ABSTRACT
Venezuelan equine encephalitis virus (VEEV) is an important human and animal alphavirus pathogen transmitted by mosquitoes. The virus is endemic in Central and South America, but has also caused equine outbreaks in southwestern areas of the United States. In an effort to better understand the molecular mechanisms of the development of immunity to this important pathogen, we performed transcriptional analysis from whole, unfractionated human blood of patients who had been immunized with the live-attenuated vaccine strain of VEEV, TC-83. We compared changes in the transcriptome between naïve individuals who were mock vaccinated with saline to responses of individuals who received TC-83. Significant transcriptional changes were noted at days 2, 7, and 14 following vaccination. The top canonical pathways revealed at early and intermediate time points (days 2 and 7) included the involvement of the classic interferon response, interferon-response factors, activation of pattern recognition receptors, and engagement of the inflammasome. By day 14, the top canonical pathways included oxidative phosphorylation, the protein ubiquitination pathway, natural killer cell signaling, and B-cell development. Biomarkers were identified that differentiate between vaccinees and control subjects, at early, intermediate, and late stages of the development of immunity as well as markers which were common to all 3 stages following vaccination but distinct from the sham-vaccinated control subjects. The study represents a novel examination of molecular processes that lead to the development of immunity against VEEV in humans and which may be of value as diagnostic targets, to enhance modern vaccine design, or molecular correlates of protection.
KEYWORDS: biomarker, gene expression, microarray, transcriptome, Venezuelan equine encephalitis virus, vaccination
Introduction
Venezuelan equine encephalitis virus (VEEV) is a single-stranded, positive sense RNA virus and a member of the Alphavirus genus of the family Togaviridae. Among the New World alphaviruses, VEEV is considered to be one of the most pathogenic for humans.1 VEEV is classified as a Category B biological threat agent by the Centers for Disease Control (CDC) and has reportedly been developed as a biological weapon in the past.2,3 The virus is highly infectious by the aerosol or inhalational route, and incidental infection has been problematic to laboratory personnel due to accidental exposures.4,5 Typical disease cases present with flu-like symptoms, including fever, chills, headache, and malaise.4 Encephalitis occurs in a small percentage of cases, and most often in children; additional symptoms of severe disease include severe headache, photophobia, ataxia, disorientation, and convulsions.6
Currently, there is no FDA-approved vaccine or therapeutic available for the prevention or treatment of Venezuelan equine encephalitis (VEE). However, there are 2 investigational new drug (IND) vaccines that are available for at-risk laboratory personnel.7 The first vaccine, TC-83, is a live-attenuated virus developed in 1961 by serial passage of the virulent Trinidad strain of VEEV though tissue culture in fetal guinea pig heart cells.8 The second, C-84, is a formalin-inactivated version of the TC-83 strain.9 Live-attenuated TC-83 has been used extensively in humans and has demonstrated high protective data as measured by the production of neutralizing antibodies; however, the rate of NonResponders is approximately 20–25%, as measured by the failure to produce neutralizing antibodies against VEEV following immunization.7 In addition to naturally-occurring nonresponse, there have been demonstrations of immune interference contributing to the lack of neutralizing antibody production for individuals receiving sequential alphavirus immunizations, including circumstances when individuals received eastern equine encephalitis virus (EEEV), western equine encephalitis virus (WEEV), or Chikungunya virus (CHIK) prior to immunization with VEEV.7,10
In recent years, the reemergence of VEEV has prompted public health concern and highlighted the persistent need to develop modern vaccines which can achieve FDA-approval or to develop effective therapeutics which can be licensed. Additionally, high rates of primary vaccine failure as well as evidence of sexually dimorphic responses to vaccination are compelling reasons that there is a current need to develop modern, rational vaccines against VEEV. However, there are few studies to date which have been conducted to assess the molecular responses to VEEV. Host transcriptional responses to VEEV have been reported in a small number of animal model systems (mice and cynomolgus macaques) and in one in vitro study of human PBMC cells.11-17 Induction of chemokine transcripts in human PBMCs infected with the live-attenuated strain of VEEV (TC-83) was noted in vitro by the increased expression of CXCL11, CCL3, CCL5, CCL7, and CCRL2 in both naïve and responder PBMC samples from human volunteers who were either VEEV vaccine-naïve or had previously presented titers in response to VEEV vaccination.17 A strong interferon-driven response was observed, with increased transcript expression noted for IFNB1, IFNG, IRF7, several forms of OAS transcripts, MX1, MX2, and STAT1.17 Other notable patterns of transcript expression that have been previously reported include widespread engagement of signaling moieties that are key players in pattern recognition receptor (PRR) detection of bacteria and viruses, including such transcripts as IL6, DDX58, TLR3, TLR7, and CASP1.17
The purpose of the present study is to examine the molecular changes that occur in humans in response to VEEV TC-83 immunization with the overarching goal to provide an in-depth analysis of the molecular events which contribute to the development of immunity, and hence may inform any attempts to design a more effective vaccine or therapeutic. Furthermore, there are significant gaps in the foundation of knowledge surrounding the host cell signaling pathways required to combat viral infection and propagation.5 To that end, we have conducted a whole transcriptome analysis of human genes which are modulated in response to VEEV immunization; samples were derived from whole, unfractionated blood at various time points, both before and after immunization, and were compared with sex- and age-matched control samples at each time point.
Results
The study consisted of 2 groups of study volunteers (n = 10 per group) who were either mock-vaccinated or vaccinated with VEEV TC-83. Blood samples were collected prior to vaccination and then serially over time after vaccination. Transcriptome analysis was conducted on RNA isolated from whole, unfractionated blood.
Overall effects of immunization with TC-83 over time
Comparison of global gene expression values across time (i.e., at 1, 4, 8 hrs, and at days 1, 2, 7, 14, 21, and 28 postvaccination), in response to treatment, and as a function of both time and treatment concomitantly, yielded results that met statistical significance criteria (cut-off p-value) at days 2, 7, and 14 postvaccination when measured against time-matched control samples (Table 1). The false discovery rate was set to the limit of 10% using the Step-up multiple test correction method. Data were then further constrained by examining the fold change of gene expression of each transcript; only transcripts with a fold change of ≥ 2 (in either direction) were included in further analyses (Fig. 1). The first time point where a difference in gene expression in the TC-83 vaccinated individuals could be detected, relative to sham-vaccinated control subjects, was at day 2 (Fig. 1). Of the 225 transcripts that were differentially expressed on day 2 with at least a 2-fold change in expression, only 32 overlapped with transcripts at both days 7 and 14. On day 7, we detected 14 differentially expressed transcripts unique to day 7; while, on day 14 postvaccination, 756 transcripts were detected and unique to this time point. The graphical interactions displayed by Venn diagramming show a clear distinction in gene expression across time; from these results, the patterns of gene expression were stratified based on early (day 2), intermediate (day 7), and late (day 14) response to vaccination (Fig. 1).
Table 1.
False discovery rate report.
| FDR Report | ||
|---|---|---|
| Significance Level: 0.1; Total number of p-values: 54675 | ||
| Method: Step Up | ||
| Variable Name | Cutoff Value | # of Significant p-values |
| p-value(Time Point) | 1.49E-02 | 8,128 |
| p-value(Treatment) | 1.10E-05 | 6 |
| p-value(Time Point * Treatment) | 1.84E-02 | 10,055 |
| p-value(0 h * Vaccine vs. 0 h * Control) | 1.83E-06 | 0 |
| p-value(1 h * Vaccine vs. 1 h * Control) | 1.83E-06 | 0 |
| p-value(4 h * Vaccine vs. 4 h * Control) | 1.83E-06 | 0 |
| p-value(8 h * Vaccine vs. 8 h * Control) | 1.83E-06 | 0 |
| p-value(day 1 * Vaccine vs. day 1 * Control) | 1.83E-06 | 0 |
| p-value(day 2 * Vaccine vs. day 2 * Control) | 6.42E-03 | 3,511 |
| p-value(day 7 * Vaccine vs. day 7 * Control) | 7.75E-04 | 424 |
| p-value(day 14 * Vaccine vs. day 14 * Control) | 3.90E-02 | 21,343 |
| p-value(day 21 * Vaccine vs. day 21 * Control) | 1.83E-06 | 0 |
| p-value(day 28 * Vaccine vs. day 28 * Control) | 1.83E-06 | 0 |
Figure 1.
