Abstract
Sex pheromones released by female moths are detected with high specificity and sensitivity in the olfactory sensilla of antennae of conspecific males. Bombykol in the silkmoth Bombyx mori was the first sex pheromone to be identified. Here we identify a male-specific G protein-coupled olfactory receptor gene, B. mori olfactory receptor 1 (BmOR-1), that appears to encode a bombykol receptor. The BmOR-1 gene is located on the Z sex chromosome, has an eight-exon/seven-intron structure, and exhibits male-specific expression in the pheromone receptor neurons of male moth antenna during late pupal and adult stages. Bombykol stimulation of Xenopus laevis oocytes expressing BmOR-1 and BmGαq elicited robust dose-dependent inward currents on two-electrode voltage clamp recordings, demonstrating that the binding of bombykol to BmOR-1 leads to the activation of a BmGαq-mediated signaling cascade. Antennae of female moths infected with BmOR-1-recombinant baculovirus showed electrophysiological responses to bombykol but not to bombykal. These results provide evidence that BmOR-1 is a G protein-coupled sex pheromone receptor that recognizes bombykol.
Keywords: baculovirus, bombykal, bombykol, olfactory receptor, Xenopus laevis oocyte
Insects use a unique class of chemical signals called pheromones as cues to recognize other members of the same species and as a means to induce a particular behavior in other members of the same species (1–4). The silkmoth, Bombyx mori, possesses the simplest pheromone system in which full sexual behavior of male moths is initiated by one achiral compound, (E,Z)-10,12-hexadecadien-1-ol (bombykol) released from the pheromone gland of female moths (2–4). Thereby, bombykol is thought to be the sole sex pheromone in B. mori. Bombykal, an oxidized form of bombykol, is also released by female moths, but it does not elicit orientating behavior in male moths (5). Two specialized chemosensory neurons in the long sensilla trichodea on the male antennae are fine-tuned to detect either bombykol or bombykal (5). This remarkable sensitivity and specificity is thought to be achieved by olfactory receptors (ORs) expressed in individual chemosensory neurons.
The OR gene families in insect species have been identified by comprehensive analysis of genome sequences (6–14). In Drosophila antennae, expression of a total of 31 conventional ORs was reported (9), and 24 of these appear to show responses to general odorants with a broad and overlapping ligand-spectrum (15). Therefore, insects use a combinatorial coding strategy to discriminate various odorants as has been previously addressed in mammalian species (16, 17). Receptors for insect pheromones, however, remain unidentified. Pheromone receptors should possess a fine-tuned ligand spectrum with a high discriminatory power. Further, it is reasonable to suggest that a sex pheromone receptor should be expressed specifically in male moths, but not in female moths. In this regard, male-specific OR genes have recently been identified in the genome database of tobacco budworm Heliothis virescens (14).
In the present study, we have undertaken a differential screening strategy to isolate the male-specific B. mori OR (BmOR) gene and cloned one gene designated BmOR-1. Using a Xenopus laevis oocyte expression system, we obtained functional evidence that BmOR-1 encoded an OR that specifically recognized bombykol. Further, ectopic expression of BmOR-1 in female antennae produced electrophysiological responses to bombykol. Finally, none of other male-specific ORs among 29 putative OR genes encoded in the B. mori genome showed any response to bombykol. Functional characterization of BmOR-1 in both heterologous and homologous systems and the results from comprehensive genome database mining most likely identify BmOR-1 as a single sex pheromone receptor in B. mori.
Materials and Methods
Synthesis of Bombykol and Bombykal. (E,Z)-10,12-hexadecadien-1-ol (bombykol) was synthesized stereospecifically starting from 1-pentyne (Sigma-Aldrich) and 10-undecyn-1-ol (Tokyo Kasei, Tokyo) following the protocol described (18). Distillation gave pure bombykol, and the stereochemistry was confirmed by the 1H-NMR spectrum at 300 MHz. (E,Z)-10,12-hexadecadien-1-al (bombykal) was prepared by Dess-Martin oxidation (19) of bombykol.
Animals and Chemicals. Silkmoths B. mori were kept at 25°C on a 12 h:12 h (light/dark) light cycle. Dissected tissues were frozen immediately in liquid nitrogen and stored at –80°C until use. Odorants used in this study were kindly provided by T. Hasegawa (Tokyo) and Takasago (Tokyo) or purchased from Wako and Sigma-Aldrich.
Differential Screenings. A B. mori male antennae cDNA library (20) was plated at low density (1,000–1,500 plaque-forming units per 140 × 100-mm plate), and phage DNA was transferred to nitrocellulose membranes in triplicate by using standard procedures. Each membrane was separately hybridized at high stringency with one of the following fluorescein-labeled probes: day-0 male antennae cDNA as a positive probe, day-0 male body (devoid of antennae and wings) cDNA or a mixture of B. mori pheromone-binding protein (PBP), general odorant-binding protein (GOBP) 1, and GOBP2 and antennal binding protein X cDNA (21) as a negative probe. A GeneImages CDP-Star detection module (Amersham Pharmacia) was used to identify 112 clones (from among ≈3,000 clones) that hybridized with the antennae cDNA probe, but not with either the body or OBP cDNA probes. The nucleotide sequences of 62 of these clones were determined on an ABI310 genetic analyzer (PerkinElmer). One independent BmOR-1 clone was isolated and identified as a putative OR gene in B. mori based on sequence similarity to insect ORs by using the blastx algorithm (www.ncbi.nlm.nih.gov/blast).
