Abstract
Ion mobility mass spectrometry of integral membrane proteins provides valuable insights into their architecture and stability. Here we show that due to their lower charge the average mobility of native-like membrane protein ions is approximately 30 % lower than that of soluble proteins of similar mass. This has implications for drift time measurements, made on travelling wave ion mobility mass spectrometers, which have to be calibrated to extract collision cross sections (Ω). Common calibration strategies employ unfolded or native-like soluble protein standards with masses and mobilities comparable to the protein of interest. We compare Ω values for membrane proteins, derived from standard calibration protocols using soluble proteins, to values measured using an RF-confined drift tube. Our results demonstrate that, while common calibration methods underestimate Ω for native-like or unfolded membrane protein complexes, higher mass soluble calibration standards consistently yield more accurate Ω values. These findings enable us to obtain directly structural information for highly charge-reduced complexes by travelling wave ion mobility mass spectrometry.
Keywords: Native mass spectrometry, charge state modulation, collision cross section, protein unfolding, travelling wave ion mobility spectrometry
Introduction
The use of ion mobility mass spectrometry (IMS) to study the gas-phase conformations of proteins is well established.1–3 Gas-phase protein ions are separated according to their mobility inside a He- or N2-filled drift cell, and the rotationally averaged collision cross section (Ω) can be calculated from the time taken to transit the cell (the drift time), indicating the absolute size of the ion. Ω measurements can be used to obtain information about the architecture of biomolecular complexes as well as their folding states and conformational dynamics.2 Recent advances in the mass spectrometry of membrane proteins4 have extended the approach to this more challenging class of protein, allowing insights into their oligomeric states and the effects of lipids on conformational stability.5,6
The simplest ion mobility designs use a drift tube (DT), a device that imposes a small potential difference across the cell to differentially pull the ions through a field of neutral gas molecules. In this setup, the relationship between Ω and mobility (K) of an ion is described by the Mason-Schamp equation:7
| (1) |
where N is the drift gas number density, m and M are the is the molecular mass of the ion and drift gas, respectively, k the Boltzmann constant, T the drift gas temperature and e the elementary charge. With all other factors constant, the mobility of an ion scales according to charge (z) and 1/Ω. In DTIMS, the Ω of an ion can be calculated directly from the measured drift time. However, instead of the linear-field separation in DTIMS, many commercial instruments use ion mobility separation based on a travelling-wave system (TWIMS). In place of a constant electric field TWIMS uses a continuously changing electric field, which can be considered as waves of direct current that push packets of ions through the mobility cell.8 While the drift time depends upon many of the same parameters as for DTIMS, it also depends on how an ion responds to the travelling waves, and consequently the relationship between drift time and mobility (or Ω) is non-linear.9 This non-linear function is not defined, which complicates the calculation of accurate Ω values from TWIMS.9 Nevertheless, it is possible to obtain Ω values through the use of calibration protocols using ions of known (DTIMS-measured) Ω values.10–13
For the analysis of small proteins and denatured proteins, the use of chemically or thermally unfolded protein standards generates highly accurate calibrations.11,12 For small molecules, the relationship between arrival time and Ω is linear in TWIMS.10 For native-like proteins, native-like protein standards are recommended as calibrants because they cover the desired mobility ranges.11,14 In practice, these calibrants are commercially available protein standards with similar molecular weights and mobilities (whose Ω are known from DTIMS measurements).11,12 For these types of ion, a power law offers a statistically justified better fit. Clearly, with this relationship changing in nature for different ions, calibrations require closely similar analyte and calibrant ions to avoid extrapolation of what is an unknown non-linear function.9,11
Here, we investigate the use of TWIMS to determine Ω values of membrane proteins. We find that the lower charge states of native-like membrane protein ions reduce their mobility compared to soluble proteins, and therefore, TWIMS calibrations for membrane proteins require much larger calibrant ions to avoid erroneous extrapolation of the calibration function.
Experimental Methods
Sample preparation
All chemicals, avidin, β-lactoglobulin, concanavalin A, cytochrome C, alcohol dehydrogenase, and pyruvate kinase were purchased from Sigma. Proteins were dissolved in 200 mM ammonium acetate, pH 7.5, to a final concentration of approximately 10 µM (monomer). Prior to analysis, 5-μL aliquots were diluted to 0.2 mg/mL and a final volume of 50 μL, and buffer-exchanged using Micro Bio-Spin 6 columns (Bio-Rad, Hercules, CA) equilibrated with 200 mM ammonium acetate, pH 7.5. Recombinant ammonia channel B (AmtB) from E. coli, multi antimicrobial extrusion protein (MATE) from P. furiosus and aquaporin Z (AqpZ) from E. coli were purified as described.5,15 Membrane protein samples were exchanged into detergent-containing solutions using a Superdex GL 10/300 column (GE Healthcare) equilibrated with 200 mM ammonium acetate, pH 7.5 containing 2×critical micelle concentration (0.5% v/v) C8E4 (Anatrace, Maumee, OH). Peak fractions were concentrated to a final protein concentration of approximately 10 μM (monomer) using a 30 kDa MWCO device (Amicon Ultra, Merck, Darmstadt, Germany) and immediately subjected to MS analysis.