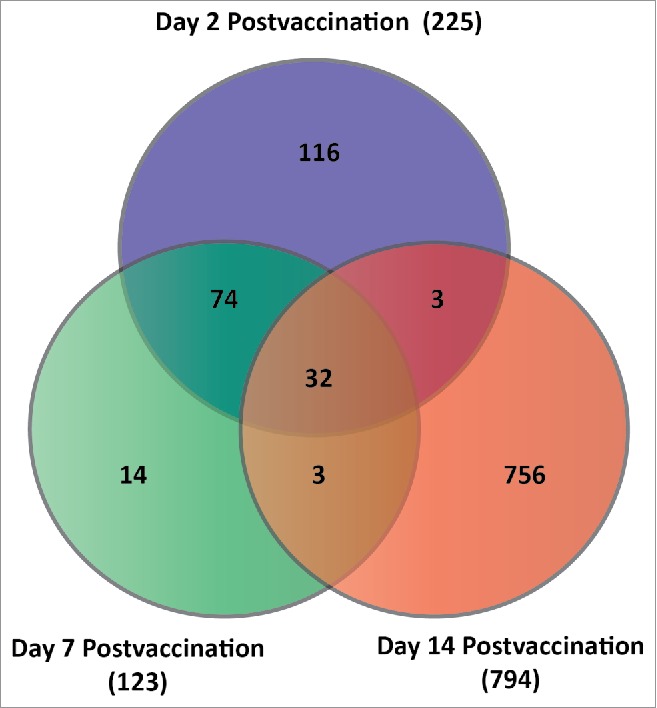
Venn diagram depicting the number of transcripts that were differentially expressed at day 2, day 7, and day 14 post-immunization. The common and unique transcripts shown are indicative of those that were statistically significant (FDR-corrected Step-up p-value ≤ 0.1) as well meeting a minimum criteria of a 2-fold change in gene expression (either up or down) over baseline levels of expression.
Cellular pathway analysis for changes in the transcriptome induced by immunization with TC-83 in humans
We conducted pathway analysis using Ingenuity Pathway Analysis software (Ingenuity, Redwood City, CA) to better understand the scope and function of the molecular responses generated in humans in response to TC-83 vaccination. The observed host responses covered a variety of pathways involved in disease processes, molecular and cellular functions, and physiology system development and function (Table 2). On day 2 postvaccination, representing the early transcriptional response, there was noted involvement of specific transcripts which were indicative of a strong antimicrobial and inflammatory response, as well as transcripts that were characteristic of infectious disease, infection mechanisms, and organismal injury (Table 2). The molecular functions related to transcripts which were differentially expressed on day 2 represented cellular movement and development, cellular signaling, post-translational modification, and protein folding. These molecular functions routinely participate in the systemic organization of hematological function, immune cell trafficking, tissue development, muscular-skeletal development, and hematopoiesis. The most prominent signaling pathways induced upon VEEV vaccination included the interferon signaling pathway, activation of interferon-response factors by cytosolic pattern recognition receptors, involvement of pattern recognition receptors in the recognition of viruses and bacteria, the RIG1-like receptors as part of a classical innate antiviral immune response (i.e., the inflammasome), and the IL-6 signaling pathway. The responses observed at day 7 postvaccination were similar to those seen at day 2 with regard to a clear induction of infectious, inflammatory, and antimicrobial responses. In addition to induction of molecular and cellular functions (e.g., post-translational modification), cellular development and protein folding functions were also observed at day 2. In contrast, by day 7 postvaccination, molecular functions expanded to transcripts related to lipid metabolism and molecular transport. Similarly, overlapping physiological system functions relating to hematological development, hematopoiesis, immune cell trafficking, and muscular-skeletal development continued to be top factors through day 7 postvaccination. However, an evolving response was evident by the induction of transcripts involved with endocrine system development and function. Likewise, the top canonical pathways that were observed on day 7 postvaccination were predominantly similar to those seen at day 2 (i.e., interferon signaling, pattern recognition receptor activation of interferon-response factors, inflammasome-related transcripts) but also included transcripts which were involved in the pathogenesis of multiple sclerosis. Two parameters characterize a dramatic shift in the results from day 14 postvaccination: First, the molecular and cellular processes observed primarily involved those of nucleic acid metabolism, cell to cell signaling, cellular compromise, gene expression, and molecular transport; and secondly, the top canonical pathways shifted from a strong interferon-driven response to one characterized by oxidative phosphorylation transcripts, protein ubiquitination, RAN signaling, T cell receptor signaling, and regulation of eIF4 and p70S6K signaling (Table 2).
Table 2.
Summary of ingenuity pathway analysis.
| TOP BIO FUNCTIONS | VEE Day 2 | VEE Day 7 | VEE Day 14 |
|---|---|---|---|
| Diseases and Disorders | Antimicrobial Response | Organismal Injury and Abnormalities | Immunological Disease |
| Inflammatory Response | Antimicrobial Response | Hematological Disease | |
| Organismal Injury and Abnormalities | Inflammatory Response | Cancer | |
| Infection Mechanism | Infection Mechanism | Reproductive System Disease | |
| Infectious Disease | Genetic Disorder | Genetic Disorder | |
| Molecular & Cellular Function | Cellular Movement | Post-Translational Modification | Nucleic Acid Metabolism |
| Cellular Development | Protein Folding | Cell-to-Cell Signaling and Interaction | |
| Cell-to-Cell Signaling and Interaction | Cellular Development | Cellular Compromise | |
| Post-Translational Modification | Lipid Metabolism | Gene Expression | |
| Protein Folding | Molecular Transport | Molecular Transport | |
| Physiology System Development & Function | Hematological System Development and Function | Endocrine System Development and Function | Tissue Development |
| Immune Cell Trafficking | Hematological System Development and Function | Tumor Morphology | |
| Tissue Development | Hematopoiesis | Immune Cell Trafficking | |
| Skeletal and Muscular System Development and Function | Skeletal and Muscular System Development and Function | Nervous System Development and Function | |
| Hematopoiesis | Immune Cell Trafficking | Organ Morphology | |
| TOP CANONICAL PATHWAYS | Interferon Signaling | Interferon Signaling | Oxidative Phosphorylation |
| Activation of IRF by Cytosolic Pattern Recognition Receptors | Activation of IRF by Cytosolic Pattern Recognition Receptors | Protein Ubiquitination Pathway | |
| Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses | Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses | Regulation of eIF4 and p70S6K Signaling | |
| Role of RIG1-like Receptors in Antiviral Innate Immunity | Pathogenesis of Multiple Sclerosis | RAN Signaling | |
| IL-6 Signaling | Role of RIG1-like Receptors in Antiviral Innate Immunity | T Cell Receptor Signaling | |
| Top Molecules - UP | RSAD2, IFI44L, IFIT1, AMPK2, ISG15, LAMP3, IFI44, HERC5, MX1, OAS3 (includes EG:4940) | IFI27, RSAD2, IFI44L, IFI44, ISG15, CMPK2, IFIT1, OAS3 (includes EG:4940), HERC5, OAS1 | IFI27, IGJ, IGL@, IFI44, IGHM, IFI44L, RSAD2, TNFRSF17, TXNDC5, IGHA1 |
| Top Molecules - DOWN | FCER1A, IL8, ITM2A, SGK1, GRAMD1C, IRS2, CLC, THBD, IGF1R, FAM101B | PI3, TUBB2A, EPB42, SLC4A1, SNCA, IGF1R, MARCH8, CPA3, FAM101B, CCR3 | PI3, EPB42, TNS1, SLC4A1, TUBB2A, SELENBP1, SNCA, GMPR, KRT1, BLVRB, |
The involvement of specific canonical pathways in this temporal study of transcriptional expression allowed us to compare and describe 3 distinct phases of human VEEV infection in vivo. We employed Ingenuity Pathway Analysis (IPA) to describe the involvement of individual transcripts and canonical pathways in the development of immunity following TC-83 immunization. During the earliest phase (day 2 postvaccination) there was a strong induction of interferon signaling genes and subsequent interferon-related factors (Table 3). Some of the most notable transcripts representing interferon signaling included IFIT1, IFIT3, MX1, OAS1, and IFI34, and many of these transcripts continued to display increased expression through day 7 as well. However, by day 14 most interferon signaling transcripts had returned to baseline levels (Table 3). Similarly, activation of interferon related factors was evident by day 2, including genes comprising the inflammasome (RIG1, also known as DDX58; MDA5, also known as IFIH1; LGP2, also known as DHX58; and a novel DEXD/H box helicase, DDX60), (Table 3). Increased transcription of genes involved in IL-6 signal transduction was highest at day 2 (e.g., IL1RN, SOCS1, and TNFAIP6) with noted decrease in IL-8 transcription (Table 3). Key signaling components of the JAK/STAT pathway were also noted to have increased transcription at day 2 following immunization which was sustained through day 7, but returned to baseline levels by day 14 (e.g., SOCS1, STAT1, and STAT2) (Table 3). The induction of CXCL10 and CCR1 transcripts at early-to-intermediate time points represent potential convergence of several pathways including pathogenesis of multiple sclerosis and IL-17 signaling.
Table 3.