BmOR-1 Genomic Sequences. Genomic DNA of adult male moths was extracted by using GenomicPrep cells and tissue DNA isolation kits (Amersham Pharmacia). BmOR-1 was amplified from B. mori genomic DNA by using LA Taq polymerase (Takara Shuzo, Kyoto) and primers 5′-GGATAGAATACTTCGATCCTCGC-3′ and 5′-TGTTGCCACCGTTCGAAGCATCACG-3′, which corresponded to nucleotides 25–47 or 1,326–1,350 in the BmOR-1 cDNA sequence. Amplification was performed by using the following thermal program: 94°C for 1 min; 30 cycles of 94°C for 30 s, 63°C for 30 s, and 68°C for 10 min; followed by one cycle at 72°C for 10 min. A single 9.4-kb PCR fragment was produced in the PCR. The fragment was cloned into a pGEM-T vector (Promega) and sequenced with vector- or sequence-specific primers. The ends of the BmOR-1 genome sequence that were not included in the PCR products were sequenced by using the bacterial artificial chromosome clone as described below.
Isolation of Bacterial Artificial Chromosome Clones and Mapping Sequences of BmOR-1. The B. mori bacterial artificial chromosome library was screened by PCR (22) by using two pairs of primers specific to two fragments of different lengths of BmOR-1 (610-bp fragment, 5′-AACAACTGAACGAAATACGA-3′ and 5′-TGAAGCGAAGCGGAAGAAGC-3′; 602-bp fragment, 5′-TGGGAATGTATGGATGAGAA-3′ and 5′-TATTATTATTAGGTGGTTGAG-3′). One of the isolated clones contained the B17F10E9 sequence, which is known as a marker sequence of the Z chromosome of B. mori (information available at http://sgp.dna.affrc.go.jp/BombMap/index.html).
RT-PCR of RNAs Isolated from Tissues. Total RNA was separately extracted from day-0 adult moth antennae, head, legs, wings, thorax and abdomen tissues by the acid guanidinium-phenolchloroform method (23), treated with DNase I, and reprecipitated. RNA was reverse-transcribed by using oligo(dT) adaptor primer (Takara Shuzo) and avian myeloblastosis virus reverse transcriptase (Takara Shuzo) at 42°C for 35 min. cDNA of BmOR-1, BmOR-2, PBP and B. mori actin 1 (24) was amplified by using Ex Taq DNA polymerase (Takara Shuzo) and the following primer pairs that were designed to span at least one predicted intron except for actin gene to distinguish between genomic DNA and cDNA templates (BmOR-1, 5′-CCTTCAAAGATGACAGTCGTTC-3′ and 5′-CGGTAGAAGATTGAACAGCCC-3′; BmOR-2, 5′-CGCTCACAGCATCATTAAGTTGGC-3′ and 5′-AACGTCTTCGCCTCCTCGGAAC-3′; PBP, 5′-GCGCTCATGGTCAACATGGCTGT-3′ and 5′-ACTGCTACGTCCATGCTCGGAGC-3′; and B. mori actin 1, 5′-ATGTGCAAGGCCGGTTTCGC-3′ and 5′-CGACACGCAGCTCATTGTAG-3′) under the thermal cycling of 95°C for 1 min, then 30 cycles at 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min, followed by 72°C for 10 min. Equal amounts of PCR products were separated by electrophoresis in an Agilent Bioanalyzer 2100 (Agilent Technology). For developmental analysis, total RNA was extracted from the egg, the head of fifth-instar larvae, and the antennae of pupae and adults. RT-PCR was performed as described above. No PCR products were produced when reverse transcriptase was excluded during reverse transcription, and sequence analysis confirmed the identity of cDNA products.
