Abstract
SOX9 is a pivotal transcription factor in developing and adult cartilage. Its gene is expressed from the multipotent skeletal progenitor cells and is active throughout chondrocyte differentiation. While it is repressed in hypertrophic chondrocytes in cartilage growth plates, it remains expressed throughout life in permanent chondrocytes of healthy articular cartilage. SOX9 is required for chondrogenesis: it secures chondrocyte lineage commitment, promotes cell survival, and transcriptionally activates the genes for many cartilage-specific structural components and regulatory factors. Since heterozygous mutations within and around SOX9 were shown to cause the severe skeletal malformation syndrome called Campomelic Dysplasia, researchers around the world have worked assiduously to decipher the many facets of SOX9 actions and regulation in chondrogenesis. The more we learn, the more we realize the complexity of the molecular networks in which SOX9 fulfills its functions and is regulated at the levels of its gene, RNA and protein, and the more we measure the many gaps remaining in knowledge. At the same time, new technologies keep giving us more means to push further the frontiers of knowledge. Research efforts must be pursued to fill these gaps and to better understand and treat many types of cartilage diseases in which SOX9 has or could have a critical role. These diseases include chondrodysplasias and cartilage degeneration diseases, namely osteoarthritis, a prevalent and still incurable joint disease. We here review the current state of knowledge of SOX9 actions and regulation in the chondrocyte lineage, and propose new directions for future fundamental and translational research projects.
Keywords: Cartilage, chondrocyte, posttranscriptional regulation, posttranslational modification, SOX9, transcriptional regulation
Introduction
The SOX9 gene was cloned in the early 90’s through a PCR-based screening approach designed to test whether mammalian genomes harbor close relatives of SRY, a gene then newly identified as the sex-determining region on the Y chromosome (1). The approach took advantage of the functional hallmark of the SRY protein, a DNA-binding domain resembling that present in a high-mobility group (HMG) of non-histone proteins. It led to the breakthrough discovery of a large family of SOX (SRY-related HMG box-containing) genes, of which most are now well known as master transcription factors in cell fate determination and differentiation in a variety of lineages. The importance of SOX9 itself came to light soon after its cloning when the severe, early-lethal human skeleton malformation syndrome called Campomelic Dysplasia (CD) was found to be due to heterozygous mutations within or around the SOX9 genetic locus (2). CD children are born with dwarfism and bent long bones, micrognathia, cleft palate, and other skeletal defects resulting from abnormal development of cartilage. They occasionally exhibit malformations of the heart, lung, kidney and pancreas, and two thirds of XY CD children are sex-reversed. Besides revealing the importance of SOX9 in development, CD also revealed through its SOX9 haploinsufficiency origin that precise regulation of SOX9 expression is critical for proper formation of cartilage and other structures.
The establishment of this SOX9/CD link spurred a wealth of worldwide research efforts aimed at identifying the actions and modes of regulation of SOX9 (3,4). Today, a PubMed search using SOX9 as keyword generates over 3000 hits, most being related to chondrogenesis, and the yearly count of hits keeps increasing. This is clear testimony that SOX9 has central roles in the chondrocyte lineage and that many discoveries are still unfolding regarding its modes of actions and regulation in cartilage. Milestone discoveries achieved so far and fueling further research include demonstration that (1) SOX9 is expressed in developing cartilage from the skeletogenic progenitor stage and remains expressed in chondrocytes until hypertrophy in growth plates and throughout adulthood in articular cartilage; (2) SOX9 is required to secure the commitment of skeletogenic progenitor cells to the chondrocyte lineage and to maintain this commitment throughout chondrocyte differentiation in embryonic cartilage primordia and in fetal and postnatal growth plates; (3) SOX9 is critical to ensure adult articular cartilage maintenance and function; (4) SOX9 underlies chondrocyte differentiation by transcriptionally activating many genes essential to build and maintain healthy cartilage, and by repressing at least in part through non-transcriptional actions factors and pathways that favor non-chondrocytic cell lineages; (5) SOX9 regulation at the level of its gene, RNA and protein involves various types of regulatory mechanisms; and (6), last but not least, alterations in these mechanisms likely underlie changes in SOX9 level that have been detected and could significantly contribute to skeletal malformation and degeneration diseases. Research efforts continue today and must continue until we reach full understanding of the implication of SOX9 in cartilage normal development and adult maintenance, in inherited cartilage malformations, and in cartilage degeneration diseases acquired later in life, and until we are able to fill the current, severe lack of strategies available to treat and cure cartilage diseases effectively.
We start here by reviewing current understanding and lack of understanding of SOX9 actions in chondrocytes. We then review many levels of SOX9 regulation, including epigenetic and transcriptional regulation of its gene, posttranscriptional regulation of its RNA, posttranslational modifications of its protein, and interactions of its protein with functional partners and inhibitors. Due to space constraints, we focus on milestone and most recent discoveries and apologize to all those whose findings could not be cited. We close with a perspective on progress achieved to this date and outstanding questions to address in future research.
SOX9 actions in cartilage development and adult maintenance
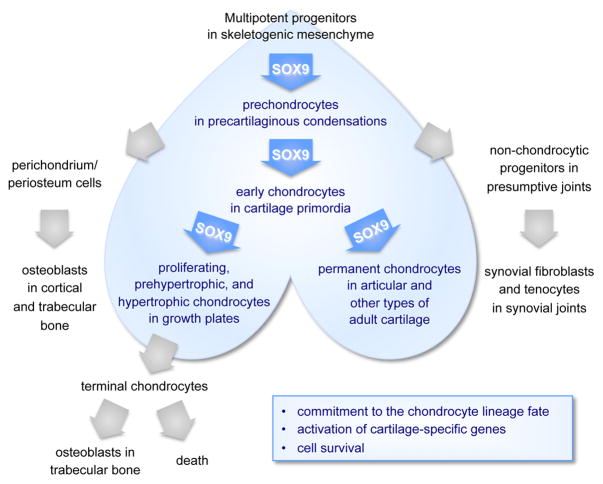
Skeletogenesis starts in the vertebrate embryo when multipotent progenitor cells settle in future skeletal sites and form skeletogenic mesenchyme (Figure 1). These cells already express SOX9, but appear to require it only at the onset of chondrogenesis. Indeed, inactivation of Sox9 in limb bud skeletogenic progenitor cells in Sox9fl/flPrx1Cre mouse embryos leads to massive cell death, but only following failure of the cells to form precartilaginous condensations and to activate early markers of the chondrocyte lineage, such as Col2a1 (encoding collagen type II) (5). Moreover, mouse embryo chimeras generated by mixing Sox9+/+ and Sox9−/− pluripotent stem cells were shown to feature intermingled wild-type and mutant cells in skeletogenic mesenchyme, perichondrium and other tissues, but to contain wild-type cells only in precartilaginous condensations (6). Thus, the first known role of SOX9 in chondrogenesis is to act in a cell-autonomous manner to enable skeletogenic cells to contribute to or survive in precartilaginous condensations.
Figure 1. SOX9 actions in the chondrocyte lineage.
Schematic of the progressive differentiation of multipotent skeletogenic cells into chondrocytes and related skeletal cell types. Blue color is used to highlight the expression domain of SOX9.
Precartilaginous condensations rapidly evolve into cartilage primordia. This developmental step occurs as prechondrocytes overtly differentiate into early-stage chondrocytes, which proliferate actively and produce abundant amounts of cartilage extracellular matrix. Inactivation of Sox9 in prechondrocytes in Sox9fl/flCol2Cre and Sox9fl/flCol11enhCre embryos demonstrated that SOX9 is required to enhance cell survival, maintain expression of cartilage markers already active (e.g., Col2a1), and activate markers of overtly differentiated chondrocytes, such as Acan (aggrecan core protein gene) (5,7). It remains unknown whether the surviving cells maintained their chondrocyte lineage commitment or switched to another fate.