Top canonical pathways and genes.
| Pathway | Gene | Day 2 | Day 7 | Day 14 | |||
|---|---|---|---|---|---|---|---|
| Interferon Signaling | Fold Change | p-value* | Fold Change | p-value* | Fold Change | p-value* | |
| IFI35 | 3.86 | 2.94E−14 | 2.84 | 9.43E−09 | 1.33 | 1.01E−01 | |
| IFIT1 | 13.11 | 1.02E−06 | 10.95 | 2.53E−05 | 2.95 | 3.88E−02 | |
| IFIT3 | 7.03 | 3.47E−08 | 7.34 | 2.57E−10 | 1.88 | 7.64E−02 | |
| IFITM1 | 2.38 | 1.66E−07 | 2.35 | 1.30E−06 | 1.61 | 6.35E−03 | |
| MX1 | 9.93 | 3.69E−09 | 6.89 | 4.17E−06 | 1.71 | 1.73E−01 | |
| OAS1 | 7.18 | 2.32E−08 | 7.42 | 6.46E−08 | 2.71 | 7.01E−03 | |
| SOCS1 | 2.43 | 5.90E−09 | 1.58 | 4.65E−02 | −1.20 | 1.19E−01 | |
| Activation of IRF by Cytosolic Factors | DDX58 | 5.00 | 2.85E−09 | 2.79 | 1.86E−03 | 1.85 | 2.69E−02 |
| DHX58 | 2.00 | 9.01E−10 | 1.50 | 4.94E−03 | 1.03 | 8.26E−01 | |
| IFIH1 | 4.50 | 7.41E−08 | 3.03 | 8.02E−04 | 2.36 | 4.12E−03 | |
| IFIT2 | 5.54 | 1.67E−12 | 4.36 | 1.81E−04 | 1.89 | 6.96E−02 | |
| IRF7 | 3.70 | 2.35E−09 | 3.68 | 1.85E−08 | 1.40 | 1.27E−01 | |
| ISG15 | 11.39 | 3.77E−11 | 11.20 | 2.76E−10 | 2.66 | 1.07E−02 | |
| ZBP1 | 3.94 | 1.32E−12 | 2.85 | 3.73E−09 | 1.72 | 3.11E−03 | |
| Role of Pattern Recognition Receptors | DDX58 | 5.00 | 2.85E−09 | 2.79 | 1.86E−03 | 1.85 | 2.69E−02 |
| in Recognition of Bacteria and | EIF2AK2 | 3.27 | 8.67E−10 | 2.96 | 1.42E−07 | 1.35 | 1.25E−01 |
| Viruses | IFIH1 | 4.50 | 7.41E−08 | 3.03 | 8.02E−04 | 2.36 | 4.12E−03 |
| IRF7 | 3.70 | 2.35E−09 | 3.68 | 1.85E−08 | 1.40 | 1.27E−01 | |
| OAS2 | 5.35 | 4.31E−12 | 4.97 | 2.57E−10 | 2.24 | 1.42E−03 | |
| OAS3 | 9.07 | 3.35E−12 | 8.20 | 1.58E−10 | 2.27 | 1.22E−02 | |
| Role of RIG1-like Receptors in Antiviral | DDX58 | 5.00 | 2.85E-09 | 2.79 | 1.86E-03 | 1.85 | 2.69E-02 |
| Innate Immunity | DDX60 | 3.53 | 1.44E-04 | 4.43 | 1.18E-05 | 3.49 | 3.78E-04 |
| DHX58 | 2.00 | 9.01E-10 | 1.50 | 4.94E-03 | 1.03 | 8.26E-01 | |
| TLR7 | 1.58 | 1.39E-06 | 1.12 | 7.93E-01 | −1.10 | 3.08E-01 | |
| Interleukin-6 Signaling | IL8 | −3.11 | 2.25E−02 | 1.36 | 8.85E−01 | 1.74 | 1.62E−01 |
| IL1RN | 3.46 | 4.00E−09 | 2.28 | 1.20E−03 | −1.19 | 5.68E−02 | |
| TNFAIP6 | 3.87 | 2.39E−06 | 2.84 | 2.31E−03 | 1.53 | 2.19E−01 | |
| Communication between Innate and | CXCL10 | 3.79 | 1.31E−12 | 2.10 | 5.95E−04 | 1.21 | 3.10E−01 |
| Adaptive Immune Cells | IL1RN | 3.46 | 4.00E−09 | 2.28 | 1.20E−03 | −1.19 | 5.68E−02 |
| TNFSF13B | 2.48 | 1.70E−05 | 1.96 | 1.27E−02 | 1.35 | 2.47E−01 | |
| JAK/STAT Signaling | SOCS1 | 2.43 | 5.90E−09 | 1.58 | 4.65E−02 | −1.20 | 1.19E−01 |
| STAT1 | 2.67 | 1.59E−05 | 1.99 | 2.68E−02 | 1.57 | 1.89E−02 | |
| STAT2 | 2.83 | 5.50E−09 | 2.19 | 1.10E−04 | 1.46 | 3.75E−02 | |
| Pathogenesis of Multiple Sclerosis | CXCL10 | 3.79 | 1.31E-12 | 2.10 | 5.95E-04 | 1.21 | 3.10E-01 |
| CCR1 | 1.80 | 1.80E+00 | 2.19 | 2.76E-02 | 1.23 | 4.23E-01 | |
| Oxidative Phosphorylation | ATP5J | −1.30 | 4.94E-01 | 1.34 | 8.34E-01 | 2.20 | 9.53E-03 |
| COX7B | −1.40 | 5.53E-01 | 1.60 | 8.10E-01 | 2.20 | 6.43E-02 | |
| COX7A2 | −1.27 | 5.13E-01 | 1.43 | 7.64E-01 | 2.04 | 1.35E-02 | |
| COX6C | −1.16 | 7.86E-01 | 1.80 | 6.65E-01 | 2.50 | 1.59E-02 | |
| UQCRB | −1.24 | 7.46E-01 | 1.67 | 8.08E-01 | 2.32 | 7.13E-02 | |
| UQCRH | −1.13 | 7.88E-01 | 1.46 | 7.79E-01 | 2.35 | 7.71E-03 | |
| PPA1 | −1.06 | 8.71E-01 | 1.44 | 6.94E-01 | 2.23 | 2.18E-03 | |
| NDUFA6 | −1.25 | 5.26E-01 | 1.48 | 7.12E-01 | 2.11 | 7.78E-03 | |
| UQCRQ | −1.18 | 7.41E-01 | 1.56 | 7.68E-01 | 2.22 | 2.44E-02 | |
| Protein Ubiquitination Pathway | PSMA3 | 1.17 | 7.02E−01 | 1.43 | 7.86E−01 | 2.42 | 5.05E−03 |
| UBR1 | −1.27 | 5.23E−01 | 1.16 | 9.30E−01 | 2.48 | 3.00E−03 | |
| USP1 | −1.74 | 7.37E−02 | 1.06 | 9.70E−01 | 2.32 | 8.32E−03 | |
| UBE3A | −1.54 | 1.91E−01 | 1.18 | 8.48E−01 | 2.07 | 7.17E−03 | |
| USP53 | −1.08 | 6.88E−01 | 1.21 | 9.10E−01 | 2.28 | 9.92E−03 | |
| PSMC6 | −1.72 | 3.37E−01 | 1.20 | 9.48E−01 | 2.71 | 2.57E−02 | |
| USP47 | −1.56 | 1.99E−01 | 1.21 | 8.97E−01 | 2.11 | 8.24E−03 | |
| USP16 | −1.64 | 1.67E−01 | 1.22 | 8.66E−01 | 2.22 | 6.24E−03 | |
| PSMA4 | 1.10 | 8.61E−01 | 1.69 | 7.09E−01 | 2.54 | 1.08E−02 | |
| HSP90AA1 | −1.02 | 9.78E−01 | 1.51 | 7.94E−01 | 2.91 | 4.74E−03 | |
| BIRC3 | −1.50 | 2.52E−01 | 1.09 | 9.63E−01 | 2.12 | 1.62E−02 | |
| BIRC2 | −1.38 | 4.13E−01 | 1.08 | 9.70E−01 | 2.22 | 5.80E−03 | |
| Regulation of eIF4 and p70S6K | ITGB1 | −1.23 | 5.79E-01 | 1.13 | 9.43E-01 | 2.38 | 4.12E-03 |
| Signaling | RPS6KB1 | −1.43 | 5.41E-02 | 1.09 | 9.15E-01 | 1.49 | 8.51E-03 |
| NRAS | −1.24 | 4.26E-01 | 1.18 | 8.84E-01 | 2.08 | 1.65E-03 | |
| RRAS2 | −1.39 | 2.32E-01 | 1.17 | 8.92E-01 | 2.31 | 4.53E-04 | |
| PIK3C2A | −1.57 | 1.79E-01 | −1.03 | 9.88E-01 | 2.03 | 9.01E-03 | |
| EIF1AX | −1.34 | 3.85E-01 | 1.20 | 8.99E-01 | 2.13 | 5.97E-03 | |
| EIF4A2 | −1.41 | 2.65E-01 | 1.17 | 9.07E-01 | 2.07 | 4.48E-03 | |
| EIF3E | −1.63 | 2.45E-01 | 1.10 | 9.65E-01 | 2.10 | 2.54E-02 | |
| EIF4E | −1.28 | 5.05E-01 | 1.13 | 9.43E-01 | 2.10 | 1.05E-02 | |
| ITGA4 | −1.28 | 4.94E-01 | −1.00 | 1.00E+00 | 2.09 | 1.08E-02 | |
| Ran Signaling | CSE1L | −1.28 | 3.47E-01 | 1.26 | 8.05E-01 | 2.00 | 2.11E-03 |
| RANBP2 | −1.48 | 2.48E-01 | −1.04 | 9.81E-01 | 2.09 | 7.33E-03 | |
| RAN | −1.45 | 3.05E-01 | 1.30 | 8.51E-01 | 2.60 | 2.11E-03 | |
| ERK5 signaling | YWHAQ | −1.25 | 4.08E−01 | 1.20 | 8.27E−01 | 2.27 | 4.00E−04 |
| IL6ST | −1.43 | 2.98E−01 | 1.24 | 8.51E−01 | 2.07 | 8.76E−03 | |
| RPS6KB1 | −1.58 | 1.82E−01 | 1.09 | 9.15E−01 | 2.11 | 7.34E−03 | |
| NRAS | −1.24 | 4.26E−01 | 1.21 | 7.60E−01 | 2.08 | 1.65E−03 | |
| RRAS2 | −1.39 | 2.32E−01 | 1.17 | 8.92E−01 | 2.31 | 4.53E−04 | |
| ATF2 | −1.48 | 2.86E−01 | 1.04 | 9.27E−01 | 2.32 | 5.48E−03 | |
| Natural Killer Cell Signaling | FYN | −1.45 | 1.65E−01 | 1.22 | 7.18E−01 | 2.09 | 1.44E−03 |
| PRKCI | −1.31 | 3.57E−01 | 1.26 | 7.35E−01 | 2.04 | 3.45E−03 | |
| NRAS | −1.24 | 4.26E−01 | 1.21 | 7.60E−01 | 2.08 | 1.65E−03 | |
| RRAS2 | −1.39 | 2.32E−01 | 1.17 | 8.92E−01 | 2.31 | 4.53E−04 | |
| PIK3C2A | −1.57 | 1.79E−01 | 1.23 | 6.55E−01 | 2.03 | 9.01E−03 | |
| KLRK1 | −1.11 | 7.89E−01 | 1.21 | 8.78E−01 | 2.01 | 8.80E−03 | |
| B Cell Development | IL7R | nd | nd | nd | nd | 2.13 | 4.13E−03 |
| IGKC | −1.02 | 9.57E−01 | 1.04 | 9.79E−01 | 2.44 | 6.13E−04 | |
| IGL@ | 1.30 | 8.19E−02 | 1.29 | 8.52E−01 | 6.01 | 2.87E−12 | |
| IGHM | −1.48 | 2.48E−01 | −1.28 | 8.48E−01 | 4.99 | 7.60E−04 | |
| T Cell Receptor Signaling | FYN | −1.45 | 1.65E-01 | 1.13 | 9.13E-01 | 2.09 | 1.44E-03 |