In Situ Hybridization. Coding regions of the BmOR-1 cDNA (nucleotide positions 61–555) or the PBP cDNA (nucleotide positions 136–353) were subcloned into the pGEM-T Easy vector (Promega). Digoxigenin (DIG)-labeled BmOR-1 and fluorescein-labeled PBP RNA probes were synthesized from linearized recombinant pGEM-T Easy vectors by using an SP6/T7 transcription kit (Roche) according to the manufacturer's instructions. Whole-mount in situ hybridization was performed according to Tautzs and Pfeiffle (25) with minor modifications. In brief, antennae of pupae on days 1–2 before eclosion were dissected from the animal, cut into pieces, fixed with 4% paraformaldehyde/PBS overnight at 4°C, treated with 50 μg/ml proteinase K/PBS for 1 h at 37°C, and then fixed for an additional 5 min in 4% paraformaldehyde/PBS. Samples were washed three times for 5 min each time in 0.1% Tween 20/PBS (PBST) at room temperature, followed by an incubation for 5 min in 1:1 PBST/hybridization buffer (50% formamide/5× SSC/50 μg/ml heparin/0.1% Tween 20/100 μg/ml herring sperm DNA). Hybridization reactions were carried out for 16 h at 60°C by using the BmOR-1 probe at a concentration of 500 ng/ml. Hybridized antennae were washed for 10 min at 60°C in hybridization buffer and 10 min in 1:1 PBST/hybridization buffer, followed by three washes in PBST for 10 min each. Hybridization was detected by using alkaline phosphatase-conjugated anti-DIG Ab (Roche; 1:5,000 in PBST) and stained with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate. Then, the antenna was embedded in Epon-82 resin (TAAB Laboratories Equipment, Reading, United Kingdom) and cut into 2-μm plastic sections.
For fluorescent in situ hybridization, day-0 adult moth antennae were fixed in 4% paraformaldehyde/PBS overnight at 4°C, dehydrated, embedded in paraffin, and cut into 10-μm sections. Tissue sections were incubated for 16 h at 60°C in 100 μl of hybridization buffer containing 500 ng/ml of both DIG-labeled BmOR-1 and fluorescein-labeled PBP antisense RNA probes. Sections were washed as described above; the hybridization signal was amplified by using the tyramide signal amplification plus fluorescence system (PerkinElmer), and signal detection was carried out according to the manufacturer's instructions. DIG-labeled probes were visualized by using horseradish peroxidase-conjugated anti-DIG Ab (Roche; 1:100) with tetramethyl-rhodamine tyramids as a substrate, whereas fluorescein-labeled probes were visualized by using horseradish peroxidase-conjugated antifluorescein Ab (Roche; 1:100) with fluorescein tyramides as a substrate.
Gene Expression in X. laevis Oocytes and Electrophysiological Recording. Stage V–VII oocytes were treated with 2 mg/ml collagenase type I (Sigma-Aldrich) in Ca2+-free saline solution (82.5 mM NaCl/2 mM KCl/1 mM MgCl2/5 mM Hepes, pH 7.5) for 1–2 h at room temperature. Oocytes were then microinjected with 50 ng of BmOR-1 cRNA and 25 pg of BmGαq cRNA synthesized from pSPUTK-BmOR-1 and pSPUTK-BmGαq, respectively (26). Oocytes injected with 50 ng of BmOR-1 cRNA or 25 pg of BmGαq cRNA were used as negative controls. Injected ooctyes were incubated for 5–7 days at 18°C in Barth's solution (88 mM NaCl/1 mM KCl/0.3 mM Ca(NO3)2/0.4 mM CaCl2/0.8 mM MgSO4/2.4 mM NaHCO3/15 mM Hepes, pH 7.6) supplemented with 10 μg/ml penicillin and streptomycin.
Whole-cell currents were recorded with a two-electrode voltage clamp (OC-725, Warner) (27). Data acquisition and analysis were carried out by using digidata1322a and pclamp software (Axon Instruments, Foster City, CA). Ca2+-dependent Cl– current was monitored by applying 200-msec depolarizing pulses every 2 sec of +60 mV from the holding potential of –80 mV. At the fifth pulse, 40 μl of concentrated stock-solution of bombykol or bombykal was applied to the recording bath (160-μl volume) to give the indicated concentrations.
Generation of BmOR-1 Recombinant Baculovirus. The entire protein-coding sequence of BmOR-1 was subcloned into the transfer vector pVL1393 to create pVL-BmOR-1, which was designed to express the native BmOR-1 protein. Spodoptera frugiperda IPLB-Sf21-AE (Sf21) cells were cotransfected with 2 μg of resultant transfer vector DNA and 1 μg of the linearized hybrid nuclear polyhedrosis virus (HyNPV) (28) genomic DNA, which is a host range-expanded Autographa californica NPV by using a lipofectin reagent (Invitrogen). The recombinant baculovirus was plaque-purified and amplified to titer of 1 × 107 plaque-forming units/ml. The recombinant baculovirus resulting from pVL-BmOR-1 was named HyBmOR-1.
Baculovirus-Infected Female Moths. At 4 days before eclosion, 50 μl of virus solution containing 5 × 105 plaque-forming units of the recombinant or WT baculovirus was percutaneously injected into the fifth abdominal segment of female pupae by using a 1-ml plastic syringe with a 26-gauge needle. Inoculated pupae were reared at 25°C until eclosion.
