The C-terminal domain (residues 780–1122) of the ATPase motor Fun30 from S. cerevisiae has been crystallized and the structure was determined at a resolution of 1.95 Å. A helix-bundle insertion in the ATPase domain was identified to be specific to the Fun30 subfamily.
Keywords: Fun30, Snf2, structure, remodeller, ATPase
Abstract
Fun30 (Function unknown now 30) is a chromatin remodeller belonging to the Snf2 family. It has previously been reported to be a regulator of several cellular activities, including DNA repair, gene silencing and maintenance of chromatin structure. Here, the crystal structure of the Fun30 ATPase-C domain (the C-lobe of the ATPase domain) is reported at 1.95 Å resolution. Although the structure displays overall similarities to those of other Snf2 family members, a new structural module was found to be specific to the Fun30 subfamily. Fun30 ATPase-C was shown be monomeric in solution and showed no detectable affinity for dsDNA.
1. Introduction
The tightly packed structure of the chromosome is an obstacle to many cellular activities, including transcription, duplication and DNA repair. A subset of Snf2 family proteins act as remodellers to change the local structure of chromatin and facilitate the access of regulators to the DNA sequence. Fun30 was originally identified by whole-genome sequencing of budding yeast (Clark et al., 1992 ▸), and was classified as a chromatin-remodelling enzyme (remodeller) based on sequence alignment (Flaus et al., 2006 ▸). Consistent with the taxonomic identification, the function of Fun30 was reported to be confined to chromosome-associated activities such as chromosome stability, integrity and segregation (Ouspenski et al., 1999 ▸). Specifically, recombinant Fun30 shows an ATPase activity that is stimulated by DNA and chromatin in vitro, and exhibits ATP-dependent chromatin-remodelling and histone dimer-exchange activity (Awad et al., 2010 ▸). Additionally, Fun30 is involved in gene silencing (Neves-Costa et al., 2009 ▸; Byeon et al., 2013 ▸), point centromere function (Durand-Dubief et al., 2012 ▸) and DNA double-strand break (DSB) repair (Chen et al., 2012 ▸; Costelloe et al., 2012 ▸). Fun30 is evolutionarily and functionally conserved in eukaryotes. Fft3 and SMARCAD1, the orthologues of Fun30 in fission yeast and human, respectively, are involved in heterochromatin structure maintenance (Strålfors et al., 2011 ▸; Yu et al., 2011 ▸; Steglich et al., 2015 ▸; Rowbotham et al., 2011 ▸) and DSB repair (Costelloe et al., 2012 ▸).
Fun30 contains 1131 amino acids, with an ATPase domain located in the C-terminal part (residues 570–1131) as well as a region of unknown function (residues 1–570) at the N-terminus. The Fun30 ATPase domain possesses common characteristics of the Snf2 family proteins and is the structural basis for its chromatin-remodelling function (Awad et al., 2010 ▸; Flaus et al., 2006 ▸). Snf2 enzymes, a subgroup of the helicase family, possess seven helicase-related motifs (I, Ia and II–VI) in the ATPase domain. The extra sequence blocks (blocks A–N), which are conserved only in the Snf2 family, differentiate the Snf2 family from other helicases (Flaus et al., 2006 ▸). The ATPase domain, the central architectural component of Snf2 proteins, has been well studied biochemically, and several structures have been reported (Dürr et al., 2005 ▸; Thomä et al., 2005 ▸; Hauk et al., 2010 ▸; Shaw et al., 2008 ▸; Xia et al., 2016 ▸). According to these structures, the ATPase domain is divided into two parts, ATPase-N and ATPase-C, which are connected by a linker. The former is the ATP-binding site and the latter participates in tracking along the DNA (Dürr et al., 2005 ▸). In contrast to the high homology in sequence and global architecture of Snf2 proteins, the local regions between blocks C and K vary greatly, with various domain/motif insertions. These regions are therefore called major insertion sites (Flaus et al., 2006 ▸). These insertions increase the structural variety of Snf2 enzymes, enabling them to fulfil diverse cellular activities.
The Snf2 family members are classified into 24 subfamilies based on the similarity in sequence in the ATPase domain. The Fun30 subfamily has a distant relationship to the other members. In this study, we determined the structure of Fun30 ATPase-C at 1.95 Å resolution. Structure comparison and sequence alignment with other Snf2 proteins revealed that insertion II, which is composed of five α-helices and two 310-helices, is specific to the Fun30 subfamily. Several biophysical approaches have been used to elucidate the oligomeric state of the ATPase-C domain of Fun30 and its DNA-binding properties.