Growth plates are cartilage primordia-derived structures that have two main functions: on one hand, they single-handedly ensure skeletal growth, and on the other hand, they crosstalk with perichondrium cells to prepare the ultimate remodeling of cartilage into bone, a process called endochondral ossification. Growth plate chondrocytes proliferate actively and line up in longitudinal columns, and subsequently undergo terminal maturation. This maturation process occurs in a staggered manner, starting in the center of the cartilage elements and progressing towards the epiphyseal ends, one cell layer at the time. It sequentially entails cell growth arrest, prehypertrophy, hypertrophy, and either cell death or conversion into osteoblasts. The analysis of Sox9fl/flCol11promCre and Sox9fl/flAcanCre embryos, which inactivated Sox9 in growth plate chondrocytes, and Col10a1SOX9 transgenic embryos, which overexpressed SOX9 in hypertrophic cells, revealed that SOX9 is required to permit chondrocyte proliferation and hypertrophy, which are the drivers of skeletal growth, and to inhibit chondrocyte prehypertrophy and ultimate death or osteoblastic transformation, which permit endochondral ossification (7–9). These studies were paradigm shifting because it was thought until then that SOX9 inhibited chondrocyte hypertrophy. The latter notion was mainly based on the fact that chondrocytes abruptly lose SOX9 RNA as they undergo hypertrophy, and it was supported by data suggesting that SOX9 might directly repress Col10a1 and other genes expressed in hypertrophic chondrocytes (see later). In addition to showing that SOX9 is necessary for chondrocyte hypertrophy, these recent studies showed that the SOX9 protein outlives its mRNA in hypertrophic chondrocytes and likely has direct actions in these cells, including activation of Col10a1. The premature death or osteoblastic conversion of SOX9-depleted prehypertrophic chondrocytes and the extension of the hypertrophic zone in SOX9-overexpression studies are very interesting in view of the recent finding that a significant proportion of terminal chondrocytes do not die upon reaching the end of the wild-type growth plate, but undergo osteoblastogenesis and participate in endochondral ossification (10–12). Together, these findings thus strongly argue that SOX9 is necessary for growth plate activity and maintenance, and achieves this function by safeguarding the chondrocyte lineage fate, differentiated activity, and survival throughout hypertrophy.
While growth plates are temporary and very dynamic structures during fetal and postnatal growth, all other types of cartilage are permanent and undergo limited remodeling in adult life. The most studied of them is articular cartilage, because its degeneration is a main feature of osteoarthritis, a prevalent disease in the later half of life. Sox9 inactivation in articular chondrocytes in Sox9fl/flAcanCreER adult mice was shown to result in severe loss of cartilage-specific proteoglycans, but did not appear to affect the tissue collagen framework (13). Based on evidence that this framework is built for life, unlike the proteoglycan gel that it entraps, this finding indicated that SOX9 is necessary to ensure the normal turnover of adult cartilage. It is thus believed that the drastic repression of SOX9 that occurs in chondrocytes responding to inflammatory cytokines could significantly contribute to the inability of osteoarthritic chondrocytes to maintain and repair their tissue properly. A role for SOX9 in maintaining the lineage fate of adult articular chondrocytes is likely based on findings in development, but has not been verified yet.
Molecular actions of SOX9 in chondrocytes
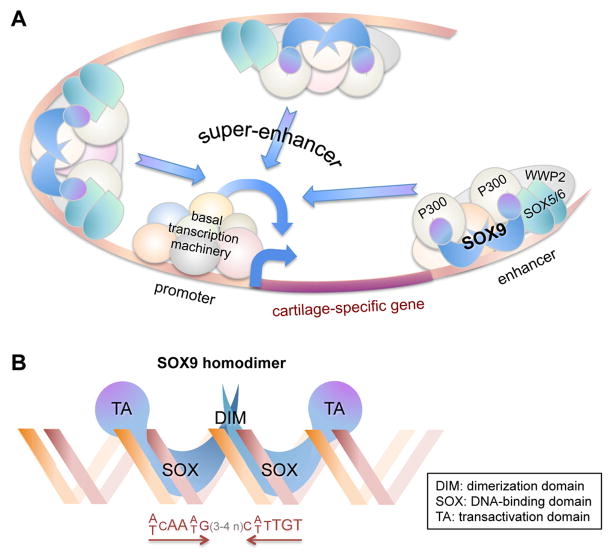
Structural and functional studies carried out over the years by multiple teams using more and more sophisticated approaches concur that SOX9 fulfills most aspects of its pivotal chondrogenic functions as a transcriptional activator. DNA-binding, reporter and other in vitro assays traditionally used to probe one gene at a time have recently been complemented with genome-wide high-throughput sequencing assays for immunoprecipitated chromatin (ChIP-seq) (3,4,14–16). Together, these approaches have provided solid evidence that SOX9 directly targets genes encoding key cartilage-specific extracellular matrix components (such as the collagen types II, IX, and XI, aggrecan, and link protein), regulators of these components (such as chondroitin 4-sulfotransferase CHST11), its own partners in transcription (such as SOX5, SOX6, and WW domain-containing protein-2, WWP2), as well as key signaling pathway mediators in cartilage (such as the fibroblast growth factor receptor-3, FGFR3). A main revelation from ChIP-seq and complementary studies is that SOX9 binds enhancers within and around a few thousands of genes and that many of these genes owe their specific and high expression in cartilage not to one or a few SOX9-bound enhancers, but to clusters of SOX9-bound enhancers. These clusters are called super-enhancers and they are located within genes and often at large distances upstream of genes (Figure 2A). These studies have consolidated evidence that SOX9 directly binds to enhancers in the chondrocyte genome by homodimerizing on pairs of 7-bp SOX-domain DNA recognition sites that face each other and are separated by 3 or 4 nucleotides (Figure 2B) (15, 16). In addition, it was noted that a small fraction of SOX9 genomic contacts were occurring in the proximal promoter of broadly expressed genes, but in the absence of SOX9-specific DNA recognition sites. These contacts are thus deemed to involve interactions of SOX9 with the basal transcription machinery rather than with DNA. Their functional consequence could be to enhance the expression of these otherwise SOX9-independent genes. It is important to note that ChIP-seq and most other studies on SOX9 actions have been conducted so far using non-hypertrophic growth plate chondrocytes. Findings may therefore be only partially relevant to the genomic actions of SOX9 in progenitor cells and in early-stage, hypertrophic, and articular chondrocytes. SOX9 may indeed target distinct genes and distinct enhancers at different cell differentiation stages and in different cartilage types to account for differential gene expression profiles.
Figure 2. SOX9 molecular actions in chondrocytes.
(A) Schematic of the genetic locus of an arbitrary cartilage-specific gene. A super-enhancer is presented as a cluster of three enhancers located upstream and downstream of the gene. Each enhancer is bound by a complex of proteins that include SOX9 and co-factors, namely SOX5/SOX6, P300, WWP2 and other yet-to-be identified proteins. Synergy between the enhancers and the basal transcription machinery, based at the promoter, results in high and cartilage-specific expression of the gene. (B) Schematic of the binding of SOX9 to DNA. Two molecules of SOX9 homodimerize upon binding in the minor groove of a DNA sequence featuring two SOX recognition sites facing each other and separated by 3 or 4 nucleotides.
In addition to being a potent transcriptional activator, SOX9 has been proposed to have other transcriptional activities. One of them is to repress undesired genes, such as markers of non-chondrocytic cells or markers of chondrocytes at different differentiation stages. It was suggested, for instance, that non-hypertrophic chondrocytes might use SOX9 to directly repress hypertrophic chondrocyte markers, such as Col10a1 (17). However, these findings are challenged by the fact that ChIP-seq studies performed to this date found strong association of SOX9 with active enhancers and promoters, but not with genomic regions presenting transcriptional repression marks. Thus, indirect mechanisms are currently favored, such as SOX9 activation of transcriptional repressors. Another transcriptional activity proposed for SOX9 in chondrogenic cells is that of a pioneer factor, i.e., a factor that binds condensed chromatin and induces the epigenetic modifications that are necessary to prime cartilage-specific genes for transactivation at the time of chondrocyte differentation (18). This assumption is based on the absolute requirement of SOX9 to specify and maintain the lineage fate of chondrocytes. However, conclusive data remain to be provided to demonstrate if and how SOX9 is a chondrogenic pioneer factor.