Electroantennogram (EAG) Recording. Antennae of the day-0 adult moths were excised at the base, and a few segments at the tip were clipped off. Antennae were immediately used for EAG by attaching an indifferent electrode to the tip of the antenna and a recording electrode to the base of the antenna. Electrodes were filled with 0.1 M NaCl, and signals from a DC preamplifier (MEZ-7101, Nihon Kohden) were plotted on a chart recorder (WX1200, Graphtech). Bombykol or bombykal (40 μl) in 1% dimethyl sulfoxide (DMSO) at 100 ng/μl were loaded on 1.2 cm2-filter paper (Advantec no. 2, Toyoroshi, Tokyo) and inserted into a plastic pipette tip. A charcoal-purified airstream was passed through the pipette tip, through Tygon tubing (inner diameter, 4 mm), and directed on the antenna from a distance of ≈5 mm. The airflow rate was adjusted at 350 ml/min, unless otherwise mentioned, and the stimulus was applied by manually opening or closing valves.
Results and Discussion
We hypothesized that the bombykol receptor is a male antennae-specific OR because bombykol elicits electrophysiological signals only in the male moth antennae. Thus, we used the differential cloning strategy to identify the bombykol receptor. A cDNA clone (designated BmOR-1) encoding a 430-aa protein with seven putative transmembrane domains that are characteristic to the G protein-coupled receptor superfamily (Fig. 1a) was isolated through differential screening of a cDNA library of a B. mori male antennae. The amino acid sequence deduced from the BmOR-1 clone showed significant sequence similarity (up to 42%) with ORs in insect species such as Drosophila melanogaster (6–9), Anopheles gambiae (10–12), H. virescens (13, 14), and D. simulans (29). BmOR-1 exhibited the closest proximity to a putative OR in H. virescens, HR13 (14), according to phylogenetic analysis (Fig. 1a Lower). The BmOR-1 gene consists of eight exons and seven introns in 9,870 bp of genomic DNA (Fig. 1b). The genomic DNA sequence was screened against a bacterial artificial chromosome library from B. mori (22) and mapped to the Z chromosome, one of the two sex chromosomes (Z and W). B. mori males are the homogametic sex, ZZ, whereas the females are the heterogametic sex, ZW (30). Southern blot analysis showed that BmOR-1 is a single copy gene in the genome (data not shown). In addition to BmOR-1, we cloned the BmOR-2 gene by using the known insect ORs with a homology-based method. BmOR-2 shows 65–87% amino acid identity with ORs that belong to the insect Or83b gene family (31) (Fig. 1a).
Fig. 1.
Amino acid sequence and genomic structure of BmOR-1. (a Upper) Deduced amino acid sequence of BmOR-1. Red boxes, putative transmembrane regions predicted by sosui (http://sosui.proteome.bio.tuat.ac.jp/cgi-bin/sosui.cgi?/sosui_submit.html) and tmpred (www.ch.embnet.org/software/TMPRED_form.html). Arrowheads, intron–exon boundaries. (a Lower) Phylogenetic tree of the amino acid sequences of candidate ORs from various insect species. Protein names starting with Or, GPR, HR, and BmOR are ORs of D. melanogaster, A. gambiae, H. virescens, and B. mori, respectively. Branch lengths are proportional, and the scale of distance is indicated. BmOR names are in red type. (b) Schematic representation of the genomic structure of BmOR-1. The position and relative size of exons and introns are drawn to scale as indicated. Structure of the BmOR-1 gene and locations of start/stop codons are shown.
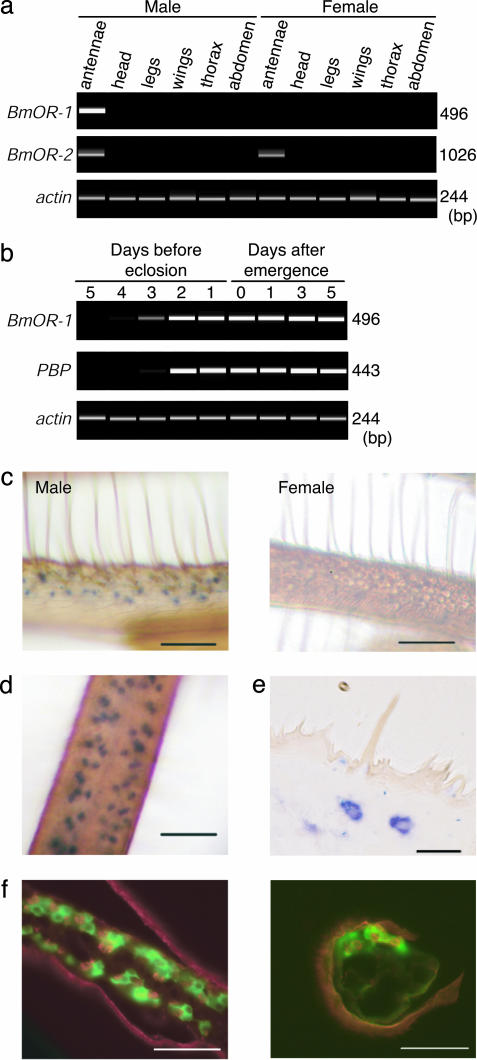
RT-PCR experiments demonstrated that BmOR-1 is expressed only in the antennae of male B. mori (Fig. 2a); the BmOR-1 gene transcript was first detected 4 days before eclosion, increased during the late pupal stages, and was continuously detected in the male antennae during the adult stage (Fig. 2b Top). BmOR-1 mRNA was not detected in the head of fifth-instar larvae nor in eggs (data not shown). Whole-mount in situ hybridization studies showed that BmOR-1 mRNA was localized to the specific cells in the epithelium of the male antennae on the side carrying chemosensory hairs (Fig. 2c Left), and no hybridization signal was detected in the female antennae (Fig. 2c Right). Distribution patterns and density of the labeled cells in the male antennae (Fig. 2d) were similar to those in pheromone-sensitive long sensillum trichodeum (32). Further, when viewed in 2-μm plastic sections, BmOR-1-reactive cells were slightly ellipsoidal and were located ≈5–10 μm beneath the antennal cuticle (Fig. 2e) where OR neurons reside. No labeling was observed with sense RNA probes (data not shown).