2. Materials and methods
2.1. Macromolecule production
The DNA fragment coding for Fun30 ATPase-C (residues 780–1122) was amplified from the Saccharomyces cerevisiae genome and inserted into the pET-21a expression vector between BamHI and XhoI sites with a 6×His tag at the C-terminus. The recombinant plasmid was verified by DNA sequencing and then transformed into Escherichia coli Rosetta (DE3) cells for protein expression.
For large-scale expression, the E. coli cells were cultured in LB medium at 310 K. When the optical density (OD600) of the cell cultures reached 0.8, the cultures were cooled to 289 K. Protein expression was induced by adding 0.3 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). The cells were harvested 16 h later, resuspended in buffer A (20 mM Tris pH 8.0, 300 mM NaCl, 20 mM imidazole, 10% glycerol, 1 mM PMSF) and then disrupted by sonication. The bacterial cell lysate was centrifuged and the supernatant was applied onto an Ni–NTA column (GE Healthcare) pre-equilibrated with buffer A. The column was washed with buffer B (20 mM Tris pH 8.0, 300 mM NaCl, 50 mM imidazole, 10% glycerol) and eluted with buffer C (20 mM Tris pH 8.0, 300 mM NaCl, 300 mM imidazole, 10% glycerol). Samples were further purified by anion-exchange chromatography using a Resource S column (GE Healthcare). Elution was conducted with a linear gradient of NaCl concentration (0.08–1 M) in 25 mM HEPES pH 7.0 and the fractions were analyzed by SDS–PAGE. Fractions containing the target protein were pooled, concentrated and loaded onto a HiLoad Superdex 200 10/300 gel-filtration column pre-equilibrated with buffer D (25 mM HEPES pH 7.0, 150 mM NaCl). Fractions containing Fun30 were collected, concentrated and stored at 193 K. Macromolecule-production information is summarized in Table 1 ▸.
Table 1. Macromolecule-production information.
The restriction-enzyme cleavage sites are underlined in the primers and the additional residues expressed by the vector are underlined in the amino-acid sequence.
| Source organism | S. cerevisiae |
| DNA source | Genomic DNA |
| Forward primer | 5′-GAGGATCCATGCCGCTATTAGCGCAAGAAGCCA-3′ |
| Reverse primer | 5′-GCGCTCGAGTCATCATAAATTATATCCTCC-3′ |
| Cloning vector | pET-21a |
| Expression vector | pET-21a |
| Expression host | E. coli Rosetta (DE3) |
| Complete amino-acid sequence of the construct produced | MASMTGGQQMGRGSMPLLAQEAITRAKTMMKPFILRRRKDQVLKHLPPKHTHIQYCELNAIQKKIYDKEIQIVLEHKRMIKDGELPKDAKEKSKLQSSSSKNLIMALRKASLHPLLFRNIYNDKIITKMSDAILDEPAYAENGNKEYIKEDMSYMTDFELHKLCCNFPNTLSKYQLHNDEWMQSGKIDALKKLLKTIIVDKQEKVLIFSLFTQVLDILEMVLSTLDYKFLRLDGSTQVNDRQLLIDKFYEDKDIPIFILSTKAGGFGINLVCANNVIIFDQSFNPHDDRQAADRAHRVGQTKEVNITTLITKDSIEEKIHQLAKNKLALDSYISEDKKSQDVLESKVSDMLEDIIYDELEHHHHHH |
2.2. Crystallization
An initial crystallization screen was performed using the hanging-drop vapour-diffusion method at 289 K. Crystals were obtained using a reservoir solution consisting of 0.1 M sodium malonate pH 5.0, 12% PEG 3350. The crystals were soaked in a cryoprotectant consisting of 20% PEG 3350 in the reservoir and then flash-cooled in liquid nitrogen. Crystallization information is summarized in Table 2 ▸.