SOX9 has also been proposed to have non-transcriptional actions of importance in chondrocytes. For instance, competition between SOX9 and β-catenin in mesenchymal progenitors was shown to be critical to specify chondrocytic and non-chondrocytic fates, respectively, at the onset of skeletogenesis. This competition was tracked down to physical interaction occurring between the two proteins and resulting in proteasomal degradation of the protein complex and thus in elimination of the less abundant of the two proteins (19,20). SOX9 was also proposed to physically interact with RUNX2 and to refrain thereby the critical actions of this RUNT-domain transcription factor in driving chondrocyte terminal maturation and osteoblast differentiation (21). These non-transcriptional mechanisms could explain some of the repressive activities of SOX9.
In conclusion, SOX9 has key transcriptional actions in chondrocytes as an activator of cartilage-specific genes, and possibly also as a pioneer and repressor. SOX9 may also have key non-transcriptional actions. Because SOX9 is necessary for chondrogenesis, it is often referred to as “the master chondrogenic factor”. However, this appellation is premature at the current state of our knowledge, and it is also likely an overstatement. It remains indeed unknown whether SOX9 is a pioneer in the early specification of chondrogenic progenitors, and it is also clear from many studies, which are described later in this review, that SOX9 alone is not sufficient for chondrogenesis. Many factors must contribute to activate the SOX9 gene and control the SOX9 protein activity. SOX9 is thus “a master chondrogenic factor”, working downstream, together with and upstream of other master chondrogenic regulators.
Regulation of the SOX9 gene
Considering its highly consequential activities in chondrocytes, it is believed and has already been well documented that SOX9 is the subject of many types of regulatory mechanisms. Gene expression is obviously the first level of SOX9 regulation. It is also a key one. The SOX9 gene indeed exhibits a highly specific spatial and temporal expression pattern. In the fetus, for instance, SOX9 RNA is abundant in chondrocytes and a few other discrete cell types, but not detectable in other cell types. In addition, SOX9 expression must be tightly controlled quantitatively, as demonstrated by the fact that SOX9 haploinsufficiency causes CD.
Chondrogenesis is determined by the interplay of numerous signaling pathways initiated by growth factors and other biophysical or biochemical cues. Accordingly, many pathways have been shown to affect SOX9 expression in this process (see 4 for a detailed review). On the positive side, for instance, Hedgehog, bone morphogenetic protein (BMP) and fibroblast growth factor (FGF) signals are required to initiate and maintain SOX9 expression in embryonic chondrogenesis, and hypoxia and mechanical loading critically enhance SOX9 expression in developing and adult cartilage. On the negative side, Notch signaling downregulates SOX9 expression in chondrocyte progenitors and canonical WNT signaling efficiently shuts off SOX9 expression in skeletogenic progenitors committing to non-chondrocytic fates. On a yet unclear side, transforming growth factor-β (TGFβ) signaling is important in skeletogenic progenitors, but possibly not for SOX9 expression. Further showing that Sox9 expression is dynamically, the circadian clock was induce oscillation of Sox9 RNA levels throughout the day in the mouse (22). Each signaling pathway relies functionally on a series of intracellular mediators, among which transcription factors directly effect their actions at the genomic level. A prerequisite to demonstrating that such transcription factors directly control SOX9 expression requires identification and validation of the cis-elements involved in SOX9 transcription.
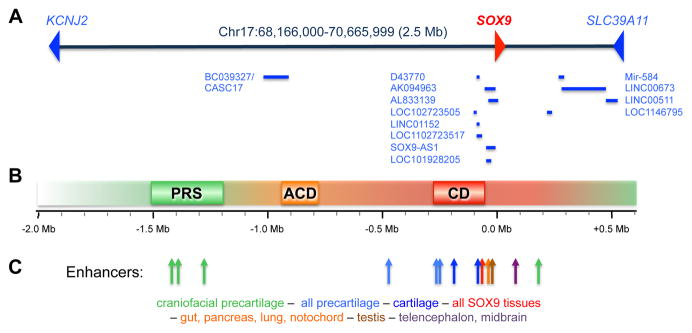
Multiple studies have used in vitro approaches to suggest that the SOX9 promoter harbors key cis-elements. These elements include binding sites for the hypoxia-inducible factor-1α (HIF1α; at −400bp relative to the transcription start site) (23), nuclear factor kappa B member RELA (at −250bp) (24), and Notch signaling mediator RBPj (at −3kb) (25). They also include binding sites for the cyclic-AMP response element-binding protein CREB and zinc-finger transcription factor SP1 (within −200bp), whose positive actions on SOX9 expression may be blocked in response to the inflammatory cytokine interleukin-1 (26). Methylation of promoters at specific CpG sites is a powerful way to repress gene expression. Accordingly, canonical WNT signaling was proposed to repress SOX9 via methylation of its promoter by the de novo DNA methyltransferase DNMT3, an effect that could be countered by FGF signaling (27). While these studies concur that the SOX9 promoter may participate critically in poising, activating or inhibiting SOX9 expression, several lines of evidence also indicate that enhancers critically needed for SOX9 expression exist outside of the SOX9 promoter. First, independent groups found that the SOX9 promoter region was sufficient to activate transgenic reporters in vivo in the spinal cord and hindbrain of mouse embryos, but was totally insufficient to activate these reporters in other sites, including skeletogenic progenitors and chondrocytes (28,29). Second, SOX9 is surrounded by very large domains (1.9 Mb upstream and 0.5 Mb downstream) that lack coding genes and that are the targets of chromosomal translocations and deletions causing SOX9-dependent diseases (Figure 3). Campomelic dysplasia (CD) is most often caused by alterations happening 50–375kb upstream of SOX9; acampomelic dysplasia (ACD, a mild form of CD) by alterations happening 789 to 932kb upstream of SOX9; and Pierre Robin sequence (PRS, craniofacial malformations similar to those seen in CD) by alterations occurring 1.2–1.5 Mb upstream of SOX9 (30–32). It must be noted that multiple cases of these diseases have also been associated with alterations occurring essentially anywhere between 1.5 Mb upstream and 1.5 Mb downstream of SOX9. SOX9 thus appears to be under the control of upstream, downstream and promoter cis-acting elements that are altogether spread over as much as 3 Mb of genomic DNA.
Figure 3. SOX9 genetic locus, disease-causing chromosomal alterations, and enhancers.
(A) Schematic of the human chromosomal segment containing SOX9 and its flanking upstream and downstream domains. The coding genes that are SOX9’s neighbors are shown (KCNJ2 and SLC39A11), as well as non-coding genes (RefSeq and GeneBank, CCDS, Rfam and tRNA, as listed on the UCSC genome browser). (B) Schematic of the domains upstream of SOX9 that are prone to chromosomal alterations causing PRS, ACD and CD disease. The shaded background illustrates that other domains are also subject to disease-causing alterations. (C) Location of tissue-specific enhancers identified upstream and downstream of SOX9. The position of each enhancer is indicated with an arrow. The main domain of activity of each enhancer is indicated and represented with colors.
To date, several long-range enhancers of SOX9 have been proposed based on histone modification marks and reporter activities in transgenic embryos (Figure 3B) (28–33). The CD/ACD-prone region includes a −70kb enhancer active in multiple cell types expressing SOX9; −84 and −196kb enhancers primarily active in growth plate chondrocytes; and a −250kb enhancer specific to precartilaginous condensations (29,33). The PRS-prone region contains multiple enhancers active in overlapping domains in the craniofacial region. Based on these data and evidence that many more regions in the SOX9 flanking domains are highly conserved in vertebrate genomes and show histone modifications typical of enhancers in samples containing cartilage, the current model is that SOX9 expression is brought about by a large series of enhancers whose activities are spatially and temporally overlapping and dependent upon various signaling pathways. Genomic deletion of these enhancers will be necessary in future research to assess the actual contribution of each enhancer to SOX9 expression.