Fig. 2.
Expression pattern of the BmOR-1 gene. (a) Tissue- and sex-specific expression of BmOR-1, BmOR-2, and B. mori actin genes (24), a positive control. RT-PCR products by using RNA isolated from various tissues of male and female moths, as indicated, were separated by electrophoresis. (b) Developmental analysis of BmOR-1 and PBP expression in the male antennae, with the B. mori actin gene as a positive control. RT-PCR products were separated by electrophoresis. A very faint band that corresponded to BmOR-1 product was detected at 4 days before eclosion. (c) Whole-mount in situ hybridization of BmOR-1 in the antennae of male (Left) and female (Right) moths. Reactive blue cells visualized by using an anti-DIG Ab were found on the side with chemosensory hairs. (d) A whole-mount in situ section of the male antenna viewed from olfactory sensilla side. (e) Whole-mount in situ labeling of a 2-μm plastic section of the male antenna. Labeled cells were found only in the antennal surface that carries olfactory sensilla. (f) Two-color fluorescent in situ hybridization of BmOR-1 (red) and PBP (green). Double-labeling was performed for a longitudinal (Left) and cross (Right) sections of the male antenna by using DIG-labeled BmOR-1 and fluorescein-labeled PBP antisense RNA. [Scale bar: 50 μm(c, d, and f), 20 μm(e).]
The developmental expression profile of PBP, which is thought to be involved in pheromone detection (33, 34), was similar to that of BmOR-1 (Fig. 2b Middle), except that PBP was also detected, although at lower levels, in the antennae of female adults and pupae (data not shown). Double labeling of the male adult antennae with BmOR-1 and PBP antisense RNAs demonstrated that BmOR-1-labeled cells (red) were localized on the sensory epithelium side and surrounded by the PBP-labeled cells (green) in both longitudinal and cross sections (Fig. 2f). These results are consistent with the previous observation that PBP mRNA was expressed in supporting cells that surround the pheromone receptor neurons (32). These results suggest that BmOR-1 is expressed exclusively in the pheromone receptor neurons in the antennae of male moths.
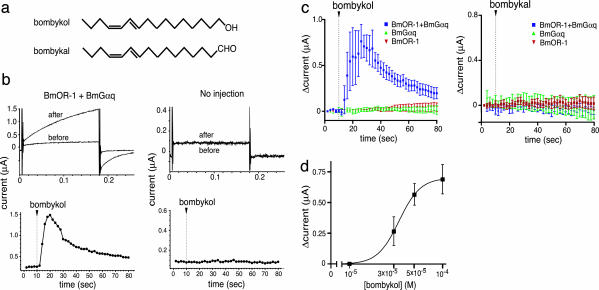
We hypothesized that BmOR-1 is involved in the detection of the sex pheromone in the male antennae and tested the ability of BmOR-1 to recognize bombykol by using two-electrode voltage clamp recordings of Xenopus oocytes with depolarizing step pulses from a holding potential of –80 mV. Robust Ca2+-dependent Cl– currents were observed upon stimulation with bombykol, but not with bombykal, when BmOR-1 was coexpressed with BmGαq (G protein α-subunits in B. mori belonging to the Gαq family) (Fig. 3). Oocytes injected with cRNA of BmOR-1 or BmGαq alone did not respond to bombykol (Fig. 3c). Additionally we tested ligand specificity of BmOR-1 by using 41 odorants that have previously been reported to elicit electronic responses in B. mori adult antennae (35). However, none of the odorants tested at 100 μM produced any response to the oocytes coexpressing BmOR-1 with BmGαq and showing response to bombykol (data not shown). These results demonstrate that BmOR-1 specifically recognizes bombykol, and subsequently couples to BmGαq, resulting in activation of Ca2+-dependent Cl– channels in oocytes. Dose-dependent response curves (Fig. 3d) show that relatively high concentrations of bombykol are necessary to fully activate BmOR-1 expressed in Xenopus oocytes, probably because of the low level of BmOR-1 expression and inefficient signal transduction mediated by BmGαq in oocytes. The EC50 value of bombykol for BmOR-1 of 34 μM was ≈14-fold lower than that of a ligand for Drosophila Or43a, a conventional odorant receptor (36). Responses were observed in 10–15% of the injected oocytes, but preliminary results suggest that coexpression with BmOR-2 improved the yield of oocytes responding to bombykol by greatly enhancing membrane translocation of BmOR-1 (T. Nakagawa and K.T., unpublished results). Nonetheless, these results provide evidence that BmOR-1 functions as a bombykol receptor in a heterologous cell system.