Table 2. Crystallization.
| Method | Hanging-drop vapour diffusion |
| Temperature (K) | 289 |
| Protein concentration (mg ml−1) | 10 |
| Buffer composition of protein solution | 25 mM HEPES pH 7.0, 150 mM NaCl |
| Composition of reservoir solution | 0.1 M sodium malonate pH 5.0, 12% PEG 3350 |
| Volume and ratio of drop | 1 µl protein solution mixed with 1 µl reservoir solution |
| Volume of reservoir (µl) | 500 |
2.3. Data collection and processing
The X-ray diffraction data were collected on the BL-1A beamline at the Photon Factory (PF), Tsukuba, Japan. The data set was processed with HKL-2000 (Otwinowski & Minor, 1997 ▸). Data-collection and processing statistics are given in Table 3 ▸.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | BL-1A, PF |
| Wavelength (Å) | 1.1000 |
| Temperature (K) | 100 |
| Detector | PILATUS 2M-F |
| Crystal-to-detector distance (mm) | 177.61 |
| Rotation range per image (°) | 1.0 |
| Total rotation range (°) | 360 |
| Exposure time per image (s) | 0.2 |
| Space group | C2 |
| a, b, c (Å) | 114.43, 118.39, 111.58 |
| α, β, γ (°) | 90, 107.28, 90 |
| Mosaicity (°) | 0.3 |
| Resolution range (Å) | 50–1.95 (1.98–1.95) |
| Total No. of reflections | 697715 |
| No. of unique reflections | 103556 |
| Completeness (%) | 100.0 (100.0) |
| Multiplicity | 6.7 (6.8) |
| 〈I/σ(I)〉 | 20.73 (2.13) |
| R meas | 0.089 (0.698) |
| Overall B factor from Wilson plot (Å2) | 25.88 |
2.4. Structure solution and refinement
The initial phase was obtained by molecular replacement with Phaser (McCoy et al., 2007 ▸) as implemented in CCP4 (Winn et al., 2011 ▸). The structure of Chd1 (PDB entry 3mwy; Hauk et al., 2010 ▸) was modified for use as a search model. The initial model was built with AutoBuild in PHENIX (Adams et al., 2010 ▸) and then modified and refined using alternate rounds of Coot (Emsley & Cowtan, 2004 ▸) and phenix.refine (Afonine et al., 2012 ▸). The final structure was refined to 1.95 Å resolution. Details of the refinement statistics are shown in Table 4 ▸. Atomic coordinates and structure factors have been deposited in the Protein Data Bank (http://www.pdb.org) as entry 5gn1. Structural figures were prepared with PyMOL (v.1.5; Schrödinger).
Table 4. Structure solution and refinement.
Values in parentheses are for the outer shell.
| Resolution range (Å) | 35.47–1.95 (2.02–1.95) |
| Completeness (%) | 100.0 |
| No. of reflections, working set | 103156 (10276) |
| No. of reflections, test set | 1993 (198) |
| Final R cryst | 0.184 (0.228) |
| Final R free | 0.214 (0.265) |
| No. of non-H atoms | |
| Protein | 10165 |
| Water | 1243 |
| R.m.s. deviations | |
| Bonds (Å) | 0.011 |
| Angles (°) | 1.04 |
| Average B factors (Å2) | |
| Overall | 30.58 |
| Protein | 31.43 |
| Water | 37.79 |
| Ramachandran plot | |
| Most favoured (%) | 98.5 |
| Allowed (%) | 1.5 |
2.5. Size-exclusion chromatography coupled with light scattering
We measured the mass of Fun30 ATPase-C in solution using a Wyatt Eclipse 3 separation system combined with a static light-scattering detector (DAWN HELEOS II, Wyatt) and a differential refractive-index detector (Optilab rEX detector, Wyatt). Protein sample (50 µl at a concentration of 1.4 mg ml−1) was injected onto a Superdex 75 10/300 GL column (GE Healthcare) with column buffer consisting of 20 mM HEPES pH 7.5, 150 mM NaCl. Data were analyzed with ASTRA 6.1 (Wyatt)
2.6. Analytic ultracentrifugation
Sedimentation-velocity experiments were performed using a Beckman Coulter ProteomeLab XL-I analytical ultracentrifuge equipped with an An-60 Ti rotor (four holes) and conventional double-sector aluminium centrepieces of 12 mm optical path length loaded with 380 µl sample and 400 µl buffer (20 mM Tris pH 8.0, 200 mM NaCl). The experiment was carried out at 20°C and 50 000 rev min−1 using an absorption optical scanner (ABS) at 280 nm. Data were analyzed using SEDFIT (https://sedfitsedphat.nibib.nih.gov/software) and the continuous c(s) distribution model.
