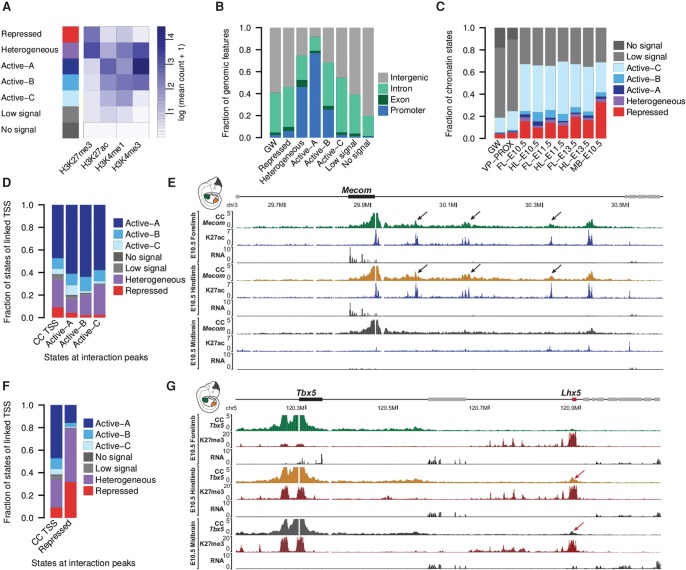
To evaluate in which type of functional chromatin domains chromatin interactions form, we generated ChIP-seq profiles for H3K27me3, H3K27ac, H3K4me3, and H3K4me1 from the same tissues and time points as described above. We used the software EpiCSeg (Mammana and Chung 2015) to segment the genome into seven chromatin states (Fig. 2A; Supplemental Fig. S3). EpiCSeg assigned each genomic region to one out of the seven states based on the abundance and co-occurrence of histone marks. The ChIP-seq signatures of the states were identified in unsupervised learning and afterward manually assigned to functional classes. Three states associated predominantly with active marks and were labeled accordingly. Active-A and Active-B with high H3K4me3 and H3K27ac occurred frequently at promoters, whereas Active-C was characterized by low H3K4me3, and high H3K27ac and H3K4me1 levels. All three states were found in intergenic and intronic regions, suggesting that they all might mark putative enhancer regions (Fig. 2B). A state solely enriched for H3K27me3 is here referred to as repressed. Interestingly, repressed regions were not strongly biased toward promoter regions but also frequently found in intergenic domains. A small fraction of the genome was found to be covered by both active and repressive marks and is here referred to as a heterogeneous state. The heterogeneous state is strongly enriched at promoter regions and associates with mild gene expression, in contrast to Active-A and -B, which are found at very active promoters, and repressed ones, which are found at lowly expressed genes (Fig. 2B; Supplemental Fig. S4; Supplemental Table S2). The heterogeneous state is likely to represent the cell heterogeneity of our in vivo samples, rather than bivalent regions, as some genomic regions might be active in some cells and repressed in others. Indeed, limb and midbrain tissues contain tens to hundreds of different cell types with individual transcriptional signatures (Zeller et al. 2009; La Manno et al. 2016). Accordingly, we observed that heterogeneous regions are enriched in TSSs (transcriptional start sites) of genes with highly localized expression such as Hand2 or Hoxd10, which are restricted to the posterior side of the developing limb bud, or Col2a1, which is expressed solely in chondrocytes (Supplemental Table S2; Bell et al. 1997; Andrey et al. 2013; Osterwalder et al. 2014). Finally, two states, low signal and no signal, cover the large majority of the genome, i.e., 63% and 18%, respectively. While low signal segments are depleted of chromatin marks probed in this study, the no signal state represents mainly regions of low mappability (Supplemental Fig. S5).
Figure 2.
Interactions associate with functional chromatin states. (A) EpiCSeg segmentation from H3K4me3, H3K4me1, H3K27ac, and H3K27me3 generates three types of active states, one repressed state, a heterogeneous state, and two types of states depleted from the surveyed chromatin marks. (B) Distribution of genomic features across chromatin states in hindlimb at E10.5, as well as their genome-wide distribution. (C) Functional chromatin domains cover 19% of the genome-wide (GW) and 25% of regions proximal to the CC viewpoints (±2 Mb) (VP-PROX). More than sixty percent of interaction sites (99th percentile) were found in functional chromatin segments. (D) States of viewpoints’ TSSs linked to active interaction peaks (data were aggregated from all tissues) (see Supplemental Fig. S7A). (E) At the Mecom gene, interaction peaks form specifically over acetylated regions (black arrows) and disappear in the midbrain, where these regions are not acetylated. The RNA-seq tracks show that the Mecom gene is expressed at high level in limb tissues, whereas its expression is much lower in midbrain. (F) States of viewpoints’ TSSs linked to repressed interaction peaks (data were aggregated from all tissues) (see Supplemental Fig. S7B). (G) Tbx5 promoter forms a long-range interaction (red arrow) with an H3K27me3 domain (Lhx5) only when it is covered by the same mark and repressed (see RNA tracks) in hindlimb and midbrain.
The called CC interaction peaks strongly associate with the functional states active, heterogeneous, and repressed, and are depleted from the low signal or no signal state in comparison to the genome-wide or viewpoint proximal (±2 Mb) frequency of each state (Fig. 2C; Supplemental Fig. S6).
As 33%–55% of interaction peaks occurred in active segments, we asked whether they are linked to viewpoints with similar or different patterns of chromatin modifications. We therefore categorized the interacting regions according to their chromatin states and determined the chromatin state at the linked TSSs. First, interactions of active chromatin sites are more frequently linked to active TSSs than to repressed ones, in comparison to the overall distribution of states at captured TSSs (Fig. 2D). These observations suggest that active chromatin sites preferentially interact with each other and that changes in active chromatin domains during tissue differentiation and development could accompany changes in 3D contacts. An illustrative example is the Mecom gene, where several interactions in forelimb and hindlimb at E10.5 co-occur with high acetylation (Fig. 2E). In contrast, these interactions were not observed in midbrain, where the Mecom gene is 10 times less expressed and the interaction sites are not acetylated.