Fig. 3.
Bombykol responses of Xenopus oocytes expressing BmOR-1 and BmGαq. (a) Structure of bombykol and bombykal. (b) The activation of BmOR-1 by bombykol was monitored by an increase in Ca2+-dependent Cl– current. (Upper) Current traces of oocytes injected with cRNAs of BmOR-1 and BmGαq(BmOR-1 + BmGαq) or noninjected control oocytes (no injection) before and after application of 100 μM bombykol. (Lower) Time course of the response plotted as the current amplitude at the end of each depolarizing pulse. Bombykol (100 μM) was applied at the time indicated by arrowheads. The data are representative of bombykol responses in 20 X. laevis oocytes from five different animals. (c) Time course of current recordings of oocytes expressing BmOR-1 + BmGαq, or BmOR-1 and BmGαq separately. Bombykol (Left) or bombykal (Right) (100 μM) was applied at the time indicated by arrowheads. Data ± SE (n = 3–4). (d) Dose-dependent increases in amplitudes of bombykol-induced currents. Current recordings at different doses were measured on different oocytes. Currents of oocytes showing responses were averaged for each concentration: 30 μM, n = 3; 50 μM, n = 3; 100 μM, n = 9.
A search of the Silkworm Genome Research Program database (http://kaikoblast.dna.affrc.go.jp) for sequences similar to BmOR-1 and other known insect OR sequences identified an additional 29 putative OR gene sequences. RT-PCR experiments revealed that 23 of these sequences were expressed in the antennae. Two of these were expressed specifically in the male antennae but not in the female antennae (data not shown). None of these receptors showed a response to bombykol in the oocyte expression system (data not shown), suggesting that BmOR-1 is most likely a single receptor for bombykol.
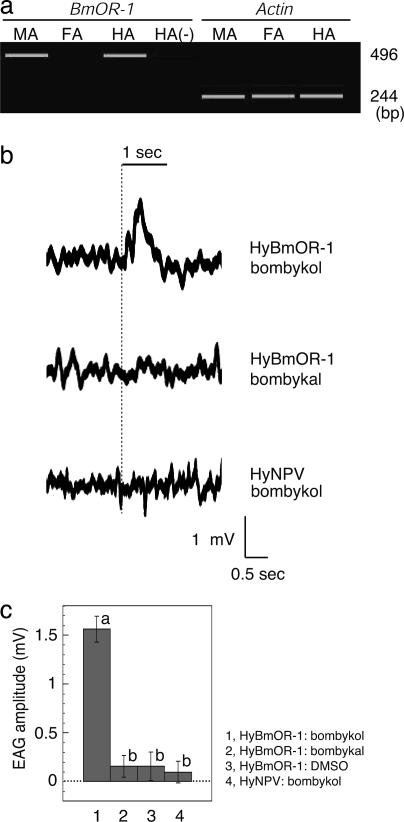
We next examined whether the bombykol response observed in the male antennae could be reconstituted in the female antennae by ectopically expressing BmOR-1 by using recombinant baculovirus carrying BmOR-1-coding sequence (HyBmOR-1). When B. mori female pupae were infected with HyBmOR-1 4 days before eclosion, BmOR-1 transcripts were detected in the antennae of the infected day-0 female adult moths (Fig. 4a). Next, we measured electrophysiological responses of the antennae of female moths ectopically expressing BmOR-1 when exposed to the sex pheromone. We recorded EAG responses of the antennae under an air stream containing bombykol or bombykal (4 μg of each on a filter paper) at a flow rate of 350 ml/min. HyBmOR-1-infected female antennae responded to bombykol, but not to bombykal (P < 0.05 by Scheffé's test, Fig. 4 b and c), indicating the strong specificity for bombykol. None of WT host range-expanded baculovirus (HyNPV)-infected female antennae (Fig. 4 b and c) nor uninfected female antennae (data not shown) responded to bombykol or bombykal, whereas responses to general odorants such as linalool were detected by the infected animals at the same sensitivity as in uninfected animals (data not shown). The amplitude of the HyB-mOR-1-infected female antennae response was ≈8-fold lower (1.56 ± 0.133 mV SEM, n = 10, Fig. 4c) than that of the male moth antennae (12.8 ± 1.51 mV SEM, n = 8, data not shown) to the same dose of bombykol (4 μg on a filter paper at a flow rate of 350 ml/min). The reduced sensitivity in the infected female antennae is likely attributed to the low efficiency of HyBmOR-1 infection into the receptor neurons. Access of the virus to OR neurons has been reported to be limited for antennae of Manduca sexta.‡‡ In conclusion, these results demonstrate that ectopic expression of BmOR-1 in female antennae conferred responsiveness to bombykol but not to bombykal, indicating that BmOR-1 functions as a highly specific receptor for bombykol in the silkmoth antennae.