2.7. Electrophoretic mobility shift assay (EMSA)
The oligonucleotides 5′-6-FAM-AAAAAAATTGCCGAAGACGAAAAAA-3′ and 5′-TTTTTTTCGTCTTCGGCAATTTTTT-3′ were synthesized and annealed to form the substrate dsDNA. The 6-FAM label was added at the 5′ end for detection. 1 µM dsDNA was pre-incubated with Fun30 ATPase-C (residues 780–1122) or ATPase-N (residues 572–794) at concentrations ranging from 0.5 to 10 µM. Fun30 ATPase-N was expressed in E. coli and purified using Ni–NTA column and size-exclusion chromatography. The EMSA binding buffer consisted of 25 mM HEPES pH 7.5, 150 mM NaCl, 2 mM DTT. After incubation at 289 K for 30 min, the reaction products were loaded onto a 6% nondenaturing polyacrylamide gel and resolved by electrophoresis at 277 K and 10 V cm−1 in 0.5× TBE buffer. Gels were visualized using a Typhoon FLA 7000.
3. Results and discussion
3.1. Overall structure of Fun30 ATPase-C
A Fun30 fragment spanning residues 780–1122 was expressed, purified and crystallized. The crystals grew to full size after 2 d and diffracted to about 1.95 Å resolution at a synchrotron (Table 1 ▸). Our construct includes the ATPase-C domain and the linker between the two lobes, whereas the linker (residues 780–808) is not visible in the electron-density map. The crystal belonged to space group C2 with four molecules per asymmetric unit, and the structure was determined using the molecular-replacement method with the structure of the Chd1 ATPase-C domain (PDB entry 3mwy) as the initial search model. Superimposition of the four molecules in the asymmetric unit shows little difference, with a root-mean-square deviation (r.m.s.d.) of about 0.5 Å.
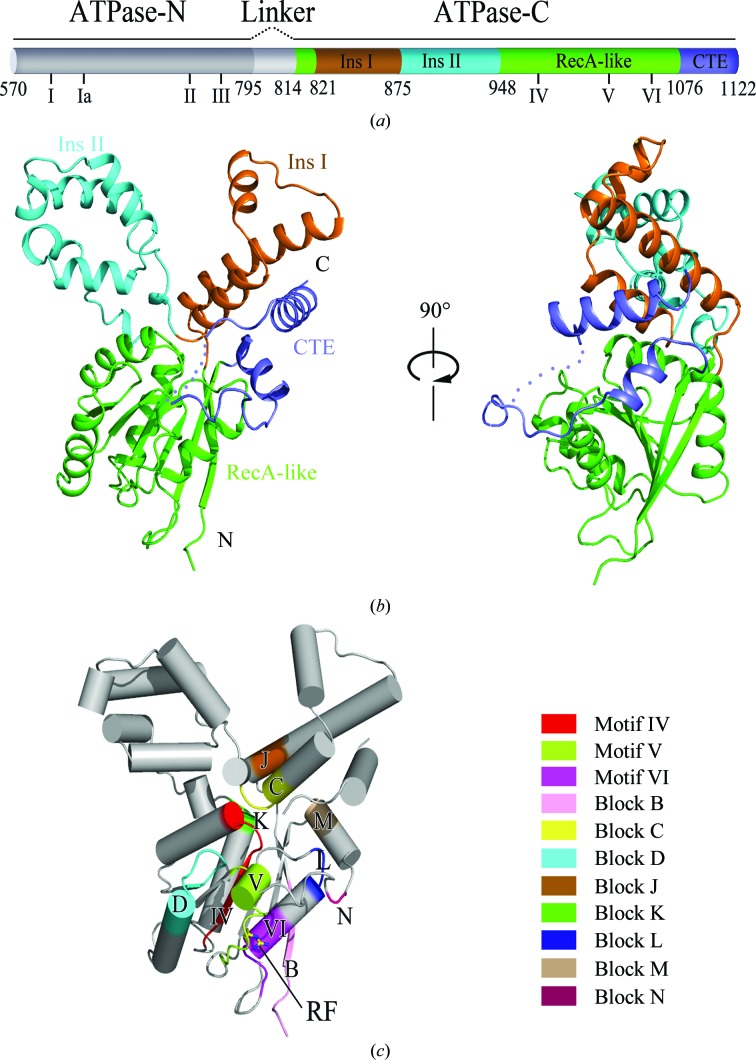
As shown in Fig. 1 ▸, the overall structure of Fun30 ATPase-C shows a cloverleaf pattern and is composed of four parts: a conserved RecA-like fold as the core structure (residues 814–820 and 948–1075), an Snf2-specific helix insertion (insertion I; residues 821–874), a nonconserved helix-bundle insertion (insertion II; residues 875–947) and a C-terminal extension (CTE; residues 1076–1122). The RecA-like fold, which includes motifs IV, V and VI and blocks B, K, D and L, forms an α/β structure characterized by a central β-sheet flanked by five α-helices (Fig. 1 ▸ c). Residue Arg1058, also called the arginine finger, is conserved in the helicase family and participates in the formation of the catalytic centre. Insertion I containing blocks C and J is composed of three tandem α-helices, two of which are almost antiparallel, with the middle helix nearly perpendicular to the other two helices. Insertion II between blocks C and K is composed of five α-helices and two 310-helices. This inserted helix bundle is not conserved in the whole Snf2 family, but shows higher conservation within its subfamily (Fig. 2 ▸ e). The CTE packed in the cleft between insertion I and the RecA-like fold contains blocks M and N, and comprises two α-helices linked by a 23-residue loop. Part of the loop (residues 1098–1103) could not be modelled owing to its high flexibility.