Similarly, we observed that repressed interaction peaks interact with TSSs enriched for the repressed state and depleted from the active ones (Fig. 2F; Supplemental Fig. S7). The methylation turnover at these repressed sites could here again associate with shifts in the 3D interactions. An example of this phenomenon is observed at the Tbx5 locus. Tbx5 is specifically active in forelimb but repressed in hindlimb and in midbrain. In the latter tissues, the Tbx5 promoter is covered with the repressive H3K27me3 mark, and one can observe a distal interaction with the first H3K27me3-repressed telomeric gene Lhx5. Both the interaction and the repressive marks are absent in Tbx5-expressing forelimb tissues (Fig. 2G).
In summary, these findings suggest that chromatin domains interact dynamically and preferentially in a homotypic fashion: Active promoters contact more often active chromatin sites than expected by the genome-wide distribution of chromatin states, and vice versa, repressed promoters contact repressed chromatin sites more often, in line with the findings reported for the HoxD cluster (Noordermeer et al. 2011, 2014; Vieux-Rochas et al. 2015).
Identification of putative enhancers and their associated target genes
A fraction of the detected interactions was found in enhancer like-domains (Active-A, Active-B, and Active-C chromatin segments). We used this co-occurrence to assign potential enhancer regions to their associated promoters by filtering for interacting regions with high H3K27ac and H3K4me1 levels. We decreased the threshold for interactions to the 95th percentile, as we hypothesized that enhancer-promoter interactions can be labile. Moreover, in some cases, the interaction might be limited to a subset of cells in the tissues leading to a diluted signal. We did not exclude putative enhancers annotated in promoter regions bearing similar levels of H3K27ac and H3K4me1, as enhancers might also be located in close proximity to active promoters. Overall, we observed that only 20% of all called interactions show histone modification patterns of enhancers (Supplemental Table S3).
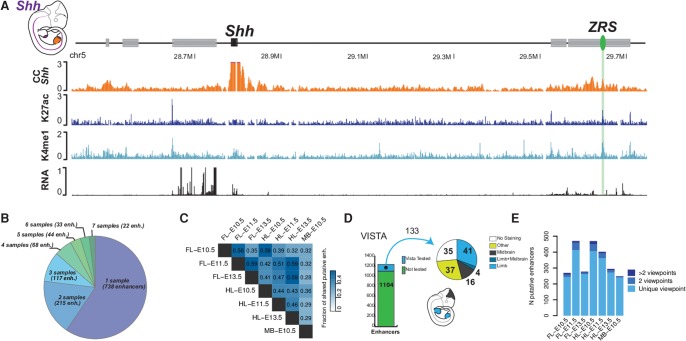
Using this approach, we were able to identify the ZPA Regulatory Sequence (ZRS) as the only limb enhancer of sonic hedgehog (Shh), despite the highly restricted expression of Shh in the limb bud, thereby demonstrating the high sensitivity of our approach (Fig. 3A). At the Ptch1 and HoxD loci, we identified previously described enhancers but also new putative regulatory regions (Supplemental Fig. S8A,B).
Figure 3.
Annotation of enhancers at limb-associated loci. (A) The ZRS (green oval) is the only limb enhancer called at the Shh locus. The expression pattern of Shh is indicated in purple in the embryo at the upper left corner of the scheme. (B) Number of tissues and/or time points (samples) where a specific putative enhancer is identified. Most of the putative enhancers are found solely in one sample and only a limited fraction is found in all of them. (C) Pairwise comparisons of enhancers’ reproducibility between tissues. The numbers represent the fraction of enhancers predicted in both tissues and/or time points. (D) Approximately 10% of the predicted limb enhancers have been tested in a LacZ assay by the VISTA consortium. (E) Some enhancers interact with different viewpoints, suggesting a shared regulatory activity.
We found that the large majority of putative enhancer regions were identified only in a single tissue and time point and only a minority of regions showed stable interactions along with H3K4me1 and H3K27ac coverage throughout different stages and tissues (Fig. 3B). When the candidate enhancer list of each tissue and time point was compared in a pairwise manner, we observed, as expected, similarities between forelimb and hindlimb samples and more differences between the limb and midbrain tissues. In line with the transcriptome analysis, we observed that forelimb and hindlimb of the same stages were more similar to each other than limb tissues of different developmental stages (Figs. 1B, 3C).
By combining and merging the results of the enhancer prediction for fore- and hindlimb as well as midbrain, we generated a list of 1237 nonoverlapping putative enhancer regions (Supplemental Table S4). We then compared these candidates to regions tested in the VISTA enhancer database and found that, out of the 1237 regions, 133 had been tested in a transgenic assay (Visel et al. 2007). In these assays, 41 were positive for limb staining, 16 for midbrain staining, and four for both limb and midbrain staining at embryonic stage E11.5 (Fig. 3D). Next, we inspected the enhancer prediction in specific samples, i.e., forelimb and hindlimb at E11.5 and midbrain at E10.5. As expected, we observed that predicted limb enhancers frequently triggered a limb staining, and midbrain enhancers were more likely to trigger a midbrain staining, thus further pointing to a functional role of these elements (Supplemental Fig. S9).