Fig. 4.
EAG responses of the HyBmOR-1-infected female adult antennae to bombykol. (a) Ectopic expression of BmOR-1 in day-0 female adult antennae infected with BmOR-1 recombinant baculovirus (HyBmOR-1) at 4 days before eclosion. RT-PCR products by using RNA isolated from the antennae of the moths were separated by electrophoresis. MA, male antennae; FA, female antennae; HA, HyBmOR-1-infected female antennae. Minus sign indicates RT-PCR was performed without reverse transcriptase. (b) EAG recordings of HyBmOR-1- or HyNPV-infected female antennae exposed to bombykol or bombykal at 500 ml/min. EAG traces of the same HyBmOR-1-infected female antenna to bombykol (Top) or bombykal (Middle) are shown. Stimulus was applied starting at the time indicated by the dotted line for the duration of 1 s, as indicated by the solid line above the recording profiles. In these recordings, a reference electrode was placed at the tip of antenna, and thereby, the response was shown as a positive signal. (c) Quantitative analysis of EAG responses of HyBmOR-1-infected female antennae to bombykol. Amplitudes of responses of HyBmOR-1-infected female antennae to bombykol (lane 1) or bombykal (lane 2) (4 μg on filter paper) or to 1% DMSO as a control (lane 3) with a flow rate of 350 ml/min (n = 10 each) were quantified. EAG recordings to these three stimuli were measured on the same antenna. WT HyNPV was used as a negative control (lane 4) (n = 25). Error bars represent ± SEM. Average values marked by different letters are significantly different (Scheffé's test: P < 0.05). The EAG response of HyBmOR-1 to bombykol (lane 1) was significantly larger than to bombykal (lane 2) or DMSO (lane 3). The other three responses (lanes 2–4) were not significantly different.
The discovery of BmOR-1 as an insect sex pheromone receptor sheds light on mechanisms underlying highly sensitive and specific detection of sex pheromones and subsequent signal integration by the brain. Identification and functional analyses of sex pheromone receptors from other insect species will provide insight into the molecular evolution of species-specific pheromones and their receptors, as well as contribute to the development of unique methodologies for controlling plant and after harvest pests.
Acknowledgments
We thank J. Takabayashi, M. Ozaki, R. Yoshimura, M. Yamao, H. Watanabe, H. Kataoka, Y. Kubo, C. Kitamura, and K. Ohta for technical assistance and discussion. This work was supported in part by grants from Ministry of Education, Culture, Sports, Science, and Technology of Japan (T. Nishioka and K.T.) and by the Program for Promotion of Basic Research Activities for Innovative Biosciences, Japan (H. Mori and K.T.). T.S. and K.T. are recipients of grants from the Naito Foundation. K.T. is the recipient of grants from the Uehara Memorial Foundation, the Kato Memorial Bioscience Foundation, and the Novartis Foundation.
Abbreviations: OR, olfactory receptor; BmOR, Bombyx mori OR; PBP, pheromone-binding protein; DIG, digoxigenin; EAG, electroantennogram; HyNPV, hybrid nuclear polyhedrosis virus.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AB059431, AB100454, AB105070, and AB101293 for cDNA sequences of BmOR-1, BmOR-2, and BmGαq and genomic DNA sequence of BmOR-1, respectively).
Footnotes
Rogers, M. E., Peterlin, Z. A., Chesler, A. T. & Firestein, S. (2002) Chem. Senses 27, A72 (abstr.).
References
- 1.Karlson, P. & Lüscher, M. (1959) Nature 183, 55–56. [DOI] [PubMed] [Google Scholar]
- 2.Butenandt, V. A., Beckmann, R., Stamm, D. & Hecker, E. (1959) Z. Naturforsch. 14, 283–284. [Google Scholar]
- 3.Schneider, D. (1969) Science 163, 1031–1037. [DOI] [PubMed] [Google Scholar]
- 4.Kaissling, K.-E. (1987) R. H. Wright Lectures on Insect Olfaction, ed. Colbow, K. (Simon Fraser Univ., Burnaby, Canada).