Along with identifying SOX9 enhancers, an important task is to find the factors that bind to these enhancers and poise, activate or repress SOX9 expression. The homeobox transcription factor MSX1 has been proposed to target the first enhancer identified in the PRS region (31). SOX9 was previously shown to positively control its own gene in chondrocytes, but evidence of a direct effect was lacking (34). It is thus significant that SOX9 was found to bind and activate the cartilage enhancers present in the CD-prone region (29,33). Long non-coding RNAs (LncRNAs, typically >200 nucleotides) are a recently discovered class of molecules that can have profound effects on gene expression through cis- or trans-acting effects. Among them, the LncRNA DA125942 (on 12q) was proposed to interact with PTHLH (encoding the parathyroid hormone-like protein; on 12p) in cis and with SOX9 (on 17q) in trans (35). As PTHLH and SOX9 are both essential in chondrogenesis, this LncRNA was proposed to facilitate the establishment of a chondrogenic transcriptional factory that could enhance the expression of both genes. Several LncRNAs have been detected in chondrocytes and several others may originate in the large domains flanking SOX9. These few examples of transcription factors and LncRNAs currently known stress the importance of pursuing research to identify the whole sets of such players that are expressed in the chondrocyte lineage and to elucidate their importance in SOX9 expression and chondrogenesis.
Regulation of the SOX9 mRNA
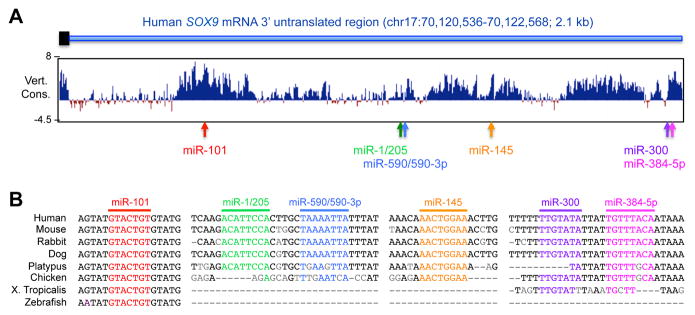
Once its gene is transcribed, SOX9 can be next regulated at the level of its mRNA. To our knowledge, the SOX9 mRNA is not subjected to differential splicing or intracellular localization under physiological conditions, but there is evidence that it can be targeted by microRNAs, i.e., tiny (approximately 20 nucleotides) non-coding RNAs that typically hybridize with specific sequences in the 3′ untranslated region (3′UTR) of mRNAs to induce either mRNA degradation or translation blockade. Sequence analysis indicates that the 3′UTR of the human SOX9 mRNA features recognition sites for six distinct miRNAs: miR-101, miR-1/205, miR-590/590-3p, miR-145, miR-300, and miR-384-5p (Figure 4A). None of these miRNAs figures among the miRNAs detected in human embryonic growth plate cartilage (36), but miR-145 was reported expressed in vivo in bovine articular cartilage and in vitro in human and mouse mesenchymal stem cells, articular chondrocytes and chondrosarcoma cells (37–40). Its binding to the SOX9 RNA was shown to reduce the level of SOX9 protein. In addition, miR-101, whose recognition site is the most conserved among those of the six miRNAs (Figure 4B), was reported expressed in rat articular chondrocytes in vitro and capable of reducing the levels of SOX9 RNA and protein (41). Interestingly, it was shown to be upregulated in chondrocytes in response to IL1β and proposed to mediate the negative effect of the inflammatory cytokine on the RNA levels of SOX9 and SOX9 targets. Functional links between the other miRNAs and SOX9 in chondrocytes or other cell types remain unknown. It is interesting to note that, in addition to being a target of microRNAs in cartilage, SOX9 directly activates expression of miR-140, whose important roles in cartilage development and adult homeostasis have been demonstrated in the mouse (42). Additional studies are thus warranted to further uncover the miRNAs that directly impact the level or translation of the SOX9 RNA, and those that indirectly counter SOX9 activity by binding to its target gene RNAs.
Figure 4. Identification microRNA binding sites in the human SOX9 mRNA.
(A) A schematic of the 3′ end of the SOX9 coding sequence (black rectangle) and entire 3′ untranslated region (3′UTR; blue line) is shown above a conservation plot for 100 vertebrate genomes downloaded from the UCSC genome browser (www.genome.ucsc.edu). The chromosome coordinates of the 3′UTR are indicated. The position of recognition sites for six microRNAs is shown with arrows. (B) Alignment of the miRNA binding sites and flanking sequences in eight vertebrate genomes.
Regulation of the intracellular distribution of the SOX9 protein
To function as a transcription factor, SOX9 must be located in the cell nucleus. Interestingly, evolution has chosen to link this event to the prominent functional domain of SOX proteins by embedding and highly conserve nuclear import and export signals for SOX9 and other SOX proteins in their SOX domain. These signals consist of an N-terminal bipartite nuclear localization (NLS) signal binding calmodulin, a central nuclear export signal (NES) binding exportin-1, and a C-terminal basic cluster NLS, binding importin-α or -β (Figure 5; 43,44). Nuclear trafficking of SOX9 is critical during mammalian sex differentiation: SOX9 is primarily cytoplasmic in the undifferentiated gonad of both sexes, and becomes transcriptionally active once translocated to the nucleus at the onset of Sertoli cell differentiation in males (45). To our knowledge, no definitive studies have yet been published showing whether the intracellular localization of SOX9 changes during chondrocyte differentiation or in specific physiological or pathological conditions in vivo. This is, however, likely since a change in SOX9 localization from the cytoplasm to the nucleus was seen in chondrosarcoma cells subjected to cyclic tensile strain in vitro (46) This change was accompanied with an increase in TGFβ signaling that allowed SMAD2/3 to cooperate with SOX9 in gene transactivation. Moreover, a similar change was observed when human articular chondrocytes were treated with a Sirtuin deacetylase cofactor in vitro. This change was thought to be possibly relevant to preliminary observation that SOX9 could be more often cytoplasmic than nuclear in osteoarthritic than in healthy articular chondrocytes (47). Clarifying the trafficking of SOX9 and its regulation in the chondrocyte lineage is thus a warranted topic for future research.
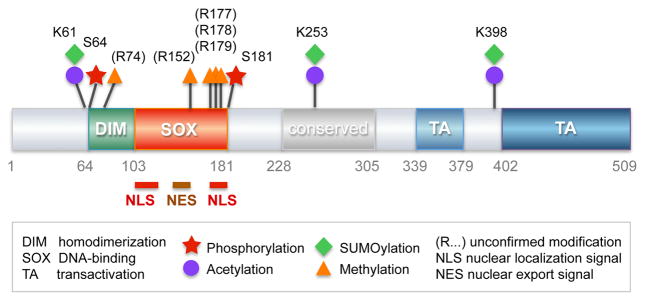
Figure 5. SOX9 protein domain organization and posttranslational regulation.
Schematic of the 509-residue-long human SOX9 protein. Functional and highly conserved domains are shown with various colors and their residue boundaries are given in grey. Posttranslational modifications and nuclear trafficking domains are represented as indicated.
Post-translational regulation of the SOX9 protein
Post-translational modifications exist in different flavors and with various consequences on protein structure, stability, and function. With 509 residues encompassing highly conserved dimerization, DNA-binding, transactivation and other domains (Figure 5), the SOX9 protein could undergo many types of regulatory modifications. Multiple studies support this concept.