Among the surveyed tissues and time points, we found 3%–15% of putative enhancers associate with more than one promoter and thus act like shared putative regulatory elements (Fig. 3E; Supplemental Table S5). Among these putative enhancers, a majority interacts with clustered genes such as the HoxD, HoxC, HoxA, or Dlx5/6, which are regulated via shared enhancers (Birnbaum et al. 2012; Lonfat and Duboule 2015). Furthermore, we found that several alternative promoters at the Tcf4, Fgf10, and Rara loci also interact with the same putative regulatory region. This observation suggests that the various transcriptional isoforms, which originate from different promoters, benefit from a partially overlapping regulatory activity. Surprisingly, we found some putative enhancers are shared between apparently unrelated genes. Some of these latter genes showed a positive correlation of their transcriptional dynamics across tissues and time points (Supplemental Table S5). This was observed for insulin-like growth factor 2 (Igf2) and troponin I, skeletal, fast 2 (Tnni2) (Pearson's correlation coefficient: 0.88), which interact with a set of putative enhancers located between both loci in various tissues and stages. In this particular case, both genes are involved in muscle development and could benefit from a common synchronized regulation (Florini et al. 1996; Londhe and Davie 2011). Even though an overlap of putative shared elements could also occur by coincidence and a strong correlation is only observed in some cases, it is possible that some genes benefit from a shared regulation, synchronizing their transcriptional activity in a specific tissue and/or at a certain time point.
Chromatin interactions associate with CTCF and RAD21
CTCF and Cohesin have been shown to associate with chromatin folding and formation of distal interactions (Merkenschlager and Nora 2016). To examine the association of these proteins with interactions identified in the CC profiles, we produced binding profiles for CTCF and the Cohesin subunit RAD21 for all tissues and time points. In line with other studies, we found that 80% of RAD21 peaks associated with CTCF binding sites but, vice versa, that only 30% of CTCF sites associated with RAD21 (Supplemental Fig. S10; Nitzsche et al. 2011).
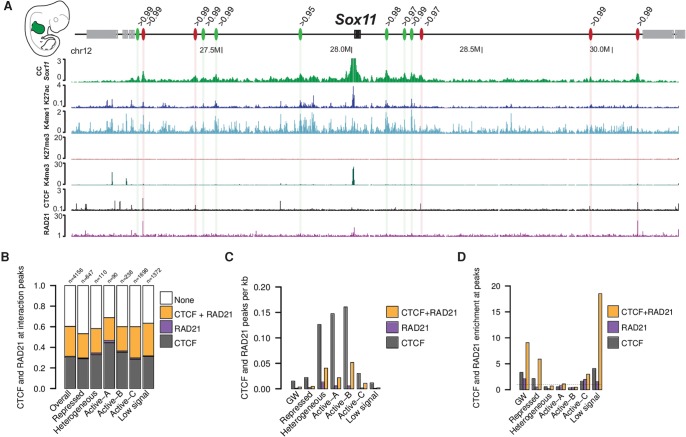
As exemplified at the Sox11 locus, we observed that several interactions are established at CTCF and RAD21 binding sites, whereas others were depleted for the binding of these proteins (Fig. 4A). Across our data set, we found that 60% of the interaction peaks were associated with CTCF/RAD21, CTCF only, or RAD21 only, independent of their association to a specific chromatin state (Fig. 4B). This finding also indicates that at least 40% of interactions could not be related to a CTCF/RAD21-based mechanism.
Figure 4.
Association of interactions with CTCF and RAD21. (A) Several distal interactions are formed at the Sox11 locus in the forelimb at E11.5. As some appear to be linked to CTCF/RAD21 (red ovals), others do not form at CTCF/RAD21 binding sites (green ovals) but rather at H3K27ac regions. The number above each oval indicates the percentile of the interaction intensity. (B) Fraction of interactions in which a peak for CTCF, RAD21, or both was called. “Overall” refers to all identified interactions, independently of the associated chromatin state in which they occur. These data are derived from the combined analysis from all tissues and time points. “n” indicates the number of interactions found in every chromatin state independently. (C) Number of CTCF/RAD21 peaks per kilobase genome-wide and for each chromatin state independently. These data are aggregated from all tissues and time points. (D) Peaks for CTCF, RAD21, and cobound CTCF/RAD21 sites were counted at interaction peaks and divided by the size of the peak regions. To calculate enrichment, we divided this density by the genome-wide and state-specific density shown in C. These data are aggregated from all tissues and time points.
To compute the enrichment of CTCF and RAD21 at interaction peaks, we first calculated the binding density of CTCF/RAD21, CTCF only, and RAD21 only genome-wide and within the different chromatin states. We found that the genome-wide binding density was relatively low, but higher in Active-A, Active-B, and heterogeneous chromatin regions, possibly because these states are frequently found at promoter-like regions, which are known to be enriched for CTCF (Fig. 4C; Barski et al. 2007). Next, we computed the enrichment of CTCF/RAD21 at interaction peaks with respect to the underlying chromatin state. We could observe that genome-wide cobinding of CTCF/RAD21 is more frequent at interaction peaks than expected by chance (Fig. 4D). This was especially prominent for interactions occurring in states with generally low CTCF/RAD21 density, such as low signal or repressed regions, showing an 18- and sixfold enrichment, respectively. In contrast, we observed slight depletion of CTCF/RAD21 at interaction peaks that occur in Active-A and Active-B and heterogeneous segments. This depletion is likely the result of the high background CTCF/RAD21 density found at these chromatin states (Fig. 4C). Altogether, we observed a strong association of CTCF and RAD21 binding with interaction peaks.