| CD28 | −1.57 | 1.72E-01 | 1.05 | 9.79E-01 | 2.57 | 9.73E-04 | |
| CAMK4 | −1.86 | 5.56E-02 | 1.04 | 9.83E-01 | 2.10 | 4.89E-03 | |
| RRAS2 | −1.39 | 2.32E-01 | 1.17 | 8.92E-01 | 2.31 | 4.53E-04 | |
| PI3KC2A | −1.57 | 1.79E-01 | −1.03 | 9.88E-01 | 2.03 | 9.01E-03 | |
| RASGRIP1 | −1.48 | 1.70E-01 | 1.23 | 8.50E-01 | 2.44 | 3.48E-04 | |
| CD3D | −1.14 | 7.40E-01 | 1.50 | 7.02E-01 | 2.24 | 4.84E-03 | |
| ITK | −1.44 | 1.83E-01 | 1.15 | 9.04E-01 | 2.12 | 1.44E-03 | |
p-values shown are FDR-corrected Step-up p-values.
nd = not detected
Transcripts which are both statistically significant and meet a 2-fold change of expression are indicated in bold text.
In sharp contrast, the canonical pathways that are highly represented by transcripts with increased expression by day 14 post immunization include that of oxidative phosphorylation (e.g., COX7A2, COX16, UQCRB, UQCRH, PPA1), the protein ubiquitination pathway (e.g., UBR1, USP1, PSMA3, PSMC6, BIRC2, BIRC3, HSP90AA1), the ERK5 Signaling pathway (e.g., IL6ST, NRAS, RRAS2, ATF2), the Natural Killer Cell Signaling pathway (e.g., KLRC2, FYN, PRKC1, KLRK1, KLRC3, RRAS2, NRAS), and the B-Cell Development pathway (e.g., IL7R, IGKC, IGL@, IGHM) (Table 3). The complete ANOVA results are available in Supplemental Data Table 1.
Biomarker analysis and identification using ingenuity pathway analysis
The temporal transcriptional responses from TC-83 vaccinated subjects and mock vaccinated control subjects were evaluated using IPA's biomarker prediction algorithm to identify potential biomarkers following immunization. Biomarker prediction is a useful tool in the transcriptome profile analysis process as an avenue to further diagnostic target prediction, biomarker validation, and potential use as a biological signature. In the analysis, we employed filters to enrich for transcripts that have been shown in published literature to be present in biological fluids (e.g., blood, sera, plasma, and urine). The biomarker assessment tool in IPA produced groups of differentially expressed transcripts that we clustered into early (day 2), intermediate (day 7), and late (day 14) expression biomarkers (Table 4). Each of these groups of biomarkers were either uniquely expressed at each stage of immune development or were common across all days following immunization. The biomarkers displayed in Table 4 were selected by restricting the analysis to the top 10 transcripts showing the greatest change in transcript expression and for consistency among all potential probe sets which correspond to each transcript. We also included MicroRNA prediction using Ingenuity Pathway Analysis to suggest microRNA which may have a role in the complex regulation of gene expression in response to vaccination (Supplemental Data Table 2).
Table 4.
Top biomarkers.
| Gene symbol | Gene Name | Cellular Location | Fold Change | Day 2 | Day 7 | Day 14 | Common |
|---|---|---|---|---|---|---|---|
| GBP4 | guanylate binding protein 4 | Cytoplasm | 3.5 | X | |||
| MT1X | metallothionein 1X | unknown | 3.2 | X | |||
| ANKRD22 | ankyrin repeat domain 22 | Nucleus | 3.2 | X | |||
| CCL2 | chemokine (C-C motif) ligand 2 | Extracellular Space | 3.1 | X | |||
| BST2 | bone marrow stromal cell antigen 2 | Plasma Membrane | 3.0 | X | |||
| LIPA | lipase A, lysosomal acid, cholesterol esterase | Cytoplasm | 2.8 | X | |||
| TRIM14 | tripartite motif containing 14 | Cytoplasm | 2.3 | X | |||
| CCR1 | chemokine (C-C motif) receptor 1 | Plasma Membrane | 2.2 | X | |||
| PSMA5 | glucuronidase, β pseudogene | unknown | 2.2 | X | |||
| PPM1K | protein phosphatase, Mg2+/Mn2+ dependent, 1K | Cytoplasm | 2.1 | X | |||
| SHISA5 | shisa homolog 5 (Xenopus laevis) | Nucleus | 2.0 | X | |||
| C18orf49 | chromosome 18 open reading frame 49 | unknown | 2.0 | X | |||
| IGJ | immunoglobulin J polypeptide, linker protein for immunoglobulin α and mu polypeptides | Extracellular Space | 6.8 | X | |||
| IGL@ | immunoglobulin lambda locus | Nucleus | 6.0 | X | |||
| IGHM | immunoglobulin heavy constant mu | Plasma Membrane | 5.0 | X | |||
| TNFRSF17 | tumor necrosis factor receptor superfamily, member 17 | Plasma Membrane | 4.1 | X | |||
| TXNDC5 | thioredoxin domain containing 5 (endoplasmic reticulum) | Cytoplasm | 4.0 | X | |||
| NDUFA5 | NADH dehydrogenase (ubiquinone) 1 α subcomplex, 5 | Cytoplasm | 3.4 | X | |||
| CMPK2 | cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial | Cytoplasm | 3.5 to 11.9 | X | |||
| RSAD2 | radical S-adenosyl methionine domain containing 2 | Cytoplasm | 4.2 to 29.2 | x | |||
| DDX60 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 | unknown | 3.5 to 4.4 | X | |||
| EPSTI1 | epithelial stromal interaction 1 (breast) | unknown | 4.1 to 8.2 | X | |||
| HERC5 | hect domain and RLD 5 | Cytoplasm | 2.2 to 10.2 | X | |||
| LY6E | lymphocyte antigen 6 complex, locus E | Plasma Membrane | 2.4 to 6.5 | X | |||
| RTP4 | receptor (chemosensory) transporter protein 4 | Plasma Membrane | 2.2 to 4.3 | X | |||
| XAF1 | XIAP associated factor 1 | Nucleus | 2.3 to 6.2 | X |
HLA phenotype and postvaccination titer
All study subjects were assessed for the development of neutralizing antibodies against live attenuated TC-83 at 28 d postvaccination; production of neutralizing antibody in response to vaccination is currently the gold standard measure of an immunity correlate of protection and denotes successful primary vaccination. Results of neutralizing antibody production were compared with HLA phenotype to describe the potential contribution of MHC haplotype to the immunological response induced by the vaccine (Table 5). Control study subjects receiving only a saline injection were also included in this portion of the study to demonstrate the lack of antibody response as a result of mock vaccination. Table 5 displays a subset of MHC Class II haplotypes (i.e., DRB1 and DQB1). A single volunteer (Vaccinee 1) who displayed the HLA DQB1*0301 allele is included in the table; however the gene expression data from that individual was removed from the microarray data analysis due to primary vaccine failure. We noted that 2 of 3 “low” responders (Day 28 postvaccination titer < 100) displayed a shared HLA haplotype (DQB1 *0302). The DQB1 *0302 phenotype was also present in one of the “high” vaccine responders (Day 28 postvaccination titer >100). Complete HLA phenotype data for study subjects, including all MHC Class I and Class II haplotypes, may be requested from the corresponding author.