Phosphorylation
While multiple phosphorylation events affect many types of proteins, including SOX proteins (www.phosphosite.org; 48,49), the only phosphorylation events reported to date for SOX9 are targeting of the residues S64 and S181 by the cAMP-dependent protein kinase A, PKA (Figure 5) (50,51). The highest level of SOX9 phosphorylated at S181 was detected in fetal growth plates in prehypertrophic chondrocytes, where the total level of SOX9 protein is also the highest. These cells specifically express the gene for the parathyroid hormone receptor (PTH1R), and in vitro and in vivo data led to suggest that PTH1R signaling might delay chondrocyte hypertrophy in part by stimulating SOX9 phosphorylation at S181. It is noteworthy that PKA is implicated in multiple pathways and could thus phosphorylate SOX9 in response to various stimuli. Moreover, other kinases have a similar activity. For instance, ROCK1 can directly phosphorylate SOX9 at S181 in vitro and may thereby mediate the positive effects of TGFβ and short-term dynamic compression on SOX9 activity in chondrocytes (52). SOX9 S181 can also be directly targeted by cGKII, a cGMP-dependent protein kinase type II that, unlike PTH1R signaling, promotes chondrocyte hypertrophy (52). Phosphorylation of SOX9 at S64 and S181 was shown to enhance SOX9’s ability to transactivate reporter genes in vitro (50,54). Two underlying mechanisms were proposed: increase in SOX9 binding to DNA (49) and increase in SOX9 translocation to the nucleus (54). S64 is located immediately upstream of the dimerization domain of SOX9, and S181 is located immediately downstream of the SOX domain and one of the nuclear import signals of SOX9. S64 phosphorylation might facilitate SOX9 dimerization and hence binding to DNA, and S181 phosphorylation might thus enhance SOX9 nuclear translocation and DNA-binding. Although many studies reported SOX9 phosphorylation at S181 in vivo or in vitro, it remains unknown whether these two phosphorylation events significantly affect SOX9 activity in vivo.
Methylation and acetylation
The past two decades have brought to light the crucial importance of chromatin-modifying enzymes, including histone methylases, demethylases, acetylases, and deacetylases, on epigenetic modification and thereby on gene regulation and cell fate determination. At the same time, it has become clear that many of these enzymes also target non-histone proteins, namely transcription factors, resulting in changes in protein levels and activities. The coactivator-associated arginine methyltransferase-1 CARM1 (also known as PRMT4) was shown in vitro to methylate arginine residues in the N-terminus and SOX domain of SOX9 (possibly at residues R74, R152, R177, R178 and R179) and to prevent interaction of SOX9 with β-catenin (Figure 5) (55). It remains to be determined whether these events occur in vivo and explain why mouse fetuses globally lacking or overexpressing CARM1 show reduced proliferation of growth plate chondrocytes and delayed endochondral ossification. To our knowledge, this is the only study suggesting that SOX9 can be methylated. Regarding other SOX proteins, the only report listed on the phosphosite is methylation of a residue in SOX12 (www.phosphosite.org).
The tat-interacting protein-60 TIP60 (also known as KAT5) was suggested through an RNAi approach in a chondrogenic micromass assay in vitro to promote chondrogenesis (56). It was also shown to directly acetylate SOX9 (possibly at residues K61, K253 and K398) (Figure 5) and to enhance SOX9 transactivation of a Col2a1 reporter in vitro. However, the lysine residues presumably targeted by TIP60 in SOX9 were not involved in the enhanced reporter activity. It thus remains unknown whether and how TIP60 impacts chondrogenesis in vivo. SIRT1 is a nicotinamide adenine dinucleotide (NAD)-dependent class III histone deacetylase involved in many processes and diseases (57). It is best known for its histone deacetylase activity that causes transcriptional repression, but it also targets non-histone proteins. Sirt1+/− mice are growth-delayed and show early signs of osteoarthritis by middle age, and Sirt1−/− pups die before weaning age with skeletal malformations. SIRT1 is able to deacetylate SOX9 in vitro and may thereby enhance ACAN and COL2A1 expression (58,59). Relevant to osteoarthritis, pro-inflammatory conditions inactivate SIRT1 through cathepsin-mediated cleavage, a mechanism that may thus contribute to loss of SOX9 activity (60). While the lysine(s) targeted by SIRT1 in SOX9 remain elusive, it was proposed that SIRT1 facilitates SOX9 activity by increasing its nuclear localization (47). Interestingly, SIRT1-mediated deacetylation of SOX2 at lysine K75 occurs during reprogramming of mouse somatic cells into pluripotent stem cells to prevent SOX2 nuclear export and degradation (61,62). SOX proteins might thus share important SIRT1-mediated regulation of intracellular localization and thereby nuclear activity. Proteomic studies have revealed that several lysine residues are acetylated in SOX9 and other SOX proteins (www.phosphosite.org), suggesting yet unknown modes of SOX protein regulation through acetylation and deacetylation.
Ubiquitination and SUMOylation
The attachment of ubiquitin or SUMO peptides to specific lysine residues can prompt protein degradation, affect protein intracellular trafficking, and modify protein interactions. The E3 ubiquitin ligase E6-AP/UBE3A can directly ubiquitinate SOX9 (at yet unknown residues) and thereby reduce its stability in vitro (63). E6-AP is present in proliferating and hypertrophic growth plate chondrocytes, but not in prehypertrophic cells, where SOX9 is most abundant. This suggests that E6-AP may limit the level of SOX9 protein in early and hypertrophic cells. Supporting this notion, primary chondrocytes isolated from E6-AP-null mice contained slightly more SOX9 protein. Hypertrophic chondrocytes matured with a mild delay in these mice, indicating that E6-AP may not critically control the SOX9 protein level, unless it acts in redundancy with other E3 ubiquitin ligases.
SOX9 sumoylation was first identified upon overexpression of mouse SOX9 in non-chondrocytic HEK293 cells (64). It occurred at a lysine equivalent to K398 in the human SOX9, slightly upstream of the main transactivation domain (Figure 5), and enhanced SOX9 transactivation of a reporter gene. It was then reported that SOX9 and SOX10, which are close relatives in the SOXE group, are sumoylated in Xenopus embryos (at residues corresponding to K61 and K398 in human SOX9) (65). Interestingly, sumoylation inhibited the neural-crest activity of SOXE proteins but promoted ear formation. The same group later reported that sumoylation changed the status of SOXE proteins from that of transcriptional activators, capable of recruiting the co-activator CREB-binding protein (CBP)/P300 histone deacetylase, to that of transcriptional repressors, capable of recruiting the Groucho co-repressor GRG4 (66). PIAS1 (protein inhibitor of activated STAT1) and other SUMO ligase family members were shown to directly bind and SUMOylate SOX9 in vitro (67,68). Altogether, these findings suggest that SOX9 ubiquitination and sumoylation may occur in chondrocytes in vivo, but the functional impacts of these events remain to be elucidated.
SOX9 functional partners
A major level of SOX9 protein regulation is achieved by physical and functional interaction with other proteins. Transcription factors typically work in enhanceosomes, i.e., complexes of proteins binding to genomic enhancer sequences and interacting with the basal transcription machinery anchored at gene promoters. Many SOX proteins synergize with transcriptional partners for DNA binding and transactivation (49). For instance, SOX2 synergizes with OCT4 in pluripotent stem cells. The two proteins physically interact with one another and thereby secure their binding to enhancers featuring recognition motifs for each protein. SOX9 is its own partner in chondrocytes, binding most efficiently to DNA by homodimerizing on pairs of inverted SOX-domain recognition motifs (Figure 2B). Many other proteins likely participate with SOX9 in chondrocyte enhanceosomes (Figure 2A). They include SOX5 and SOX6, two members of the SOX family that are co-expressed with SOX9 in chondrocytes. SOX5 and SOX6 are very closely related to each other, but besides sharing partial homology with SOX9 in the SOX domain, they are totally different from SOX9 (69). Loss-of-function experiments in vivo have revealed that they act redundantly in the chondrocyte lineage to potentiate the ability of SOX9 to differentiate chondrocytes, but not to secure lineage commitment, and gain-of-function experiments have shown that SOX5/SOX6 and SOX9 form a trio, often called the chondrogenic SOX trio, which is necessary and sufficient to promote chondrocyte differentiation of mesenchymal progenitor cells in vitro and in vivo (70). SOX5/SOX6 lack intrinsic transactivation domains, but by binding to recognition sites close to those of SOX9 on enhancers, they consolidate SOX9 binding to DNA and thereby potentiate SOX9 activity (16). The mechanism underlying this consolidation remains unknown, but likely involves other factors because SOX5/SOX6 appear unable to physically interact with SOX9. WWP2 is a NEDD4-like HECT domain-containing ubiquitin ligase that physically interacts with SOX9 and that was proposed to enhance chondrocyte-specific gene transactivation by participating in SOX trio-containing enhanceosomes (71). Its gene is a target of the SOX trio in chondrocytes (16) and its importance in skeletogenesis was revealed by showing that its knockdown in the zebrafish resulted in defective palatogenesis (71). Other proteins proposed to participate with SOX9 in transcriptional activation include the transcriptional co-activator p300/CBP (72) and the TGFβ transcriptional mediator SMAD3 (73). MEF2C is a possible partner of SOX9 in the activation of Col10a1 and possibly other genes in prehypertrophic and hypertrophic chondrocytes (8). The genome-wide analysis of transcription factor recognition motifs enriched around the binding sites of the SOX trio on chondrocyte-specific enhancers has suggested the possibility that forkhead, RUNT-domain, NFAT, zinc finger and AP1 family members might partner with the chondrogenic SOX trio (15,16). Alternatively, these factors could precede the SOX trio to poise or inhibit enhancers or could succeed to the SOX trio to prolong or inhibit enhancer activities. As of today, the identity and roles of these factors in chondrocyte enhancers remain speculative. Thus, more research is necessary to reveal the whole sets of factors that directly participate in mastering chondrogenesis along with SOX9 at all steps of chondrocyte differentiation.