Stable and variable interactions associate with CTCF/RAD21 and functional chromatin, respectively
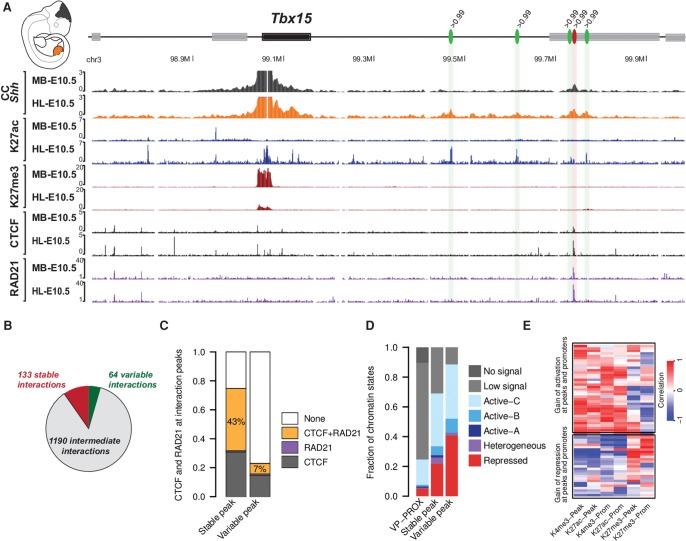
From the previous observations, we concluded that interactions associate with functional chromatin states and CTCF/RAD21 binding. However, it is unclear if these two types of associations relate to different chromatin interaction regimes. A first indication came from the observation that chromatin interactions change across tissues and developmental stages. At the Tbx15 locus, for example, one single interaction appeared stable in midbrain and in hindlimb at E10.5, whereas four other interactions were present solely in the hindlimb (Fig. 5A). Interestingly, the single stable interaction overlapped with a strong CTCF/RAD21 cobinding site, whereas the others were located at dynamic chromatin segments, which were specifically acetylated in the hindlimb and depleted of CTCF/RAD21 binding.
Figure 5.
Stable and variable interaction sites associate with CTCF/RAD21 and functional chromatin states, respectively. (A) At the Tbx15 locus, strong interaction peaks are formed with H3K27ac domains in early hindlimbs but are not present in the midbrain (green ovals), while another interaction seems stable and associates with CTCF/RAD21 (red oval). All the green peaks bear a signal strength >99th percentile in the hindlimb at E10.5; the red peak bears a signal strength >99th percentile in hindlimb and in midbrain at E10.5. (B) Number of stable, intermediate, and variable interactions. (C) Overlap of stable and variable interaction peaks with CTCF/RAD21 peaks. The percentage represents the fraction of cobinding CTCF/RAD21. (D) Distribution of chromatin states at stable and variable interaction peaks. (E) Heat map representing the Spearman correlation between interaction intensities at variable sites and corresponding H3K4me3, H3K27ac, H3K27me3 coverage at the linked promoter as well as the interaction site itself (see different columns).
To assess if CTCF/RAD21 were more strongly associated with constitutive interactions than variable ones, we classified interactions according to their stability. We defined stable interactions as interactions that were above the 99th percentile in all tissues and time points. In contrast, an interaction was considered to be variable when the interaction intensity was above the 99th percentile in at least one tissue and below the 80th percentile in at least two replicates in other tissues (Fig. 5B). Both groups define the more stringent cases of the stable and variable definition. Thus, most interaction sites are more ambiguously in between and were not considered in the following analyses.
We observed that stable interactions were more enriched with CTCF and RAD21 binding than the unstable ones. Specifically, 75% of the stable interactions but only 23% of the variable ones associated with CTCF alone, RAD21 alone, or CTCF/RAD21 (Fig. 5C). Moreover, the fraction of stable and variable interactions cobound by CTCF/RAD21 differed from 43% to 7%, respectively. In contrast, the association with functional chromatin states appeared to show an opposing trend. Here, variable interactions were more strongly associated with functional chromatin segments than stable interactions. These observations suggest that the formation of variable interactions might rely more on factors associated with specific chromatin modifications than on CTCF/RAD21 (Fig. 5D). We observed similar results when a single tissue, i.e., forelimb or hindlimb, was investigated across different developmental stages (E10.5 to E13.5) and for all tissues at the same developmental stage (E10.5). Thereby, stable and variable interactions follow the same respective associations, when they are defined by developmental stages or by tissue specificity (Supplemental Fig. S11).
To further test this hypothesis, we correlated the changes in interaction strength at variable interaction sites with the enrichment of chromatin modifications across tissues and time points. By doing so, we found two main patterns of correlation. In a first group, the interaction strength between the viewpoint and the interacting region correlated positively with the amount of active chromatin marks (i.e., H3K4me3/H3K27ac) and correlated negatively with the amount of repressive chromatin marks (i.e., H3K27me3) at the viewpoint and/or the interaction sites (Fig. 5E). In the second group, the interaction strength between the viewpoint and the interaction site correlated positively with the amount of H3K27me3 enrichment and negatively with the abundance of active marks at viewpoints and/or interaction site. Such correlations between interaction strength and amount of histone modifications relates to the observation that interactions occur in a homotypic fashion and that changes in the epigenetic status of any interaction partner associate with the interaction strength (see Fig. 2).
Altogether, we observe two distinct types of chromatin interactions and a mixture of both could be present at the intermediate interactions that were excluded from this analysis. On the one hand, stable interactions appear to be associated with CTCF/RAD21 binding, and on the other hand, variable interactions seem to associate more strongly with functional chromatin marks.
Discussion
The three-dimensional chromatin organization of the genome during embryonic development is critical for the accurate control of gene expression. In the past years, the extensive use of proximity-ligation methods to explore genomic architectures has strongly increased our knowledge of genome folding. However, the limited number of loci, cell types, or comparability between studies has made it difficult to draw general conclusions regarding locus-specific chromatin folding. In this work, we have examined the interaction dynamics at several hundreds of promoters at high resolution in a highly parallelized and quantitative manner using CC (Hughes et al. 2014). To annotate such interactions, we implemented an empirical background model and analyzed rather strong interactions to reduce the risk of false-positive interactions. By this strategy, we also miss capturing part of the complexity of chromatin folding. The background model scales individual profiles by a constant factor but does not account for local biases of the site interacting with the viewpoint, which might influence locally the propensity for capturing contacts. However, the comparison with another, recently published method (Cairns et al. 2016) recapitulates many of the identified interactions. Furthermore, we did not restrict the search for interactions to TADs to enable the annotation of inter-domain chromatin contacts. As the exact positions, sizes, and functions of TADs as well as their associated boundaries remain uncertain, we decided for a more unbiased approach and did not incorporate such annotations as prior knowledge to our model.