Table 5.
Human leukocyte antigen (HLA) phenotype and postvaccination titer.
| Treatment | HLA-DRB1 Phenotype | HLA-DQB1 Phenotype | Day 28 Post vaccination Titer |
|---|---|---|---|
| Control 1 | 0401/1501 | 0302/0602 | < 10 |
| Control 2 | 0401/0701 | 0301/0319 / 0202 | < 10 |
| Control 3 | 0301/0701 | 0201/0202 | < 10 |
| Control 4 | 0701/1302 | 0202/0302 | < 10 |
| Control 5 | 0101/0701 | 0501/0303 | < 10 |
| Control 6 | 0301/1602 | 0201/0502 | < 10 |
| Control 7 | 0401/0701 | 0302/0202 | < 10 |
| Control 8 | 0101/0701 | 0501/0303 | < 10 |
| Control 9 | 0401/0701 | 0301/0319 / 0202 | < 10 |
| Control 10 | 0302/1503 | 0402/0602 | < 10 |
| Vaccinee 1 | 1101/1302 | 0301/0319 / 0604/0634 | < 10* |
| Vaccinee 2 | 0402/0701 | 0302/0202 | 20 |
| Vaccinee 3 | 0301/0401 | 0202 / 0301/0319 | 40 |
| Vaccinee 4 | 0402/0701 | 0302/0202 | 80 |
| Vaccinee 5 | 0801/1501 | 0402/0602 | 160 |
| Vaccinee 6 | 1101/1302 | 0301/0319 / 0609 | 160 |
| Vaccinee 7 | 0401/0701 | 0202/0302 | 320 |
| Vaccinee 8 | 0701/1501 | 0202/0602 | 320 |
| Vaccinee 9 | 0701/1401 | 0303/0503 | 1280 |
| Vaccinee 10 | 1501 | 0602 | 1280 |
*Titer repeated at Day 56; Subject confirmed as vaccine NonResponder
To address the potential role or contribution of certain HLA DQB1 alleles to vaccine outcome, a second ANOVA was performed to include neutralizing titer as a variable (i.e., low titer <100, high titer >100). A False Discovery Rate report was generated to incorporate multiple test corrections (Supplemental Data Table 3). Temporal gene expression values in low- and high-titer immune response groups were compared to describe changes which could be observed between these 2 groups (Supplemental Data Table 4). While the expression of many genes met the criteria of statistical significance for differential expression, none of the significant genes met the cut off of 2-fold or higher change in expression level used in the primary analysis, suggesting that the pathways and processes that are critical for vaccine success or failure in humans are tightly regulated and may be influenced even by small changes in transcriptional expression.
Discussion
VEEV is a reemerging pathogen with potential risk as both a public health and biological threat.18 The advent and widespread application of vaccines has been hailed as one of the most profound achievements for public health in the 20th century. Successful vaccination is ideally mediated through both B and T cell mediated responses. Vaccine responses, as correlates of protection, are often measured by the ability of the vaccine to generate measurable levels of neutralizing antibodies and are usually the only correlate of protection data available in vaccination studies.19 Currently, there is no FDA-approved vaccine for human immunization against VEEV, although there is an Investigational New Drug (IND) vaccine, live-attenuated VEEV TC-83, which has been used for decades by military and at-risk laboratory personnel.7,20 The mechanism of protection induced by vaccination with TC-83 is believed to be through the production of neutralizing antibody, but other molecular mechanisms of protection are not well understood or defined.7,20 The present study explored the sequential molecular events, in vivo. which occur following human immunization with TC-83, and which lead to the development of immunity.
The early and sustained engagement of interferon signals and interferon response factors beginning on day 2 and extending to day 7 observed postvaccination are indicative of a traditional innate antiviral immune response. There is an extensive overlap between the molecules that exhibit changes in transcript expression and the canonical pathways in which they participate, particularly between genes of the interferon response, interferon-response factors, activation of pattern recognition receptors, and engagement of the inflammasome. Potent induction of expression in IFIT1 (ISG54), IFIT3, IRF7, TLR7, and OAS 1-3 represent induction of a classic type I interferon signaling mediated in response to single-stranded RNA viruses.21-24 IFIT1 has been shown to act as a molecular receptor for 5′ tri-phosphorylated RNA and consequently inhibit viral replication.24 We also observed increased transcription of IFIT3 which contributes to antiviral signaling by bridging mitochondrial antiviral signaling and TBK1.25
Early induction of the broad-spectrum innate inflammasome response was noted as a consequence of immunization, spanning days 2 through 7. Engagement of the inflammasome has been shown to be classically mediated through TLR7, DDX58 (also known as RIG-1), IFIH1 (also known as MDA5), and DHX58 (also known as LGP2).19 We found that TC-83 immunization caused transcriptional induction of DDX60, an RNA helicase related to DDX58 which has also been demonstrated in functional genomics studies to be required for RIG-1 or MDA5-dependent signaling in response to viral infection.26-27 Satoh et al.28 describe the importance of DHX58 (LGP2), an ATP-dependent RNA helicase, as a key modulator of both RIG-1 and MDA5-mediated responses ostensibly through activity which makes viral RNA more accessible to either RIG-1 or MDA5 directly or by altering the cellular location of viral ribonucleoprotein complexes for greater access. Other proteins that can initiate anti-viral responses include IFIT2 (ISG56), RSAD2 (viperin), and ISG15; our results demonstrate strongly increased transcription for each of these transcripts on both days 2 and 7 following vaccination suggesting that the type I interferon response is primarily regulated through IRF3 activation.29 Over expression of RSAD2 has been linked to expression and regulation by histone deacetylase 1 (HDAC1) which results in transcriptional repression; during VEEV-induced early engagement of the inflammasome, HDAC1 expression was not altered. Indeed, HDAC1 expression was not altered significantly until day 14 postvaccination.30 Regulation of HDAC1 has been shown to be dependent both on the cell type and influenced by the physiological environment.30
The HLA-DQB1 phenotype has previously been associated with autoimmune disorders and suggested to be involved with hyporesponsiveness to vaccination.17,31-33 A number of studies also suggest that certain combinations of the DQB1 allele play an important role in linkage disequilibrium patterns.34-36 From the current study, 9 of 10 vaccinated volunteers produced an effective immune response, as measured by the production of neutralizing antibody against VEEV. However, no trend in either HLA-DRB1 or HLA-DQB1 phenotype could be definitively determined with respect to linking the phenotype allele to an immunization outcome. We previously reported results of an in vitro assessment of changes in transcription in PBMCs from volunteers previously vaccinated with VEEV TC-83 in which it was suggested that there may be an inverse association between HLA DQB1 alleles and production of neutralizing titer.17 In that instance, either the HLA DQB1 *0301 or *0302 allele was present in the samples of volunteers with the lowest neutralizing antibody titer. Interestingly, specific alleles of the HLA DQB1 haplotypes, including DQB1*0201 and DQB1*0302, have been reported to confer up to 50% of the risk of heritable Type I diabetes.36 We noted decreased transcription of several genes related to insulin signaling, IRS2, SGK, and IGF1R, during the course of vaccination and immune development, suggesting that the insulin signaling pathway may be involved in early responses to vaccination. Additionally, within the DRB1 haplotype, the DRB1 *1501 allele has been associated with Multiple Sclerosis.37 The data suggest that the association between vaccine failure (i.e., vaccine NonResponders) and responders with low neutralizing titer may not necessarily be due to a random association with DQB1 *0301 or *0302 alleles, but rather these results prompt further study to test the hypothesis that primary vaccine failure and weak vaccine take can be explained, at least in part, by association with specific HLA haplotypes. Indeed, the answer to such questions may not ultimately rest on only one haplotype (e.g., DQB1) but may be influenced by the combination of specific DRB1 and DQB1 alleles. While the results are intriguing, it is clear that there are additional factors that affect both disease outcome and vaccination success; further work will need to be conducted to address the questions that such results inspire and with greater numbers of subjects to achieve statistical significance.