Figure 1.
Overview of the structure of Fun30 ATPase-C. (a) Schematic representation of the Fun30 ATPase motor organization. The ATPase-N is shown in grey and the ATPase-C is in colour. The RecA-like fold, insertion I (Ins I), insertion II (Ins II) and the CTE are depicted in green, orange, cyan and blue, respectively. The locations of the conserved helicase-related motifs I–VI are marked below the cartoon diagram. (b) Cartoon representation of Fun30 ATPase-C. The colours of the different segments are the same as in (a). The loop in the CTE which is not modelled is displayed with dots. (c) The conserved motifs and blocks mapped onto Fun30 ATPase-C. Motif IV (residues 968–976), motif V (residues 1024–1034), motif VI (residues 1054–1061), block B (residues 810–815), block C (residues 871–878), block D (residues 995–1005), block J (residues 826–830), block K (residues 948–950), block L (residues 1047–1049), block M (residues 1079–1082) and block N (residues 1090–1091) are coloured according to the legend. Residue Arg1058, the conserved arginine finger in motif IV, is shown as a stick model.
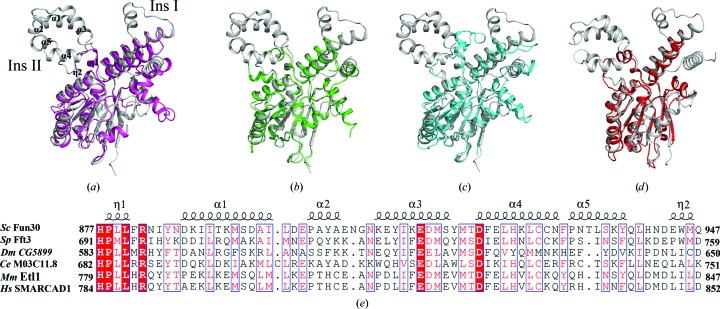
Figure 2.
Comparison of Fun30 ATPase-C with its homologues. (a–d) Superposition of Fun30 ATPase-C (grey) on SSO1653 (magenta), Chd1 (green), Rad54 (cyan) and Snf2 (red). The comparison highlights the differences in the insertion I and insertion II regions. (e) Sequence alignment of insertion II in the Fun30 subfamily, including Fft3 from Schizosaccharomyces pombe (Sp), CG5899 from Drosophila melanogaster (Dm), M03C11.8 from Caenorhabditis elegans (Ce), Etl1 from mouse (Mm) and SMARCAD1 from human (Hs). Conserved regions are shown in blue boxes. Highly conserved residues are in white and shaded red; moderately conserved residues are in red. Sequence alignment was performed using ESPript (Robert & Gouet, 2014 ▸).