There are limitations to our enhancer identification, where the chosen threshold is a compromise between sensitivity and specificity. Indeed, weaker interactions with putative enhancer regions, as exemplified at the Shh locus (Fig. 3A), could be the result of signal dilution due to the inherent cell heterogeneity found in developing limb or midbrain tissues. An important fraction of genes selected for this study displays restricted or cell-type–specific expression patterns, and the detection of active and repressive chromatin modification, i.e., heterogeneous state, at their TSS could reflect it (Zeller et al. 2009; Diez-Roux et al. 2011; La Manno et al. 2016).
Among the 1237 putative enhancers linked to the 446 promoters included in this study, we found several which were contacted by several promoters and that are likely shared by several genes. The importance of global regulations has been extensively studied at gene clusters, such as Hox or beta-globin loci (de Laat and Duboule 2013). However, less is known about the coregulation of apparently unrelated genes or of alternative promoters. In the latter case, one might suppose that the selection of some promoters can allow for the usage of alternative transcripts in the same regulatory environment, including or excluding exons. In fact, enhancers regulating the Nprl3 gene were previously shown to influence the production of alternative transcript isoforms at the locus (Kowalczyk et al. 2012). Alternatively, it is possible that partially overlapping sets of enhancers control these alternative promoters, thus affecting the expression patterns of alternative transcripts. Functional approaches will be necessary to test the validity of these putative shared enhancers and the regulatory effect they trigger.
Then, by comparing different time points of development and tissues, we identified and characterized two regimes of interactions. First, we found a category of interactions which is stable and associates with CTCF/RAD21 cobinding events. As these interactions are stable across tissues and stages during embryogenesis, they might be essential to shape the 3D genome architecture and might correspond to anchor loops, which were found to delineate contact domains or TADs (Rao et al. 2014; Sanborn et al. 2015; Tang et al. 2015). Stable interactions could also correspond to intra-TAD interactions associated with CTCF and act as a scaffold for facultative enhancer-promoter interactions (Hadjur et al. 2009; de Laat and Duboule 2013; Phillips-Cremins et al. 2013; Seitan et al. 2013; Giorgetti et al. 2014). Future work will help to understand to what extent these stable chromatin interactions contribute to 3D segmentation of the genome, thereby working as a mold for more variable interactions to form. Moreover, we could observe that a majority of these stable interactions associate with repressive or active functional chromatin states. This observation suggests that pervasive interactions might occur at regulatory regions that interact with their associated promoters independently of their activity. Such regions intuitively appear as ideal sites to evolve regulatory sequences or to spatially cluster regions with similar function.
In contrast, we identified also more dynamic interactions which are less associated with CTCF/RAD21 but are strongly linked to functional chromatin. Our data suggest that the variable interactions rely on mechanisms directly associated with the chromatin state of both interacting partners, specifically the promoter and the interaction site. In this view, stable loops might be formed by the Cohesin-CTCF interactions, as formulated in the loop extrusion model, and variable ones could be the product of a “roadblock” of the Cohesin translocating complex induced by the presence of tissue-specific transcription factors and associated protein complexes (Sanborn et al. 2015; Fudenberg et al. 2016). However, we did not observe a strong enrichment of RAD21 binding at variable interaction peaks, and thus other members of the Cohesin complex should be assayed in the future. Alternatively, previous works have shed light on protein complexes associated with functional chromatin regions, which control long-range interactions. At active/enhancer regions, the Mediator complex is thought to sustain long-range interactions with associated promoters and could account for part of these CTCF-independent interactions (Phillips-Cremins et al. 2013). At repressed regions, the PRC1 and PRC2 complex were shown to mediate long-range interactions and could account for the homotypic bridging of H3K27me3-rich chromatin regions (Denholtz et al. 2013; Joshi et al. 2015; Schoenfelder et al. 2015). The controlled spatial clustering of repressed or active regions, analogous to the domain-wide clustering of A and B compartments on a larger scale (Lieberman-Aiden et al. 2009), could be seen as another layer of gene regulation. On the one hand, clustering of repressed regions through PRC1 and PRC2 could increase the reliability of repression. On the other hand, fine-tuned enhancer-promoter communication could allow for the transformation of rather unspecific enhancer activities into more subtle transcriptional output of their target genes, as observed at several developmental loci (Montavon et al. 2011; Andrey et al. 2013; Osterwalder et al. 2014).
Taken together, this in vivo data set provides a detailed characterization of 446 limb-associated regulatory landscapes, including their 3D chromatin folding and dynamics, as well as a compendium of predicted enhancers active during limb development. Finally, the two regimes of interaction presented in this work defined a framework for understanding regulatory architecture and chromatin dynamics during embryogenesis.
Methods
SureSelect design and Capture-C library
The library of SureSelect enrichment probes was designed over a 2- to 7-kb interval at the viewpoints using the online tool of Agilent: SureDesign (https://earray.chem.agilent.com/suredesign/index.htm) (Supplemental Table S6). The 3C library was prepared according to Hagege et al. (2007), i.e., crosslinking, cell lysis, DpnII digestion, ligation, and de-crosslinking. Religated products were then sheared using a Covaris sonicator (duty cycle: 10%, intensity: 5, cycles per burst: 200, time: six cycles of 60 sec each, set mode: frequency sweeping, temperature: 4°C–7°C). Adaptors were added to the sheared DNA and amplified. The adapted-ligated library was then hybridized to the custom-designed Sure-Select beads indexed for sequencing using an indexing PCR following Agilent instructions. The libraries were sequenced 100 bp paired-end. All Capture-C experiments were performed in biological duplicates.