We queried the IPA analysis to evaluate the effects of vaccination on the microRNA population; changes in the expression of certain microRNA may represent an avenue of future investigation to suggest regulatory mechanisms for differentially expressed genes. Several microRNA factors were identified as having been effected by VEEV infection, including let-7, miR-21, miR30, miR-101, and miR-214; the presence of these microRNA suggest that the regulation of transcription of certain genes may also be influenced by microRNA. Further studies are needed to pinpoint the hypothesized involvement of specific roles these microRNA and what role each factor may play in the transcriptional regulation of genes and the timing of interaction (transcription, translation, or post-translational modification of genes) (Supplemental Data Table 3).
The study is not without limitations. We were able to detect statistically significant changes in gene expression at days 2, 7, and 14, but at no other time points in the study. This could have been in part due to the restrictive statistical parameters used (i.e., 95% power, 0.001 2-sided t-test, 2-fold change filter, 0.5 CV). Future studies should include a time point between day 7 and day 14 to bridge the changes associated with a largely interferon-driven response and the beginning of development of immunity. Follow on studies may benefit from using a larger sample size to detect more discreet changes of gene expression, a study population comprised of both males and females, and potentially determine whether a correlation between HLA DRB1 or DQB1 alleles and neutralizing antibody production could be established. This would provide further support of previously published in vitro data.17 The present study also utilized only male volunteers between the ages of 23-48 as a strategy to control confounding factors such as age and female sex hormone signaling; future studies should address potential differences in immune response due to age and gender, as well. We compared the in vivo expression data with previously published results obtained independently from the present effort. Statistically significant changes in gene expression observed in our study were in agreement with results identified in select nonhuman primate tissues infected with VEEV Trinidad and human PBMCs infected with TC-83 for the following transcripts: GBP-1, LAMP3, IFIT2, NEXN, DDX58, SAMD9L, XAF1, CD38, HSH2D, and CCRL2.12,17 Additionally, we compared the biomarkers identified in the present work (Table 4) with the ANOVA results from an independent, previously published TC-83 in vitro study to corroborate significant differential gene expression for early and common biomarkers.17 We observed similar significant transcript expression patterns in both studies for GBP4, ANDRD22, CCL2, BST2, TRIM14, CCR1, SMA5, PPM1K, CMPK2, RSAD2, DDX60, EPSTI1, HERC5, LY6E, RTP4, and XAF1.17 As the results of more transcriptional profiling studies become publicly available, it may be possible to discern which responses are indicative of a non-specific innate immune response and which are indicative of a specific bacterial or virus-induced response. Comparing the gene expression profiles generated in response to TC-83 vaccination with previously published results of Tularemia vaccine-induced gene expression, we tentatively identified several genes of the putative tularemia alarm signal cluster that appear to be common between our study and the results published by both Andersson et al. and Paranavitana et al.38-39 These transcripts were GBP1, GCH1, SAT1, TAP1. Other transcripts identified by Andersson et al., including BAG1, CASP1, CCL4, IFITM1, KPNA2, NMI, PIM1, PSME2 did not display significant or temporal changes in gene expression in response to the TC-83 vaccine (Supplemental Table 1). The data support the concept that such biomarkers may be specific markers to Tularemia vaccination or infection. Future direct comparisons between such studies, including other viral vaccination studies, may provide valuable evidence to support hypothesis-driven research on the specificity of biomarker signatures. Finally, we acknowledge that caution should be taken when extrapolating data conclusions from the live-attenuated TC-83 strain in comparison to fully virulent strains of VEEV in human disease. The present work aims to describe that in many cases the viral responses that are caused by the live-attenuated strain represent a controlled level of infection and may still find common ground with the human response to a fully virulent, pathogenic VEEV infection. Due to the similarity of the viruses, we can expect some overlap may be found between molecular responses to human pathogenic VEEV strains and the live-attenuated TC-83 VEEV strain. The changes observed from whole blood sampling of the transcriptome of subjects vaccinated with live-attenuated VEEV TC-83 provide the first glimpse of the molecular epidemiology events that contribute to the specific development of alphaviral immunity in a human host. The most profound changes were noted at days 2, 7, and 14 postvaccination and represent early, intermediate, and late transcriptional events. By day 14, it is not surprising that many of the top molecules which are differentially expressed are related to immunoglobulin genes (Table 2, Table 3, and Table 4). While the early and intermediate phases are dominated by interferon responses, driving innate anti-viral host responses, the events that occur at day 14 are among the most interesting and are represented by changes relating to oxidative phosphorylation, protein ubiquitination, MAPK-related cell signaling pathways, and both natural killer signaling and B-cell development. These changes are similar to reports of involvement of ubiquitination in other alphaviruses. Indeed, nsP2 proteins of Sindbis, Semliki Forest, and Chikungunya viruses have been shown to inhibit cellular transcription by ubiquitination of Rpb1, a catalytic subunit of the RNAPII complex, suggesting a possible mechanism utilized by Old World alphaviruses to subvert the cellular antiviral response.40 Differentially expressed transcripts for the MAPK pathway and for the pore-forming protein perforin and the family of granzymes have been suggested as a potential antiviral role in cytotoxic T lymphocyte (CTL) and natural killer (NK) cells in another positive sense RNA virus, the Japanese encephalitis virus infection.41 The exploitation of similar mechanisms by VEEV, as suggested by our results, may represent highly conserved responses.
Biomarkers which are unique to each phase or common across all stages of infection have been identified with the potential to serve as a molecular signature of infection or as molecular correlates of protection. The HLA phenotype data combined with analysis of the immunity process in humans to VEEV vaccination establish new frontiers for further evaluation of identified HLA phenotypes and induced host genes for their contribution to genomic instability of certain phenotypes and production of neutralizing antibody titers, which are currently the gold-standard in terms of correlates of immunity.17,42 Additionally, the suggested host mechanisms affected by vaccination with live-attenuated VEEV TC-83 in humans revealed potential viral subversion strategies to achieve productive infection, which could be manipulated therapeutically or in immunization intervention protocols to achieve full protection against VEEV and related alphaviruses.
Patients and methods
Selection of volunteers
The research protocol was conducted under Good Clinical Practice (GCP) quality standards, approved by the USAMRIID Institutional Review Board (IRB), and volunteers signed a written informed consent document (ICD) prior to enrollment in the study which described the purpose of the study, as well as the manner in which samples would be collected, used, and disposed. The study consisted of 20 male volunteers between the ages of 23 and 48 y. Male volunteers were selected for the study to reduce the confounding impact of hormonal variation on global gene expression. Additionally, each vaccinee was age-matched to a control volunteer. Study participants were individuals who had not previously received any alphavirus IND vaccines (i.e., against WEEV, EEEV, or VEEV). Prior to enrollment and participation in the study, all study participants were screened for antibodies by ELISA and PRNT for prior exposure to new world Alphaviruses (VEEV, EEEV, and WEEV) and demonstrated to be negative for previous exposure.17 Participants were also genotyped for Human Leukocyte Antigen (HLA) allele expression, as previously described.17 The in vivo study, conducted under Good Clinical Practice quality standards and approved human use protocol FY06-17, included 10 vaccinees who received 0.5 ml of live-attenuated TC-83 VEEV (NDBR-102 vaccine) [roughly equivalent to 1.7 × 105 plaque forming units (PFU) of the virus] administered subcutaneously (SC) in the upper outer aspect of the arm, as well as 10 control subjects who were administered 0.5 ml saline via the same procedure. Whole, unfractionated blood was collected at specific time points immediately prior to (0 h) and following vaccination (1, 4, 8 hrs, and days 1, 2, 7, 14, 21, and 28). On day 56 postvaccination, serum was drawn from volunteers to assess development of neutralizing antibody titer against VEEV. The dataset is comprised of expressed transcripts from 9 responder vaccinees and 10 mock vaccinated control subjects; one vaccinated subject was removed due to primary vaccine immunization failure. Total blood RNA samples from these individuals were subjected to microarray analysis.
RNA isolation and sample preparation for microarray analysis
RNA was isolated from whole, unfractionated blood using the PAXgene Blood RNA kit according to manufacturer's instructions (Qiagen, Valencia, CA). Briefly, RNA from whole blood was collected in PAXgene Blood RNA tubes from each volunteer at each time point. Samples were subjected to quality and concentration analysis using the Agilent RNA 6000 Nano BioAnalyzer kit, according to manufacturer's instructions (Agilent, Santa Clara, CA). Total RNA samples were then prepared for hybridization to the Affymetrix Human Genome U133 plus 2.0 Gene chip arrays according to manufacturer's specifications (Affymetrix, Inc., Santa Clara, CA). The microarray hybridizations were performed at the Core Laboratory Facility at the Virginia Bioinformatics Institute (Blacksburg, VA).