- 5.Kaissling, K.-E. & Kasang, G. (1978) Naturwissenschaften 65, 382–384. [Google Scholar]
- 6.Clyne, P. J., Warr, C. G., Freeman, M. R., Lessing, D., Kim, J. & Carlson, J. R. (1999) Neuron 22, 327–338. [DOI] [PubMed] [Google Scholar]
- 7.Vosshall, L. B., Amrein, H., Morozov, P. S., Rzhetsky, A. & Axel, R. (1999) Cell 96, 725–736. [DOI] [PubMed] [Google Scholar]
- 8.Gao, Q. & Chess, A. (1999) Genomics 60, 31–39. [DOI] [PubMed] [Google Scholar]
- 9.Vosshall, L. B., Wong, A. M. & Axel, R. (2000) Cell 102, 147–159. [DOI] [PubMed] [Google Scholar]
- 10.Fox, A. N., Pitts, R. J., Robertson, H. M., Carlson, J. R. & Zwiebel, L. J. (2001) Proc. Natl. Acad. Sci. USA 98, 14693–14697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox, A. N., Pitts, R. J. & Zwiebel, L. J. (2002) Chem. Senses 27, 453–459. [DOI] [PubMed] [Google Scholar]
- 12.Hill, C. A., Fox, A. N., Pitts, R. J., Kent, L. B., Tan, P. L., Chrystal, M. A., Cravchik, A., Collins, F. H., Robertson, H. M. & Zwiebel, L. J. (2002) Science 298, 176–178. [DOI] [PubMed] [Google Scholar]
- 13.Krieger, J., Raming, K., Dewer, Y. M. E., Bette, S., Conzelmann, S. & Breer, H. (2002) Eur. J. Neurosci. 16, 619–628. [DOI] [PubMed] [Google Scholar]
- 14.Krieger, J., Grosse-Wilde, E., Gohl, T., Dewer, Y. M. E., Raming, K & Breer, H. (2004) Proc. Natl. Acad. Sci. USA 101, 11845–11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hallem, E. A., Ho, M. G. & Carlson, J. R. (2004) Cell 117, 965–979. [DOI] [PubMed] [Google Scholar]
- 16.Malnic, B., Hirono, J., Sato, T. & Buck, L. B. (1999) Cell 96, 713–723. [DOI] [PubMed] [Google Scholar]
- 17.Kajiya, K., Inaki, K., Tanaka, M., Haga, T., Kataoka, H. & Touhara, K. (2001) J. Neurosci. 21, 6018–6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyaura, N. & Suginome, H. (1983) Tetrahedron 39, 3271–3277. [Google Scholar]
- 19.Dess, D. B. & Martin, J. C. (1991) J. Am. Chem. Soc. 113, 7277–7287. [Google Scholar]
- 20.Tanoue, S., Sumida, S., Suetsugu, T., Endo, Y. & Nishioka, T. (2001) Insect Biochem. Mol. Biol. 31, 971–979. [DOI] [PubMed] [Google Scholar]
- 21.Krieger, J., von Nickisch-Rosenegk, E., Mameli, M., Pelosi, P. & Breer, H. (1996) Insect Biochem. Mol. Biol. 26, 297–307. [DOI] [PubMed] [Google Scholar]
- 22.Wu, C., Asakawa, S., Shimizu, N., Kawasaki, S. & Yasukochi, Y. (1998) Mol. Gen. Genet. 261, 698–706. [DOI] [PubMed] [Google Scholar]
- 23.Chomczynski, P. & Sacchi, N. (1987) Anal. Biochem. 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 24.Mounier, N., Gaillard, J. & Prudhomme, J.-C. (1987) Nucleic Acids Res. 15, 2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tautzs, D. & Pfeiffle, C. (1989) Chromosoma 98, 81–85. [DOI] [PubMed] [Google Scholar]
- 26.Katada, S., Nakagawa, T., Kataoka, H. & Touhara, K. (2003) Biochem. Biophys. Res. Commun. 305, 964–969. [DOI] [PubMed] [Google Scholar]
- 27.Kubo, Y., Miyashita, T. & Murata, Y. (1998) Science 279, 1722–1725. [DOI] [PubMed] [Google Scholar]
- 28.Mori, H., Nakazawa, H., Shirai, N., Shibata, N., Sumida, M. & Matsubara, F. (1992) J. Gen. Virol. 73, 1877–1880. [DOI] [PubMed] [Google Scholar]
- 29.Dobritsa, A. A., van der Goes van Naters, W., Warr, C. G., Steinbrecht, R. A. & Carlson, J. R. (2003) Neuron 37, 827–841. [DOI] [PubMed] [Google Scholar]
- 30.Hashimoto, H. (1933) Jpn. J. Genet. 8, 245–258. [Google Scholar]
- 31.Krieger, J., Klink, O., Mohl, C., Raming, K. and Breer, H. (2003) J. Comp. Physiol. A 189, 519–526. [DOI] [PubMed] [Google Scholar]
- 32.Steinbrecht, R. A., Laue, M. & Ziegelberger, G. (1995) Cell. Tissue Res. 282, 203–217. [Google Scholar]
- 33.Maida, R., Steinbrecht, R. A., Ziegelberger, G. & Pelosi, P. (1993) Insect Biochem. Mol. Biol. 23, 243–253. [Google Scholar]
- 34.Prestwich, G. D., Du, G. & LaForest, S. (1995) Chem. Senses 20, 461–469. [DOI] [PubMed] [Google Scholar]
- 35.Topazzini, A., Mazza, M. & Pelosi, P. (1990) J. Insect Physiol. 36, 619–624. [Google Scholar]
- 36.Wetzel, C. H., Behrendt, H.-J., Gisselmann, G., Störtkuhl, K. F., Hovemann, B. & Hatt, H. (2001) Proc. Natl. Acad. Sci. USA 98, 9377–9380. [DOI] [PMC free article] [PubMed] [Google Scholar]