Capture-C data processing and analysis
Generation of contact profiles
Paired-end sequencing data from Capture-C experiments was mapped and cleaned from invalid di-tags and duplicates using the HiCUP pipeline v0.5.8 (Wingett et al. 2015) (nofill:1, format: Sanger, without di-tag length restriction) and Bowtie 2.2.6 (Langmead and Salzberg 2012). All sequencing data were mapped to mouse reference genome mm9. Quality measurements of Capture-C experiments from the various HiCUP runs can be found in Supplemental Table S7. Viewpoint-specific contact profiles were generated from BAM files with custom Java code using htsjdk 1.139 (https://samtools.github.io/htsjdk/) for the processing of BAM files. A di-tag was considered only if MAPQ ≥ 30 for both mates and one mate was located in an enriched viewpoint region while the other mapped to the nonenriched part of the genome. Twenty-four viewpoints were excluded from further analyses because they yielded poor sequencing coverage or because they do not correspond to promoter regions, leading to 446 used viewpoints (Supplemental Table S8). Contact profiles were generated for each viewpoint by counting the number of reads per restriction fragment. Count data were binned into a regular grid of 1-kb intervals centered around the middle of the corresponding viewpoint region. The count value of each restriction fragment was assigned to the bin, which contains the fragment center. Count data were smoothed by averaging over a running window of five bins. To make contact profiles with different read densities comparable, each profile was scaled by the total contact count on the chromosome of the viewpoint (Sum of cis contacts/103). Additionally to the individual profiles for each replicate, replicates were also combined to merged profiles and binned, smoothed, and scaled as mentioned above.
Calling of interaction peaks: background model
Scaled contact profiles were pooled across all samples, viewpoints, and replicates to generate a simple, empirical background model, which incorporates the distance-dependent decay of the contact frequency. The positioning of the grid around the viewpoint center allows an overlay of all profiles by aligning their centers. The region considered for the analysis was ±2 Mb around the viewpoint. Now, each 1-kb bin can be described by its offset relative to the viewpoint center and has the contact information from 6244 profiles (seven tissues/stages × 446 viewpoints × two replicates). The 99th or 95th percentile of the empirical distribution was used to derive an individual threshold for each bin. A spline function was used to smooth the resulting threshold line. To avoid the calling of spurious interactions due to a lower profile density at locations distant from the viewpoint, the threshold line was fixed and set constant for all regions >0.5 Mb away from the viewpoint (Fig. 1C). A bin was selected and added to the candidate regions, when the contact value exceeded the threshold in both replicates. Within all candidate regions, the local maxima from the merged profile were used to determine peak summits. A region ±10 kb around the selected peak was excluded from the candidate regions to suppress peaks nearby. The selection of maxima and exclusion of neighborhood was repeated until no further candidates were left.
Calling of interaction peaks: CHiCAGO
The software package CHiCAGO (Cairns et al. 2016) was used to further validate interactions identified in this study by an alternative approach. CHiCAGO applies a statistical model, which accounts for viewpoint (bait)-specific biases, for biases of the interacting fragment, and for technical noise, and corrects for multiple testing. However, the characteristics of our data set deviate from those the tool was originally applied on. Instead of more than 20,000 viewpoints located on individual restriction fragments, we enriched only several hundred viewpoints spanning several restriction fragments. Furthermore, instead of a 6-bp cutter, we used a 4-bp cutter (DpnII), which yields much shorter restriction fragments, accumulating less sequencing reads. This leads to a different proportion of viewpoints and fragments for which biases have to be estimated. To compensate at least partially for this, we created virtual restriction fragments spanning five adjacent DpnII fragments. All DpnII fragments overlapping viewpoints were also merged into single virtual fragments. Both replicates were used together as input for the detection of interactions. We ran CHiCAGO with default parameters and used a score of ≥5 to filter for significant interactions. Overlaps with peaks from the background model were computed by extending interactions identified by CHiCAGO by ±5 kb and counted, when overlapping at least 1 bp.
ChIP-seq
For chromatin modifications, chromatin was prepared from the different tissues with a 1% formaldehyde crosslinking for 15 min and resuspended in buffer 3 for sonication (Lee et al. 2006). For CTCF and RAD21 ChIP-seq, we prepared chromatin as follows: Tissues were disrupted in 0.1% collagenase at 37°C and homogenized using a needle. Cells were then centrifuged and resuspended in 10% FCS, 0.2% Cs, 1% L-Glu, 0.5% Pen-Strep in DMEM:HAM's F-12 1:1 and fixed in 1% FA for 10 min on ice. Cells were then lysed in buffer 1 and 2 and resuspended in buffer 3 for sonication (Lee et al. 2006). We sheared chromatin using Bioruptor until reaching a fragment size of 200–500 bp. Ten to 15 micrograms of chromatin were then used for each replicate chromatin modification ChIP and 30 µg for CTCF and RAD21 ChIP. ChIP for H3K4me1 (Abcam: 8898), H3K4me3 (Millipore: 07-473), H3K27ac (Diagenode: C1540174), H3K27me3 (Millipore: 07-449), CTCF (Active motif: 613111), and RAD21 (Abcam: ab992) was then performed as in Lee et al. (2006). All ChIP-seq experiments were performed in biological duplicates. Libraries were prepared using the Nextera adaptors and sequenced.