Conclusion
The last two decades have opened pioneering leads in the molecular exploration of chondrogenesis. Fueled by keen scientific curiosity and by acute motivation to better understand and treat cartilage malformation and degeneration diseases, many of these leads can be traced back to the discovery that SOX9 heterozygous mutations cause Campomelic Dysplasia and that Sox9 inactivation in the mouse precludes chondrogenesis. Thanks to remarkable efforts worldwide, there is no doubt left today that SOX9 is a pivotal chondrogenic transcription factor and that a whole network of regulatory mechanisms depend on and influence SOX9 expression and activity. At the same time, it is also very clear that many questions remain unanswered and that many more may arise as research unveils new facets of SOX9 actions and regulation. For instance, the question of “which came first, the chicken or the egg?” remains largely open regarding the onset of SOX9 gene expression in progenitor cells and the specification of these cells towards chondrogenesis. Questions also remain incompletely answered on the spectrum of SOX9 actions in chondrogenesis: is SOX9 a pioneer factor before becoming a transcriptional activator and is it able to directly repress genes? Finally, questions have hardly begun to be addressed on the posttranscriptional and posttranslational regulation of SOX9 and on their functional consequences on chondrogenesis. To this day, insights into SOX9 functions in chondrogenesis have come using developmental models more often than postnatal, aging and disease models, using in vitro models more often than animal models and clinical samples, and using isolated readouts more often than global approaches in genomics, transcriptomics and proteomics. By employing the most advanced techniques, such as CRISPR/Cas-mediated genome editing, scientists should aim in the near future to expand knowledge and to validate in vivo many of the findings made in vitro on SOX9 actions and regulation from development to aging and diseases. It is important to keep in mind that subtle changes in SOX9 function and expression during development could have profound impact on cartilage aging and wear and tear later in life. Building on already acquired knowledge and on promising new technologies, one can predict that the next two decades will see an explosion of breakthrough discoveries from which new strategies will emerge to prevent, treat and cure cartilage diseases.
Footnotes
Declaration of interest
The authors report no conflicts of interest. This review was supported by research grants from the USA/Israel Binational Science Foundation to VL and MDG, the National Institutes of Health to VL (AR46249 and AR60016), the European Commission Framework 7 programme (EU FP7; HEALTH.2012.2.4.5-2, project 305815) and The Marie Curie European IRG reintegration grant (proposal 268214) to MDG.
References
- 1.Lovell-Badge R. The early history of the SOX genes. Int J Biochem Cell Biol. 2010;42:378–80. doi: 10.1016/j.biocel.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Schafer AJ, Foster JW, Kwok C, Weller PA, Guioli S, Goodfellow PN. Campomelic dysplasia with XY sex reversal: diverse phenotypes resulting from mutations in a single gene. Ann N Y Acad Sci. 1996;785:137–49. doi: 10.1111/j.1749-6632.1996.tb56252.x. [DOI] [PubMed] [Google Scholar]
- 3.Akiyama H, Lefebvre V. Unraveling the transcriptional regulatory machinery in chondrogenesis. J Bone Miner Metab. 2011;29:390–5. doi: 10.1007/s00774-011-0273-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozhemyakina E, Lassar AB, Zelzer E. A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development. 2015;142:817–31. doi: 10.1242/dev.105536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–28. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–9. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 7.Ikegami D, Akiyama H, Suzuki A, Nakamura T, Nakano T, Yoshikawa H, Tsumaki N. Sox9 sustains chondrocyte survival and hypertrophy in part through Pik3ca-Akt pathways. Development. 2011;138:1507–19. doi: 10.1242/dev.057802. [DOI] [PubMed] [Google Scholar]
- 8.Dy P, Wang W, Bhattaram P, Wang Q, Wang L, Ballock RT, Lefebvre V. Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev Cell. 2012;22:597–609. doi: 10.1016/j.devcel.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hattori T, Müller C, Gebhard S, Bauer E, Pausch F, Schlund B, Bösl MR, Hess A, Surmann-Schmitt C, von der Mark H, de Crombrugghe B, von der Mark K. SOX9 is a major negative regulator of cartilage vascularization, bone marrow formation and endochondral ossification. Development. 2010;137:901–11. doi: 10.1242/dev.045203. [DOI] [PubMed] [Google Scholar]
- 10.Yang L, Tsang KY, Tang HC, Chan D, Cheah KS. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc Natl Acad Sci U S A. 2014;111:12097–102. doi: 10.1073/pnas.1302703111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X, von der Mark K, Henry S, Norton W, Adams H, de Crombrugghe B. Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet. 2014;10:e1004820. doi: 10.1371/journal.pgen.1004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park J, Gebhardt M, Golovchenko S, Perez-Branguli F, Hattori T, Hartmann C, Zhou X, de Crombrugghe B, Stock M, Schneider H, von der Mark K. Dual pathways to endochondral osteoblasts: a novel chondrocyte-derived osteoprogenitor cell identified in hypertrophic cartilage. Biol Open. 2015;4:608–21. doi: 10.1242/bio.201411031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henry SP, Liang S, Akdemir KC, de Crombrugghe B. The postnatal role of Sox9 in cartilage. J Bone Miner Res. 2012;27:2511–25. doi: 10.1002/jbmr.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh CD, Lu Y, Liang S, Mori-Akiyama Y, Chen D, de Crombrugghe B, Yasuda H. SOX9regulates multiple genes in chondrocytes, including genes encoding ECM proteins, ECM modification enzymes, receptors, and transporters. PLoS One. 2014;9:e107577. doi: 10.1371/journal.pone.0107577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohba S, He X, Hojo H, McMahon AP. Distinct Transcriptional Programs Underlie Sox9 Regulation of the Mammalian Chondrocyte. Cell Rep. 2015;12:229–43. doi: 10.1016/j.celrep.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu CF, Lefebvre V. The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res. 2015;43:8183–203. doi: 10.1093/nar/gkv688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung VY, Gao B, Leung KK, Melhado IG, Wynn SL, Au TY, Dung NW, Lau JY, Mak AC, Chan D, Cheah KS. SOX9 governs differentiation stage-specific gene expression in growth plate chondrocytes via direct concomitant transactivation and repression. PLoS Genet. 2011;7:e1002356. doi: 10.1371/journal.pgen.1002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaret KS, Mango SE. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev. 2016;37:76–81. doi: 10.1016/j.gde.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR, McCrea PD, de Crombrugghe B. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 2004;18:1072–87. doi: 10.1101/gad.1171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Topol L, Chen W, Song H, Day TF, Yang Y. Sox9 inhibits Wnt signaling by promoting beta-catenin phosphorylation in the nucleus. J Biol Chem. 2009;284:3323–33. doi: 10.1074/jbc.M808048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou G, Zheng Q, Engin F, Munivez E, Chen Y, Sebald E, Krakow D, Lee B. Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc Natl Acad Sci USA. 2006;103:19004–9. doi: 10.1073/pnas.0605170103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dudek M, Gossan N, Yang N, Im HJ, Ruckshanthi JP, Yoshitane H, Li X, Jin D, Wang P, Boudiffa M, Bellantuono I, Fukada Y, Boot-Handford RP, Meng QJ. The chondrocyte clock gene Bmal1 controls cartilage homeostasis and integrity. J Clin Invest. 2016;126:365–76. doi: 10.1172/JCI82755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amarilio R, Viukov SV, Sharir A, Eshkar-Oren I, Johnson RS, Zelzer E. HIF1alpha regulation of Sox9 is necessary to maintain differentiation of hypoxic prechondrogenic cells during early skeletogenesis. Development. 2007;134:3917–28. doi: 10.1242/dev.008441. [DOI] [PubMed] [Google Scholar]