IM-MS
DTIMS spectra were recorded on a modified Synapt G1 (Waters, Manchester, UK) as described.5,11,16 TWIMS spectra were recorded on a commercial Synapt G1 HDMS instrument (Waters, Manchester, UK) equipped with a Z-spray source and a 32k m/z quadrupole mass filter. For membrane proteins, instrument settings were: capillary voltage 1.5 V, cone voltage 130 V, extraction voltage 4 V and transfer collision voltage 50 V. Collision voltages in the collision/trap cell were optimized for each membrane protein to yield compact conformations indicative of native-like states. Voltages were 50 V for MATE, 90 V for AqpZ, and 90 V for AmtB. For collision-induced unfolding of AmtB, the collision voltage was ramped from 90 V to 200 V in 10-V increments. For soluble proteins, instrument settings were: capillary voltage 1.5 V, cone voltage 20 V, extraction voltage 4 V, trap collision voltage 10 V and transfer collision voltage 5 V. The source pressure was maintained at 5.5 mbar for all proteins. Ion mobility settings were (or as otherwise noted): wave velocity 350 m/s and wave height 10 V, 13 V, or 15 V in the IMS cell, or wave velocity 300 m/s and wave height 13 V for collision-induced unfolding of AmtB. Transfer wave velocity was 248 m/s and wave height 13 V. Drift cell gas was N2 with a pressure of 1.6 Torr.
Measurements recorded for DTIMS Ω determination used a minimum of five different drift voltage settings from 50 V to 150 V. The drift gas was helium. The pressure and temperature were measured directly and noted for each acquisition. DTIMS spectra were processed and analyzed in PULSAR to determine Ω values.15
TWIMS drift time calibrations were carried out using a simplified version of the previously reported protocol.16 The observed drift times (td) were corrected for m/z-dependent flight times using the empirically determined enhanced duty cycle delay coefficient c:16
| (2) |
The literature Ω values in helium are corrected for reduced mass (μ) and normalized by z:
| (3) |
Since the mobility K depends only weakly on the ion mass,17 the mobility of an ion is scaled by Ω′.9 From the slope of the plot of ln(Ω′) as a function of ln(td′), the factors x and ln(A) are determined:
| (4) |
Data were analyzed and visualized using a modified version of the PULSAR software15 (see below). All calculations used Ω measured in He and are therefore effective He values.
Calibration tools within PULSAR
The tools to calibrate TWIMS data have been implemented in the ion mobility mass spectrometry analysis software PULSAR, which is available from http://pulsar.chem.ox.ac.uk/.15 These include the ability to semi-automatically process TWIMS data for calibrant ions using a customizable calibrant database. Calibrations can then be assembled using selected calibrant ions, and fitted with linear or power-law equations. These calibrations can be used to determine Ω of analytes. We find it useful, when assessing the suitability of a calibration, to compute the expected arrival times for an ion of particular mass and Ω, and overlay this on the experimental arrival time distributions. Notably, suitable calibrations will show data points in arrival time that overlay with all ions that are expected to be folded, whereas poor calibrations will show theoretical arrival times that do not match any of the experimental peaks, or transgress from lower to higher arrival times or vice versa (see Figure S3 for examples). This is a particularly easy way to assess calibration quality as deviations from the expected drift time trend can be identified by visual inspection. An appropriate calibration should not only have in itself a good fit (R2),11,12 but accurately correlate Ω values with ion drift times.