Single-end reads from ChIP-seq experiments were mapped with Bowtie 2.2.6 (Langmead and Salzberg 2012) to reference genome mm9. Mapped reads were filtered for mapping quality ≥ 10, and duplicates were removed. To compute ChIP enrichment and quality measurements, we used the SPP tools (version 1.13) and phantompeakqualtools (Kharchenko et al. 2008). Reads were extended (chromatin modifications: to 300 bp, CTCF: to 200 bp) and scaled by the total number of unique reads (total count of reads/106) to produce coverage tracks. For figure display purposes, replicate ChIP-seq tracks were merged. RAD21 ChIP-seq tracks were produced using the coverage function of the Q-PeakCaller (Hansen et al. 2015). Peaks of CTFC and Rad21 were called on merged replicates with the Q-PeakCaller. Peaks were filtered with a QES > 0.27 and > 0.3 for RAD21 and CTCF, respectively.
Chromatin segmentation
Mapped short read information from ChIP-seq experiments of histone modifications was used for chromatin segmentation by the EpiCSeg tool (Mammana and Chung 2015). The software splits the genome into a regular grid and assigns a state to each bin based on abundance and co-occurrence of histone marks. The number of states is a free parameter and was set to seven. The bin size was set to 200 bp and the shift of the single-end read position in the computation of the DNA fragment center was set to 150 bp. EpiCSeg allows the integration of replicates, as used here. Based on the matrix of histone mark counts, we manually assigned the labels repressed, heterogeneous, Active-A, Active-B, Active-C, low signal, and no signal to the states after segmentation.
Overlay of peaks and annotation of interaction peaks
Chromatin interaction peaks from different samples were considered to be overlapping when their distance was below 5 kb. The center of each peak was used to define the chromatin state associated with it.
Co-occurrence of interaction peaks and CTCF and RAD21 binding were computed by extending the center of the CC interaction peaks (±5 kb) and overlapping the region with called ChIP-seq peaks. Additionally, the density of CTCF/RAD21 peaks was determined genome-wide (overall) and within the different chromatin states separately. The enrichment was subsequently calculated as the ratio between the density measured at interaction peaks and the density observed genome-wide and for the chromatin states separately.
Putative enhancers regions were called in a window ±2 Mb around the viewpoints based on the combination of three thresholds: the 95th percentile of the empirical background of the bin for the interaction strength, the 99th percentile for H3K27ac, and the 95th percentile for H3K4me1. Percentiles of the histone modifications were computed based on genome-wide distribution of mapped reads within 5-kb windows. Annotation of genomic features, such as promoters, exons, and introns was taken from txdb.mmusculus.ucsc.mm9 package from Bioconductor (R package version 3.2.2) (http://bioconductor.org/packages/release/data/annotation/html/TxDb.Mmusculus.UCSC.mm9.knownGene.html). Promoters are defined here as region ±1 kb around transcript starts. In the case of a genomic region overlapping with more than one genomic feature, it was assigned exclusively to one class in the order promoter, exon, intron, and intergenic.
All analyses were conducted within R using packages GenomicRanges (Lawrence et al. 2013) for computing overlaps, and bamsignals (R package version 1.4.2) (http://bioconductor.org/packages/release/bioc/html/bamsignals.html) to count mapped short reads within genomic regions.
RNA-seq
RNA was extracted from the different tissues using an RNeasy kit from Qiagen, and a poly(A) preparation was performed followed by 50-bp paired-end sequencing. All RNA-seq experiments were performed in biological duplicates. We mapped paired-end reads using the STAR mapper (Dobin et al. 2013). Read counts were generated using custom R scripts. For assessing changes in expression, we used DESeq2 (Love et al. 2014) with default parameters to perform pairwise comparison between samples using both replicates each. We considered genes to be differentially expressed when the absolute value of the log2 fold change was >1 and the adjusted P-value below 0.01.
Data access
All sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE84795.
Supplementary Material
Acknowledgments
We thank Asita Stiege for technical assistance. G.A. is supported by an Advanced Postdoc.Mobility grant from the Swiss National Science Foundation. This research was supported by a grant from the Deutsche Forschungsgemeinschaft and a European Community's Seventh Framework Programme, grant agreement no. 602300 (SYBIL) to S.M. We thank Francisca Martínez Real, Lila Allou, Darío G. Lupiáñez, and other members of the Mundlos laboratory for their critical reading of the manuscript.
Author contributions: G.A., R.S., M.V., and S.M. designed the research and wrote the manuscript. G.A. and C.P. produced the Capture-C and RNA-seq experiments. G.A. and D.M.I. produced the chromatin ChIP-seq experiments. I.J. did the CTCF and RAD21 ChIP-seq. S.H. provided support for RNA-seq analysis. R.S., V.H., and G.A. analyzed the data sets. M.H. and B.T. produced the high-throughput sequencing data.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.213066.116.