3.2. Structural comparison between Fun30 and its homologues
To identify the structural features of Fun30 ATPase-C, we superposed it on four Snf2 enzymes, SSO1653 (PDB entry 1z6a; Dürr et al., 2005 ▸), Rad54 (PDB entry 1z3i; Thomä et al., 2005 ▸), Chd1 (PDB entry 3mwy; Hauk et al., 2010 ▸) and Snf2 (PDB entry 5hzr; Xia et al., 2016 ▸), that belong to distinct subfamilies. As shown in Figs. 2 ▸(a)–2 ▸(d), the greatest differences were found to be located in the insertion regions. Insertion I in Fun30 is composed of three helices, whereas its counterparts in the other four proteins consist of two helices linked by a loop. In addition, the structure of insertion II in Fun30 is much larger and more complex than the others. The sequence of the middle helix of Fun30 insertion I is not conserved according to sequence alignment (data not shown), whereas insertion II is highly conserved in the Fun30 subfamily (Fig. 2 ▸ e). Therefore, insertion II seems to be a hallmark of the Fun30 subfamily. The insertions at the major insertion site usually confer specific functions on Snf2 proteins. For example, the insertion of a RING domain into the ATPase domain of Rad5/Rad16 subfamily enables them to be a component of the E3 ubiquitin ligase in addition to a remodeller (Parker & Ulrich, 2009 ▸; Gillette et al., 2006 ▸). Thus, it is reasonable to speculate that insertion II may be related to certain conserved functions of the Fun30 subfamily. Futher biochemical data are needed to elucidate the function of the Fun30 insertion regions.
3.3. Fun30 ATPase-C is in a monomeric state
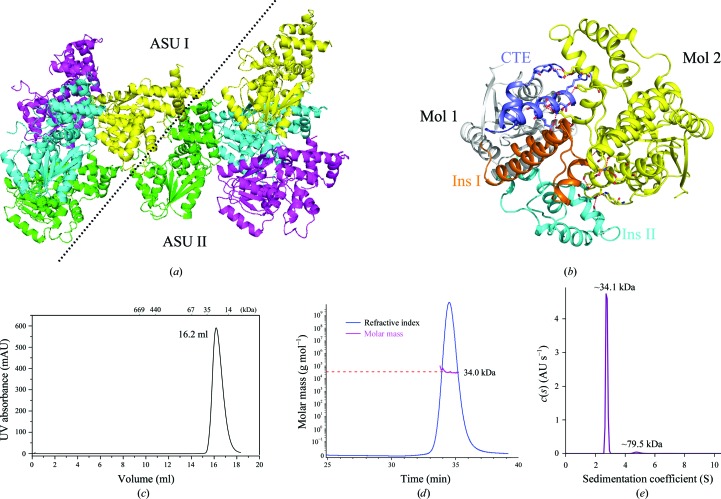
As shown in Fig. 3 ▸(a), our structural model contains four molecules arranged in an asymmetric unit; however, the interactions among these molecules are so weak that this arrangement is most likely to be a result of crystal packing. We also noticed that the molecules from two neighbouring asymmetric units form tight interactions (Fig. 3 ▸ a). Insertion I and the CTE of one molecule generate extensive hydrogen bonds with insertions I and II of the other molecule (Fig. 3 ▸ b). Furthermore, the area of the interface, as analyzed by the PISA server (Krissinel & Henrick, 2007 ▸), is 1933.1 Å2. The large area of the interface supports the formation of a dimer.
Figure 3.
Fun30 ATPase-C is a monomer in solution. (a) Monomers from neighbouring asymmetric units (ASUs) form tight interactions. The four monomers in an asymmetric unit are in different colours. (b) The interface of the dimer in the crystal. The CTE (blue), Ins I (orange) and Ins II (cyan) of one monomer interact with the other monomer (yellow). Hydrogen bonds are shown as red dashed lines. (c) Size-exclusion chromatogram of Fun30 ATPase-C. The molecular masses of the protein standards are indicated at the top. (d) The mass of Fun30 ATPase-C in solution is about 34.0 kDa, as measured by static light scattering, which corresponds to a monomeric configuration. (e) Analytical ultracentrifugation of Fun30 ATPase-C demonstrates that 95.8% of the total protein, with a molecular mass of about 34.1 kDa, is a monomer. Only 2.6% of the sample shows a molecular mass of 79.5 kDa, which may be an impurity.
However, during gel filtration using a Superdex 200 10/300 column the protein eluted at a volume of 16.2 ml, with an apparent molecular mass of about 40 kDa (Fig. 3 ▸ c), which corresponds to an ATPase-C monomer. In order to investigate the oligomeric state of Fun30 ATPase-C more precisely, we utilized size-exclusion chromatography coupled with static light scattering and analytical ultracentrifugation. The molecular weight of Fun30 ATPase-C was determined to be about 34 kDa by those two methods (Fig. 3 ▸ d and 3 ▸ e). The results both agree with the weight of a monomer. The measured molecular mass may be smaller than the calculated mass owing to a slight degradation of the N-terminus of the recombinant expressed fragment. Therefore, we propose that Fun30 ATPase-C is a monomer in solution but forms a dimer in crystal packing.