Microarray data analysis
The gene expression data (Affymetrix.CEL files) were imported into Partek Genomics Suite v6.0 software (Partek Inc., St. Louis, MO). Using the Robust Multi-array Average (RMA) algorithm, the gene expression data (Affymetrix gene probe sets) were normalized and log2 transformed.43 To detect differential expression, a 4-way ANOVA was constructed by using the restricted maximum likelihood (REML) approach to produce an unbiased estimate of variance.44 The following equation describes the partitioning of time, vaccine type, and subject variability from variability due to biological and experimental noise:
Where Yijklm represents the mth observation on the ith Scan Date, jth Time Point, kth Treatment, lth Subject. The common effect for the whole experiment is represented by μ, and εijklm represents the random error present in the mth observation on the ith Scan Date, jth Time Point, kth Treatment, lth Subject. The errors εijklm are assumed to be normally and independently distributed with mean 0 and standard deviation δ for all measurements. The symbols T, V, VT, and S(V) represent effects due to time, vaccination type, treatment-by-time interaction, and subject-nested-within-treatment, respectively. Vaccine type and time are fixed effects; scan date and subject are random effects. Using this mixed model ANOVA, gene expression data from 9 individuals from the VEE vaccine group were contrasted against those from 10 individuals of the placebo vaccination group (control group). The p-value for each condition was then corrected using the step-up false discovery rate (FDR) multiple test correction with a cut-off value of 0.1 to produce the list of significantly modulated genes (Table 1).45 Contrasts between vaccinated and control subjects at each time point were achieved using Fisher's Least Significant Difference (LSD) of Log2 transformed data and applying a further restriction of at least 2-fold change in gene expression (either up or down).46 Requests for the complete microarray data should be directed to the corresponding author.
Ingenuity pathway analysis
For the cellular pathway analysis, gene expression values for the significantly modulated genes were imported into the Ingenuity Pathway Analysis (IPA) software to identify canonical pathways associated with genes from the Ingenuity Pathways Analysis library. The genes associated with a canonical pathway were measured in 2 ways: 1) Ratio of the number of genes from the data set that map to the pathway is displayed. The ratio provides the percentage of genes in the data set that were part of a defined list of genes associated with a particular pathway. 2) Fisher's exact test was used to calculate a p-value, which expresses the probability that the association between the genes in the dataset and the canonical pathway can be explained by chance alone; highly significant p-values support an alternate hypothesis that suggests that the interaction is not due to random chance.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The following human use protocol was associated with the work described in this presentation: FY-06-17.
The authors thank Dr. Mohan Ranadive, Ms. Denise Bovenzi, Mr. Larry Korman, and Mr. Vincent Fulton for expert work with the execution of the study, Ms. Tamara Clements for completion of ELISA assays, Ms. Denise Danner for completion of PRNT assays, and Mr. William Discher for expert preparation of figures and tables.
Funding
USAMRIID work has been funded through DOD grant under Plan# 05-4-8I-052.
Opinions, interpretations, conclusions, and recommendations are those of the author and are not necessarily endorsed by the US Army.
References
- [1].Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, evolution. Microbiol Rev 1994; 58:491-562; PMID:7968923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Weaver SC, Barrett AD. Transmission cycles, host range, evolution, and emergence of arboviral disease. Nat Rev Microbiol 2004; 2:789-801; PMID:15378043; http://dx.doi.org/ 10.1038/nrmicro1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Franz DR, Jahrling PB, Friedlander AM, McClain DJ, Hoover DL, Bryne WR, Pavin JA, Christopher GW, Eitzen EM. Clinical recognition and management of patients exposed to biological warfare agents. JAMA 1997; 278(5):399-411; PMID:9244332; http://dx.doi.org/ 10.1001/jama.1997.03550050061035 [DOI] [PubMed] [Google Scholar]
- [4].Paessler S, Weaver SC. Vaccines for Venezuelan equine encephalitis. Vaccine 2009; 27:D80-D85; PMID:19837294; http://dx.doi.org/ 10.1016/j.vaccine.2009.07.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Reichert E, Clase A, Bacetty A, Larsen J. Alphavirus antiviral drug development: Scientific gap analysis and prospective research areas. Biosecur Bioterror 2009; 7:413-427; PMID:20028250; http://dx.doi.org/ 10.1089/bsp.2009.0032 [DOI] [PubMed] [Google Scholar]
- [6].Zacks MA, Paessler S. Encephalitic alphaviruses. Vet Microbiol 2010; 140(34):281-286; PMID:19775836; http://dx.doi.org/ 10.1016/j.vetmic.2009.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pittman PR, Makuch RS, Mangiafico JA, Cannon TL, Gibbs PH, Peters CJ. Long-term duration of neutralizing antibodies after administration of live-attenuated VEE vaccine and following booster vaccination with inactivated VEE vaccine. Vaccine 1996; 14(4):337-343; PMID:8744562; http://dx.doi.org/ 10.1016/0264-410X(95)00168-Z [DOI] [PubMed] [Google Scholar]
- [8].Berge TO, Gleiser CA, Gochenour WS, Miesse ML, Tigertt WD. Studies on the virus of Venezuelan equine encephalomyelitis. J Immunol 1961; 87:509-517; PMID:13867629 [PubMed] [Google Scholar]
- [9].Cole FE Jr, May SW, Eddy GA. Inactivated Venezuelan equine encephalomyelitis vaccine prepared from attenuated (TC-83 strain) virus. Appl Microbiol 1974; 27(1):150-153; PMID:4809906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].McClain DJ, Pittman PR, Ramsburg HH, Nelson GO, Rossi CA, Mangiafico JA, Schmaljohn AL, Malinoski FJ. Immunologic interference from sequential administration of live attenuated alphavirus vaccines. J Infect Dis 1998; 177:634-641; PMID:9498442; http://dx.doi.org/ 10.1086/514240 [DOI] [PubMed] [Google Scholar]
- [11].Hammamieh R, Barmada M, Ludwig G, Peel S, Koterski N, Jett M. Blood genomic profiles of exposure to Venezuelan equine encephalitis in Cynomolgus macaques (Macaca fascicularis). Virology J 2007; 4:82; PMID:17727720; http://dx.doi.org/ 10.1186/1743-422X-4-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Koterski J, Twenhafel N, Porter A, Reed DS, Martino-Catt S, Sobral B, Crasta O, Downey T, DaSilva L. Gene expression profiling of nonhuman primates exposed to aerosolized Venezuelan equine encephalitis virus. FEMS Immunol Med Microbiol 2007; 51(3):462-72; PMID:17894805; http://dx.doi.org/ 10.1111/j.1574-695X.2007.00319.x [DOI] [PubMed] [Google Scholar]
- [13].Sharma A, Bhattacharya B, Puri RK, Maheshwari RK. Venezuelan equine encephalitis virus infection causes modulation of inflammatory and immune response genes in mouse brain. BMC Genomics 2008; 9:289; PMID:18558011; http://dx.doi.org/ 10.1186/1471-2164-9-289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sharma A, Maheshwari RK. Oligonucleotide array analysis of Toll-like receptors and associated signaling genes in Venezuelan equine encephalitis virus-infected mouse brain. J Gen Virol 2009; 90:1836-47; PMID:19369408; http://dx.doi.org/ 10.1099/vir.0.010280-0 [DOI] [PubMed] [Google Scholar]
- [15].Sharma A, Bhomia M, Honnold SP, Maheshwari RK. Role of adhesion molecules and inflammation in Venezuelan equine encephalitis virus infected mouse brain. Virology J 2011; 8:197; PMID:21529366; http://dx.doi.org/ 10.1186/1743-422X-8-197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bhomia M, Balakathiresan N, Sharma A, Gupta P, Biswas R, Maheshwari RK. Analysis of microRNAs induced by Venezuelan equine encephalitis virus infection in mouse brain. BBRC 2010; 395:11-16; PMID:20303929; http://dx.doi.org/ 10.1016/j.bbrc.2010.03.091 [DOI] [PubMed] [Google Scholar]