- 24.Ushita M, Saito T, Ikeda T, Yano F, Higashikawa A, Ogata N, Chung U, Nakamura K, Kawaguchi H. Transcriptional induction of SOX9 by NF-kappaB family member RelA in chondrogenic cells. Osteoarthritis Cartilage. 2009;17:1065–75. doi: 10.1016/j.joca.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Chen S, Tao J, Bae Y, Jiang MM, Bertin T, Chen Y, Yang T, Lee B. Notch gain of function inhibits chondrocyte differentiation via Rbpj-dependent suppression of Sox9. J Bone Miner Res. 2013;28:649–59. doi: 10.1002/jbmr.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piera-Velazquez S, Hawkins DF, Whitecavage MK, Colter DC, Stokes DG, Jimenez SA. Regulation of the human SOX9 promoter by Sp1 and CREB. Exp Cell Res. 2007;313:1069–79. doi: 10.1016/j.yexcr.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar D, Lassar AB. Fibroblast growth factor maintains chondrogenic potential of limb bud mesenchymal cells by modulating DNMT3A recruitment. Cell Rep. 2014;8:1419–31. doi: 10.1016/j.celrep.2014.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bagheri-Fam S, Barrionuevo F, Dohrmann U, Günther T, Schüle R, Kemler R, Mallo M, Kanzler B, Scherer G. Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev Biol. 2006;291:382–97. doi: 10.1016/j.ydbio.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Mead TJ, Wang Q, Bhattaram P, Dy P, Afelik S, Jensen J, Lefebvre V. A far-upstream (-70 kb) enhancer mediates Sox9 auto-regulation in somatic tissues during development and adult regeneration. Nucleic Acids Res. 2013;41:4459–69. doi: 10.1093/nar/gkt140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon CT, Attanasio C, Bhatia S, Benko S, Ansari M, Tan TY, Munnich A, Pennacchio LA, Abadie V, Temple IK, Goldenberg A, van Heyningen V, Amiel J, FitzPatrick D, Kleinjan DA, Visel A, Lyonnet S. Identification of novel craniofacial regulatory domains located far upstream of SOX9 and disrupted in Pierre Robin sequence. Hum Mutat. 2014;35:1011–20. doi: 10.1002/humu.22606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benko S, Fantes JA, Amiel J, Kleinjan DJ, Thomas S, Ramsay J, Jamshidi N, Essafi A, Heaney S, Gordon CT, McBride D, Golzio C, Fisher M, Perry P, Abadie V, Ayuso C, Holder-Espinasse M, Kilpatrick N, Lees MM, Picard A, Temple IK, Thomas P, Vazquez MP, Vekemans M, Roest Crollius H, Hastie ND, Munnich A, Etchevers HC, Pelet A, Farlie PG, Fitzpatrick DR, Lyonnet S. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet. 2009;41:359–64. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
- 32.Gordon CT, Tan TY, Benko S, Fitzpatrick D, Lyonnet S, Farlie PG. Long-range regulation at the SOX9 locus in development and disease. J Med Genet. 2009;46:649–56. doi: 10.1136/jmg.2009.068361. [DOI] [PubMed] [Google Scholar]
- 33.Yao B, Wang Q, Liu CF, Bhattaram P, Li W, Mead TJ, Crish JF, Lefebvre V. The SOX9 upstream region prone to chromosomal aberrations causing campomelic dysplasia contains multiple cartilage enhancers. Nucleic Acids Res. 2015;43:5394–408. doi: 10.1093/nar/gkv426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar D, Lassar AB. The transcriptional activity of Sox9 in chondrocytes is regulated by RhoA signaling and actin polymerization. Mol Cell Biol. 2009;29:4262–73. doi: 10.1128/MCB.01779-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maass PG, Rump A, Schulz H, Stricker S, Schulze L, Platzer K, Aydin A, Tinschert S, Goldring MB, Luft FC, Bähring S. A misplaced lncRNA causes brachydactyly in humans. J Clin Invest. 2012;122:3990–4002. doi: 10.1172/JCI65508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McAlinden A, Varghese N, Wirthlin L, Chang LW. Differentially expressed microRNAs in chondrocytes from distinct regions of developing human cartilage. PLoS One. 2013;8:e75012. doi: 10.1371/journal.pone.0075012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang B, Guo H, Zhang Y, Chen L, Ying D, Dong S. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS One. 2011;6:e21679. doi: 10.1371/journal.pone.0021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Sanchez A, Dudek KA, Murphy CL. Regulation of human chondrocyte function through direct inhibition of cartilage master regulator SOX9 by microRNA-145 (miRNA-145) J Biol Chem. 2012;287:916–24. doi: 10.1074/jbc.M111.302430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mak IW, Singh S, Turcotte R, Ghert M. The epigenetic regulation of SOX9 by miR-145 in human chondrosarcoma. J Cell Biochem. 2015;116:37–44. doi: 10.1002/jcb.24940. [DOI] [PubMed] [Google Scholar]
- 40.Dunn W, DuRaine G, Reddi AH. Profiling microRNA expression in bovine articular cartilage and implications for mechanotransduction. Arthritis Rheum. 2009;60:2333–9. doi: 10.1002/art.24678. [DOI] [PubMed] [Google Scholar]
- 41.Dai L, Zhang X, Hu X, Zhou C, Ao Y. Silencing of microRNA-101 prevents IL-1β-induced extracellular matrix degradation in chondrocytes. Arthritis Res Ther. 2012;14:R268. doi: 10.1186/ar4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyaki S, Sato T, Inoue A, Otsuki S, Ito Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, Takada S, Lotz MK, Ueno-Kudo H, Asahara H. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173–85. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Südbeck P, Scherer G. Two independent nuclear localization signals are present in the DNA-binding high-mobility group domains of SRY and SOX9. J Biol Chem. 1997;272:27848–52. doi: 10.1074/jbc.272.44.27848. [DOI] [PubMed] [Google Scholar]
- 44.Gasca S, Canizares J, De Santa Barbara P, Mejean C, Poulat F, Berta P, Boizet-Bonhoure B. A nuclear export signal within the high mobility group domain regulates the nucleocytoplasmic translocation of SOX9 during sexual determination. Proc Natl Acad Sci U S A. 2002;99:11199–204. doi: 10.1073/pnas.172383099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morais da Silva S, Hacker A, Harley V, Goodfellow P, Swain A, Lovell-Badge R. Sox9 expression during gonadal development implies a conserved role for the gene in testis differentiation in mammals and birds. Nat Genet. 1996;14:62–8. doi: 10.1038/ng0996-62. [DOI] [PubMed] [Google Scholar]
- 46.Furumatsu T, Matsumoto E, Kanazawa T, Fujii M, Lu Z, Kajiki R, Ozaki T. Tensile strain increases expression of CCN2 and COL2A1 by activating TGFβ-Smad2/3 pathway in chondrocytic cells. J Biomech. 2013;46:1508–15. doi: 10.1016/j.jbiomech.2013.03.028. [DOI] [PubMed] [Google Scholar]