Since full-length Fun30 has been reported to form a homodimer in vitro and in vivo (Awad et al., 2010 ▸), we speculated that certain sequences outside Fun30 ATPase-C are responsible for the dimerization of Fun30. This speculation is supported by our research on Fun30 ATPase-N, which exhibits both monomeric and dimeric states in solution according to the results of analytical ultracentrifugation (Supplementary Fig. S1).
3.4. Fun30 ATPase-C has little affinity for dsDNA
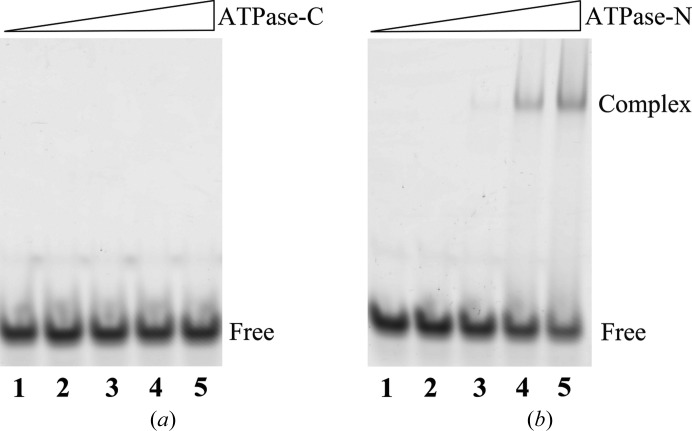
The ATPase domain of Snf2 family proteins is not only the catalytic core of ATP hydrolysis, but also the site for DNA binding. In the structure of the SSO1653–dsDNA complex (PDB entry 1z63) the DNA strands lie in the cleft between the ATPase-N and ATPase-C domains. The ATPase-N domain of SSO1653 displays comparable affinity for DNA as that of the full ATPase motor, whereas the ATPase-C domain by itself shows little affinity for DNA (Dürr et al., 2005 ▸). Since Fun30 has previously been reported to have affinity for dsDNA (Awad et al., 2010 ▸), we tested whether Fun30 ATPase-C retained the affinity for dsDNA. The results of EMSA showed that Fun30 ATPase-C alone was unable to bind dsDNA, whereas the ATPase-N domain bound dsDNA to form a supershift on native PAGE (Fig. 4 ▸). This result indicated that Fun30, just like SSO1653, uses the ATPase-N domain as the main site to interact with DNA. Fun30 may adopt a similar way of binding DNA as other Snf2 proteins.
Figure 4.
Fun30 ATPase-C has little DNA-binding affinity. The DNA-binding abilities of ATPase-C (a) and ATPase-N (b) were tested by EMSA; lane 1 shows the dsDNA substrate as a control and lanes 2–5 contain samples with increasing protein concentrations, resulting in final protein:DNA molar ratios of 0.5, 2, 5 and 10, respectively.
3.5. Conclusions
In summary, we determined the crystal structure of the C-terminal part of the Fun30 ATPase domain at 1.95 Å resolution. This is the first structural report from the Fun30 subfamily to date. Sequential and structural comparison of Fun30 ATPase-C with its homologues demonstrated that Fun30 ATPase-C has the same conserved motifs and the same structural fold as its homologues, but possesses a helix-bundle insertion that is specific to the Fun30 subfamily. Moreover, we showed that Fun30 ATPase-C is monomeric in solution and has little affinity for dsDNA. These results will aid in future biochemical analyses of Fun30, as well as other members of the Fun30 subfamily.
Supplementary Material
PDB reference: C-terminal part of Fun30 ATPase domain, 5gn1
Supplementary Figure S1.. DOI: 10.1107/S2053230X16019269/no5111sup1.pdf
Acknowledgments
We thank the staff at the BL-1A beamline, Photon Factory, Tsukuba, Japan for technical assistance during data collection. This work was supported by the Strategic Priority Research Program (XDB08010301).