- [17].Erwin-Cohen RA, Porter A, Pittman PR, Rossi CA, DaSilva L. (2012). Host responses to live-attenuated Venezuelan equine encephalitis virus (TC-83): Comparison of naïve, vaccine responder and NonResponder to TC-83 challenge in human peripheral blood mononuclear cells. Hum Vaccin Immunother 2012; 8(8):1053-65; PMID:22617845; http://dx.doi.org/ 10.4161/hv.20300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Aguilar PV, Estrada-Franco JG, Navarro-Lopez R, Ferro C, Haddow AD, Weaver SC. Endemic Venezuelan equine encephalitis in the Americas: hidden under the dengue umbrella. Future Virol 2011; 6(6):721-40; PMID:21765860; http://dx.doi.org/ 10.2217/fvl.11.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nakaya HI, Li S, Pulendran B. Systems vaccinology: Learning to compute the behavior of vaccine induced immunity. Wiley Interdiscip Rev Syst Biol Med. 2011; 4(2):193-205; PMID:22012654; http://dx.doi.org/ 10.1002/wsbm.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pittman PR, Liu CT, Cannon TL, Mangiafico JA, Gibbs PH. Immune interference after sequential alphavirus vaccine vaccinations. Vaccine 2009; 27(36):4879-82; PMID:19576665; http://dx.doi.org/ 10.1016/j.vaccine.2009.02.090 [DOI] [PubMed] [Google Scholar]
- [21].Sixtos-Alonso MS, Sanchez-Muñoz F, Sanchez-Avila JF, Martınez RA, Lopez AD, Vorackova FV, Uribe M. IFN-stimulated gene expression is a useful potential molecular marker of response to antiviral treatment with peg-IFNa 2b and ribavirin in patients with Hepatitis C virus genotype 1. Arch Med Res 2011; 42:28-33; PMID:21376259; http://dx.doi.org/ 10.1016/j.arcmed.2011.01.001 [DOI] [PubMed] [Google Scholar]
- [22].Boo KY, Yang JS. Intrinsic cellular defenses against virus infection by antiviral type I interferon. Yonsei Med J 2010; 51(1):9-17; PMID:20046508; http://dx.doi.org/ 10.3349/ymj.2010.51.1.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fensterl V, Wetzel JL, Ramachandran S, Ogino T, Stohlman SA, Bergmann CC, Diamond MS, Virgin HW, Sen GC. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog 2012; 8(5):e1002712; PMID:22615570; http://dx.doi.org/ 10.1371/journal.ppat.1002712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pichlmair A, Lassnig C, Eberle CA, Górna1 MW, Baumann CL, Burkard TR, Bürckstümmer T, Stefanovic A, Krieger S, Bennett KL, et al.. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nature Immunology 2011; 12(7):624-32; PMID:21642987; http://dx.doi.org/ 10.1038/ni.2048 [DOI] [PubMed] [Google Scholar]
- [25].Liu XY, Chen W, Wei B, Shan YF, Wang C. IFN-induced TPR protein IFIT3 potentiates antiviral signaling by bridging MAVS and TBK1. J Immunol 2011; 187:2559-68; PMID:21813773; http://dx.doi.org/ 10.4049/jimmunol.1100963 [DOI] [PubMed] [Google Scholar]
- [26].Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li GM, et al.. Systems biology of seasonal influenza vaccination in humans. Nature Immunol 2012; 12(8):786-95. ; PMID:21743478; http://dx.doi.org/ 10.1038/ni.2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H helicase, is a novel antiviral factor promoting RIG-1-like receptor-mediated signaling. Mol Cell Biol 2011; 31(18):3801-19; PMID:21791617; http://dx.doi.org/ 10.1128/MCB.01368-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Satoh T, Kato J, Kumagai Y, Yoneyama M, Sato S, Matushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-1- and MDA5-mediated antiviral responses. Proc Natl Acad Sci 2010; 107(4):1512-7; PMID:20080593; http://dx.doi.org/ 10.1073/pnas.0912986107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Khan KA, Dô F, Marineau A, Doyon P, Clément J-F, Woodgett JR. Fine-tuning of the RIG-I-like receptor/interferon regulatory factor 3-dependent antiviral innate immune response by the glycogen synthase kinase3/Β-Catenin Pathway. Mol and Cell Biol 2015; 35(17):3029-43; PMID:26100021; http://dx.doi.org/ 10.1128/MCB.00344-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nagesh PT, Husain M. Influenza A virus dysregulates host histone deacetylase 1 that inhibits viral infection in lung epithelial cells. J Virol 2016; 90(9):4614-25; advanced online publication; PMID:26912629; http://dx.doi.org/ 10.1128/JVI.00126-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Stayoussef M, Benmansour J, Al-Jenaidi FA, Nemr R, Ali ME, Mahjoub T, Almawai WY. Influence of common and specific HLA-DRB1/DQB1 haplotypes on genetic susceptibilities of three distinct Arab populations to type diabetes. Clin Vaccine Immunol 2009; 16(1):136-8; PMID:19005023; http://dx.doi.org/ 10.1128/CVI.00215-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Stayoussef M, Benmansour J, Al-Irhayim AQ, Said HB, Rayana CB, Mahjoub T, Almawai WY. Autoimmune type 1 diabetes genetic susceptibility encoded by human leukocyte antigen DRB1 and DQB1 genes in Tunisia. Clin Vaccine Immunol 2009; 16(8):1146-50; PMID:19553558; http://dx.doi.org/ 10.1128/CVI.00105-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Narwaney KJ, Glanz JM, Norris JM, Fingerlin TE, Hokanson JE, Rewers M, Hambidge SJ. Association of HLA class II genes with clinical hyporesponsiveness to trivalent inactivated influenza vaccine in children. Vaccine 2013; 31(7):1123-8; PMID:23261040; http://dx.doi.org/ 10.1016/j.vaccine.2012.12.026 [DOI] [PubMed] [Google Scholar]
- [34].Blomhoff A, Olsson M, Johansson S, Akselse HE. Linkage disequilibrium and haplotype blocks in the MHC vary in an HLA haplotype specific manner assessed mainly by DRB1*03 and DRB1*04 haplotypes. Genes Immun 2006; 7:130-40; PMID:16395395; http://dx.doi.org/ 10.1038/sj.gene.6364272 [DOI] [PubMed] [Google Scholar]
- [35].Lie BA, Thorsby E. Several genes in the extended human MHC contribute to predisposition to autoimmune diseases. Curr Opin Immunol 2005; 17:526-31; PMID:16054351; http://dx.doi.org/ 10.1016/j.coi.2005.07.001 [DOI] [PubMed] [Google Scholar]
- [36].Kallionpää H, Elo LL, Laajala E, Mykkänen J, Ricaño-Ponce I, Vaarma M, Teemu D, Laajala TD, Hyöty H, Ilonen J, et al.. Innate immune activity is detected prior to seroconversion in children with HLA-conferred type 1 diabetes susceptibility. Diabetes 2014; 63:2402-14; PMID:24550192; http://dx.doi.org/ 10.2337/db13-1775 [DOI] [PubMed] [Google Scholar]
- [37].Alcina A, del Mar Abad-Grau M, Fedetz M, Izquierdo G, Luca M, Fernandez O, Ndagire D, Catalá-Rabasa A, Ruiz A, Gayán J, et al.. Multiple sclerosis risk variant HLA-DRB1*1501 associates with high expression of DRB1 gene in different human populations. PLoS One 2012; 7(1):e29819; PMID:22253788; http://dx.doi.org/ 10.1371/journal.pone.0029819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Andersson H, Hartmanová B, Bäck E, Eliasson H, Landfors M, Näslund L, Rydén P, Sjöstedt A. Transcriptional profiling of the peripheral blood response during tularemia. Genes Immun 2006; 7:503-13; PMID:16826236; http://dx.doi.org/ 10.1038/sj.gene.6364321 [DOI] [PubMed] [Google Scholar]
- [39].Paranavitana C, Pittman PR, Velauthapillai M, Zelazowska E, DaSilva L. Transcriptional profiling of Francisella tularensis infected peripheral blood mononuclear cells: A predictive tool for tularemia. FEMS Immunol Med Microbiol 2008; 54:92-103; PMID:18680519; http://dx.doi.org/ 10.1111/j.1574-695X.2008.00456.x [DOI] [PubMed] [Google Scholar]
- [40].Akhrymuk I, Kulemzin SV, Frolova EI. Evasion of the innate immune response: the old world alphavirus nsP2 protein induces rapid degradation of Rpb1, a catalytic subunit of RNA polymerase II. J Virol 2012; 86(13):7180-91; PMID:22514352; http://dx.doi.org/ 10.1128/JVI.00541-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yang Y, Ye J, Yang X, Jiang R, Chen H, Cao S. Japanese encephalitis virus infection induces changes of mRNA profile of mouse spleen and brain. Virol J 2011; 8:80; PMID:21345237; http://dx.doi.org/ 10.1186/1743-422X-8-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Plotkin SA. Correlates of vaccine-induced immunity. Vaccines 2008; 47:401-9. [DOI] [PubMed] [Google Scholar]
- [43].Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4(2):249-64; PMID:12925520; http://dx.doi.org/ 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- [44].Thompson WA., Jr The Problem of Negative Estimates of Variance Components. Ann Math Statistics 1962; 33:273-89; http://dx.doi.org/ 10.1214/aoms/1177704731 [DOI] [Google Scholar]
- [45].Benjamini Y, Höchberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B 1995; 57:289-300. [Google Scholar]
- [46].Tamhane AC, Dunlop DD. Statistics and data analysis from elementary to intermediate. Upper Saddle River, NJ: Prentice Hall; 2000. p 473-4. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