References
- Acemel RD, Tena JJ, Irastorza-Azcarate I, Marletaz F, Gomez-Marin C, de la Calle-Mustienes E, Bertrand S, Diaz SG, Aldea D, Aury JM, et al. 2016. A single three-dimensional chromatin compartment in amphioxus indicates a stepwise evolution of vertebrate Hox bimodal regulation. Nat Genet 48: 336–341. [DOI] [PubMed] [Google Scholar]
- Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. 2013. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340: 1234167. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. [DOI] [PubMed] [Google Scholar]
- Bell DM, Leung KK, Wheatley SC, Ng LJ, Zhou S, Ling KW, Sham MH, Koopman P, Tam PP, Cheah KS. 1997. SOX9 directly regulates the type-II collagen gene. Nat Genet 16: 174–178. [DOI] [PubMed] [Google Scholar]
- Birnbaum RY, Everman DB, Murphy KK, Gurrieri F, Schwartz CE, Ahituv N. 2012. Functional characterization of tissue-specific enhancers in the DLX5/6 locus. Hum Mol Genet 21: 4930–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J, Freire-Pritchett P, Wingett SW, Varnai C, Dimond A, Plagnol V, Zerbino D, Schoenfelder S, Javierre BM, Osborne C, et al. 2016. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol 17: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W, Duboule D. 2013. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 502: 499–506. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295: 1306–1311. [DOI] [PubMed] [Google Scholar]
- Denholtz M, Bonora G, Chronis C, Splinter E, de Laat W, Ernst J, Pellegrini M, Plath K. 2013. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and Polycomb proteins in genome organization. Cell Stem Cell 13: 602–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, et al. 2011. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol 9: e1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518: 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florini JR, Ewton DZ, Coolican SA. 1996. Growth hormone and the insulin-like growth factor system in myogenesis. Endocr Rev 17: 481–517. [DOI] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. 2016. Formation of chromosomal domains by loop extrusion. Cell Rep 15: 2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, Huber W, Furlong EE. 2014. Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512: 96–100. [DOI] [PubMed] [Google Scholar]
- Giorgetti L, Galupa R, Nora EP, Piolot T, Lam F, Dekker J, Tiana G, Heard E. 2014. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell 157: 950–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, Chen CJ, Kaplan N, Chang HY, Heard E, et al. 2016. Structural organization of the inactive X chromosome in the mouse. Nature 535: 575–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. 2009. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460: 410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T. 2007. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc 2: 1722–1733. [DOI] [PubMed] [Google Scholar]
- Hansen P, Hecht J, Ibrahim DM, Krannich A, Truss M, Robinson PN. 2015. Saturation analysis of ChIP-seq data for reproducible identification of binding peaks. Genome Res 25: 1391–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JR, Roberts N, McGowan S, Hay D, Giannoulatou E, Lynch M, De Gobbi M, Taylor S, Gibbons R, Higgs DR. 2014. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat Genet 46: 205–212. [DOI] [PubMed] [Google Scholar]
- Joshi O, Wang SY, Kuznetsova T, Atlasi Y, Peng T, Fabre PJ, Habibi E, Shaik J, Saeed S, Handoko L, et al. 2015. Dynamic reorganization of extremely long-range promoter-promoter interactions between two states of pluripotency. Cell Stem Cell 17: 748–757. [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, Park PJ. 2008. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26: 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk MS, Hughes JR, Garrick D, Lynch MD, Sharpe JA, Sloane-Stanley JA, McGowan SJ, De Gobbi M, Hosseini M, Vernimmen D, et al. 2012. Intragenic enhancers act as alternative promoters. Mol Cell 45: 447–458. [DOI] [PubMed] [Google Scholar]
- La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, Borm LE, Stott SR, Toledo EM, Villaescusa JC, et al. 2016. Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167: 566–580.e519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9: e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc 1: 729–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. 2003. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12: 1725–1735. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londhe P, Davie JK. 2011. Γ interferon modulates myogenesis through the major histocompatibility complex class II transactivator, CIITA. Mol Cell Biol 31: 2854–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonfat N, Duboule D. 2015. Structure, function and evolution of topologically associating domains (TADs) at HOX loci. FEBS Lett 589(20 Pt A): 2869–2876. [DOI] [PubMed] [Google Scholar]
- Lonfat N, Montavon T, Darbellay F, Gitto S, Duboule D. 2014. Convergent evolution of complex regulatory landscapes and pleiotropy at Hox loci. Science 346: 1004–1006. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammana A, Chung HR. 2015. Chromatin segmentation based on a probabilistic model for read counts explains a large portion of the epigenome. Genome Biol 16: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M, Nora EP. 2016. CTCF and cohesin in genome folding and transcriptional gene regulation. Annu Rev Genomics Hum Genet 17: 17–43. [DOI] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. 2011. A regulatory archipelago controls Hox genes transcription in digits. Cell 147: 1132–1145. [DOI] [PubMed] [Google Scholar]
- Montavon T, Thevenet L, Duboule D. 2012. Impact of copy number variations (CNVs) on long-range gene regulation at the HoxD locus. Proc Natl Acad Sci 109: 20204–20211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitzsche A, Paszkowski-Rogacz M, Matarese F, Janssen-Megens EM, Hubner NC, Schulz H, de Vries I, Ding L, Huebner N, Mann M, et al. 2011. RAD21 cooperates with pluripotency transcription factors in the maintenance of embryonic stem cell identity. PLoS One 6: e19470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. 2011. The dynamic architecture of Hox gene clusters. Science 334: 222–225. [DOI] [PubMed] [Google Scholar]
- Noordermeer D, Leleu M, Schorderet P, Joye E, Chabaud F, Duboule D. 2014. Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci. eLife 3: e02557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder M, Speziale D, Shoukry M, Mohan R, Ivanek R, Kohler M, Beisel C, Wen X, Scales SJ, Christoffels VM, et al. 2014. HAND2 targets define a network of transcriptional regulators that compartmentalize the early limb bud mesenchyme. Dev Cell 31: 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. 2013. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153: 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. 2015. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci 112: E6456–E6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Sugar R, Dimond A, Javierre BM, Armstrong H, Mifsud B, Dimitrova E, Matheson L, Tavares-Cadete F, Furlan-Magaril M, et al. 2015. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat Genet 47: 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Faure AJ, Zhan Y, McCord RP, Lajoie BR, Ing-Simmons E, Lenhard B, Giorgetti L, Heard E, Fisher AG, et al. 2013. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res 23: 2066–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148: 458–472. [DOI] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, et al. 2015. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163: 1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieux-Rochas M, Fabre PJ, Leleu M, Duboule D, Noordermeer D. 2015. Clustering of mammalian Hox genes with other H3K27me3 targets within an active nuclear domain. Proc Natl Acad Sci 112: 4672–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. 2007. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res 35: D88–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingett S, Ewels P, Furlan-Magaril M, Nagano T, Schoenfelder S, Fraser P, Andrews S. 2015. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res 4: 1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller R, Lopez-Rios J, Zuniga A. 2009. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat Rev Genet 10: 845–858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.