- 47.Bar Oz M, Kumar A, Elayyan J, Reich E, Binyamin M, Kandel L, Liebergall M, Steinmeyer J, Lefebvre V, Dvir-Ginzberg M. Acetylation reduces SOX9 nuclear entry and ACAN gene transactivation in human chondrocytes. Aging Cell. 2016 doi: 10.1111/acel.12456. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernard P, Harley VR. Acquisition of SOX transcription factor specificity through protein-protein interaction, modulation of Wnt signalling and post-translational modification. Int J Biochem Cell Biol. 2010 Mar;42(3):400–10. doi: 10.1016/j.biocel.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 49.Kamachi Y, Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140:4129–44. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- 50.Huang W, Zhou X, Lefebvre V, de Crombrugghe B. Phosphorylation of SOX9 by cyclic AMP-dependent protein kinase A enhances SOX9’s ability to transactivate a Col2a1 chondrocyte-specific enhancer. Mol Cell Biol. 2000;20:4149–58. doi: 10.1128/mcb.20.11.4149-4158.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang W, Chung UI, Kronenberg HM, de Crombrugghe B. The chondrogenic transcription factor Sox9 is a target of signaling by the parathyroid hormone-related peptide in the growth plate of endochondral bones. Proc Natl Acad Sci U S A. 2001;98:160–5. doi: 10.1073/pnas.011393998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haudenschild DR, Chen J, Pang N, Lotz MK, D’Lima DD. Rho kinase-dependent activation of SOX9 in chondrocytes. Arthritis Rheum. 2010;62:191–200. doi: 10.1002/art.25051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chikuda H, Kugimiya F, Hoshi K, Ikeda T, Ogasawara T, Shimoaka T, Kawano H, Kamekura S, Tsuchida A, Yokoi N, Nakamura K, Komeda K, Chung UI, Kawaguchi H. Cyclic GMP-dependent protein kinase II is a molecular switch from proliferation to hypertrophic differentiation of chondrocytes. Genes Dev. 2004;18:2418–29. doi: 10.1101/gad.1224204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malki S, Nef S, Notarnicola C, Thevenet L, Gasca S, Méjean C, Berta P, Poulat F, Boizet-Bonhoure B. Prostaglandin D2 induces nuclear import of the sex-determining factor SOX9 via its cAMP-PKA phosphorylation. EMBO J. 2005;24:1798–809. doi: 10.1038/sj.emboj.7600660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ito T, Yadav N, Lee J, Furumatsu T, Yamashita S, Yoshida K, Taniguchi N, Hashimoto M, Tsuchiya M, Ozaki T, Lotz M, Bedford MT, Asahara H. Arginine methyltransferase CARM1/PRMT4 regulates endochondral ossification. BMC Dev Biol. 2009;9:47. doi: 10.1186/1471-213X-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hattori T, Coustry F, Stephens S, Eberspaecher H, Takigawa M, Yasuda H, de Crombrugghe B. Transcriptional regulation of chondrogenesis by coactivator Tip60 via chromatin association with Sox9 and Sox5. Nucleic Acids Res. 2008;36:3011–24. doi: 10.1093/nar/gkn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dvir-Ginzberg M, Steinmeyer J. Towards elucidating the role of SirT1 in osteoarthritis. Front Biosci. 2013;18:343–55. doi: 10.2741/4105. [DOI] [PubMed] [Google Scholar]
- 58.Dvir-Ginzberg M, Gagarina V, Lee EJ, Hall DJ. Regulation of cartilage-specific gene expression in human chondrocytes by SirT1 and nicotinamide phosphoribosyltransferase. J Biol Chem. 2008;283:36300–10. doi: 10.1074/jbc.M803196200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buhrmann C, Busch F, Shayan P, Shakibaei M. Sirtuin-1 (SIRT1) is required for promoting chondrogenic differentiation of mesenchymal stem cells. J Biol Chem. 2014;289:22048–62. doi: 10.1074/jbc.M114.568790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dvir-Ginzberg M, Gagarina V, Lee EJ, Booth R, Gabay O, Hall DJ. Tumor necrosis factor α-mediated cleavage and inactivation of SirT1 in human osteoarthritic chondrocytes. Arthritis Rheum. 2011;63:2363–73. doi: 10.1002/art.30279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baltus GA, Kowalski MP, Zhai H, Tutter AV, Quinn D, Wall D, Kadam S. Acetylation of Sox2 induces its nuclear export in embryonic stem cells. Stem Cells. 2009;27:2175–84. doi: 10.1002/stem.168. [DOI] [PubMed] [Google Scholar]
- 62.Mu WL, Wang YJ, Xu P, Hao DL, Liu XZ, Wang TT, Chen F, Chen HZ, Lv X, Liu DP. Sox2 Deacetylation by Sirt1 Is Involved in Mouse Somatic Reprogramming. Stem Cells. 2015;33:2135–47. doi: 10.1002/stem.2012. [DOI] [PubMed] [Google Scholar]
- 63.Hattori T, Kishino T, Stephen S, Eberspaecher H, Maki S, Takigawa M, de Crombrugghe B, Yasuda H. E6-AP/UBE3A protein acts as a ubiquitin ligase toward SOX9 protein. J Biol Chem. 2013;288:35138–48. doi: 10.1074/jbc.M113.486795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Komatsu T, Mizusaki H, Mukai T, Ogawa H, Baba D, Shirakawa M, Hatakeyama S, Nakayama KI, Yamamoto H, Kikuchi A, Morohashi K. Small ubiquitin-like modifier 1 (SUMO-1) modification of the synergy control motif of Ad4 binding protein/steroidogenic factor 1 (Ad4BP/SF-1) regulates synergistic transcription between Ad4BP/SF-1 and Sox9. Mol Endocrinol. 2004;18:2451–62. doi: 10.1210/me.2004-0173. [DOI] [PubMed] [Google Scholar]
- 65.Taylor KM, Labonne C. SoxE factors function equivalently during neural crest and inner ear development and their activity is regulated by SUMOylation. Dev Cell. 2005;9:593–603. doi: 10.1016/j.devcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 66.Lee PC, Taylor-Jaffe KM, Nordin KM, Prasad MS, Lander RM, LaBonne C. SUMOylated SoxE factors recruit Grg4 and function as transcriptional repressors in the neural crest. J Cell Biol. 2012;198:799–813. doi: 10.1083/jcb.201204161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hattori T, Eberspaecher H, Lu J, Zhang R, Nishida T, Kahyo T, Yasuda H, de Crombrugghe B. Interactions between PIAS proteins and SOX9 result in an increase in the cellular concentrations of SOX9. J Biol Chem. 2006;281:14417–28. doi: 10.1074/jbc.M511330200. [DOI] [PubMed] [Google Scholar]
- 68.Oh HJ, Kido T, Lau YF. PIAS1 interacts with and represses SOX9 transactivation activity. Mol Reprod Dev. 2007;74:1446–55. doi: 10.1002/mrd.20737. [DOI] [PubMed] [Google Scholar]
- 69.Lefebvre V. The SoxD transcription factors--Sox5, Sox6, and Sox13--are key cell fate modulators. Int J Biochem Cell Biol. 2010;42:429–32. doi: 10.1016/j.biocel.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ikeda T, Kamekura S, Mabuchi A, Kou I, Seki S, Takato T, Nakamura K, Kawaguchi H, Ikegawa S, Chung UI. The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum. 2004;50:3561–73. doi: 10.1002/art.20611. [DOI] [PubMed] [Google Scholar]
- 71.Nakamura Y, Yamamoto K, He X, Otsuki B, Kim Y, Murao H, Soeda T, Tsumaki N, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B, Postlethwait JH, Warman ML, Nakamura T, Akiyama H. Wwp2 is essential for palatogenesis mediated by the interaction between Sox9 and mediator subunit 25. Nat Commun. 2011;2:251. doi: 10.1038/ncomms1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Furumatsu T, Tsuda M, Yoshida K, Taniguchi N, Ito T, Hashimoto M, Ito T, Asahara H. Sox9 and p300 cooperatively regulate chromatin-mediated transcription. J Biol Chem. 2005;280:35203–8. doi: 10.1074/jbc.M502409200. [DOI] [PubMed] [Google Scholar]
- 73.Furumatsu T, Ozaki T, Asahara H. Smad3 activates the Sox9-dependent transcription on chromatin. Int J Biochem Cell Biol. 2009;41:1198–204. doi: 10.1016/j.biocel.2008.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]