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Awad, S., Ryan, D., Prochasson, P., Owen-Hughes, T. & Hassan, A. H. (2010). J. Biol. Chem. 285, 9477–9484. [DOI] [PMC free article] [PubMed]
- Byeon, B., Wang, W., Barski, A., Ranallo, R. T., Bao, K., Schones, D. E., Zhao, K., Wu, C. & Wu, W.-H. (2013). J. Biol. Chem. 288, 23182–23193. [DOI] [PMC free article] [PubMed]
- Chen, X., Cui, D., Papusha, A., Zhang, X., Chu, C.-D., Tang, J., Chen, K., Pan, X. & Ira, G. (2012). Nature (London), 489, 576–580. [DOI] [PMC free article] [PubMed]
- Clark, M. W., Zhong, W. W., Keng, T., Storms, R. K., Barton, A., Kaback, D. B. & Bussey, H. (1992). Yeast, 8, 133–145. [DOI] [PubMed]
- Costelloe, T., Louge, R., Tomimatsu, N., Mukherjee, B., Martini, E., Khadaroo, B., Dubois, K., Wiegant, W. W., Thierry, A., Burma, S., van Attikum, H. & Llorente, B. (2012). Nature (London), 489, 581–584. [DOI] [PMC free article] [PubMed]
- Durand-Dubief, M., Will, W. R., Petrini, E., Theodorou, D., Harris, R. R., Crawford, M. R., Paszkiewicz, K., Krueger, F., Correra, R. M., Vetter, A. T., Miller, J. R., Kent, N. A. & Varga-Weisz, P. (2012). PLoS Genet. 8, e1002974. [DOI] [PMC free article] [PubMed]
- Dürr, H., Körner, C., Müller, M., Hickmann, V. & Hopfner, K. P. (2005). Cell, 121, 363–373. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Flaus, A., Martin, D. M., Barton, G. J. & Owen-Hughes, T. (2006). Nucleic Acids Res. 34, 2887–2905. [DOI] [PMC free article] [PubMed]
- Gillette, T. G., Yu, S., Zhou, Z., Waters, R., Johnston, S. A. & Reed, S. H. (2006). EMBO J. 25, 2529–2538. [DOI] [PMC free article] [PubMed]
- Hauk, G., McKnight, J. N., Nodelman, I. M. & Bowman, G. D. (2010). Mol. Cell, 39, 711–723. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Neves-Costa, A., Will, W. R., Vetter, A. T., Miller, J. R. & Varga-Weisz, P. (2009). PLoS One, 4, e8111. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Method Enzymol. 276, 307–326. [DOI] [PubMed]
- Ouspenski, I. I., Elledge, S. J. & Brinkley, B. R. (1999). Nucleic Acids Res. 27, 3001–3008. [DOI] [PMC free article] [PubMed]
- Parker, J. L. & Ulrich, H. D. (2009). EMBO J. 28, 3657–3666. [DOI] [PMC free article] [PubMed]
- Robert, X. & Gouet, P. (2014). Nucleic Acids Res. 42, W320–W324. [DOI] [PMC free article] [PubMed]
- Rowbotham, S. P., Barki, L., Neves-Costa, A., Santos, F., Dean, W., Hawkes, N., Choudhary, P., Will, W. R., Webster, J., Oxley, D., Green, C. M., Varga-Weisz, P. & Mermoud, J. E. (2011). Mol. Cell, 42, 285–296. [DOI] [PubMed]
- Shaw, G., Gan, J. H., Zhou, Y. N., Zhi, H. J., Subburaman, P., Zhang, R. G., Joachimiak, A., Jin, D. J. & Ji, X. H. (2008). Structure, 16, 1417–1427. [DOI] [PMC free article] [PubMed]
- Steglich, B., Strålfors, A., Khorosjutina, O., Persson, J., Smialowska, A., Javerzat, J. P. & Ekwall, K. (2015). PLoS Genet. 11, e1005101. [DOI] [PMC free article] [PubMed]
- Strålfors, A., Walfridsson, J., Bhuiyan, H. & Ekwall, K. (2011). PLoS Genet. 7, e1001334. [DOI] [PMC free article] [PubMed]
- Thomä, N. H., Czyzewski, B. K., Alexeev, A. A., Mazin, A. V., Kowalczykowski, S. C. & Pavletich, N. P. (2005). Nature Struct. Mol. Biol. 12, 350–356. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Xia, X., Liu, X., Li, T., Fang, X. & Chen, Z. (2016). Nature Struct. Mol. Biol. 23, 722–729. [DOI] [PubMed]
- Yu, Q., Zhang, X. & Bi, X. (2011). J. Biol. Chem. 286, 14659–14669. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: C-terminal part of Fun30 ATPase domain, 5gn1
Supplementary Figure S1.. DOI: 10.1107/S2053230X16019269/no5111sup1.pdf