Results and Discussion
Native-like membrane protein ions have reduced mobility compared to soluble proteins
The charge states of membrane protein ions formed by nano-electrospray ionization (nESI) are generally lower than those of soluble proteins with a similar surface area, which may be due to the removal of the detergent micelle following desolvation.18 As with soluble proteins, charge reduction helps to maintain native-like conformations after detergent stripping,19 and consequently charge-reducing PEG-based detergents that can be removed at low activation energies are particularly suitable for intact MS of membrane protein complexes.4,20
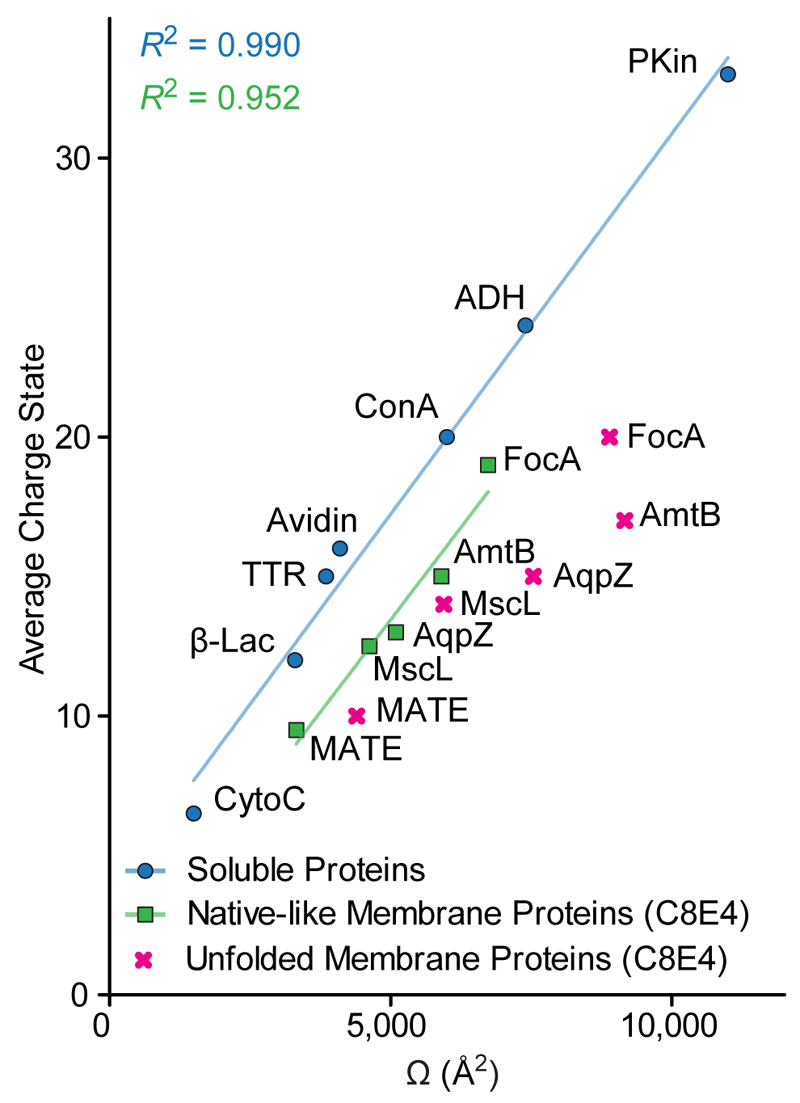
Since the mobility of an ion is directly proportional to its charge (eq. 1), we reasoned that nESI of membrane proteins under conditions optimized for stabilizing native-like states produces ions with a greatly reduced mobility than those of soluble proteins with the same mass. Comparing the average charge-states and DTIMS-measured Ω values of membrane proteins (in the PEG detergent C8E4) to commonly used TWIMS calibrants, revealed a consistently lower mobility for membrane proteins due to their higher Ω/z ratios (Figure 1). The average mobility of membrane protein ions is approximately 30 % lower than that of soluble proteins of similar mass that are commonly used for TWIMS calibration (Tables S1 and S2).
Figure 1.
Average charge state and Ω of membrane proteins and commonly used soluble calibrant proteins measured by DTIMS. Under nESI-MS and detergent conditions optimized to preserve native-like conformations, membrane proteins exhibit significantly lower charge states than soluble proteins with comparable Ω. It should be noted that gas-phase unfolding exacerbates this difference. The non-database21 Ω, for transthyretin (TTR), Mycobacterium tuberculosis mechanosensitive channel of large conductance (MscL), and E. coli formate channel (FocA), were taken from the literature.5,20,29 A plot with non-averaged data is provided in Figure S1.
Effects of calibrant ion selection for native-like membrane protein complexes
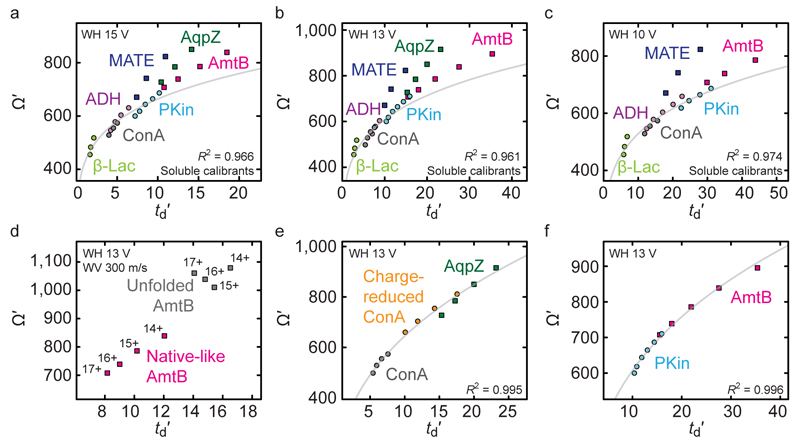
To assess how low ion charge affects Ω measurements by TWIMS, we compared four soluble calibrant proteins spanning a mass range from 36 to 239 kDa with three membrane protein complexes solubilized in C8E4 ranging from 50 to 126 kDa, for which Ω values have been determined by DTIMS (Figure 2, Tables S1 and S2)5,20,21. As outlined above, calibrant ions are frequently chosen to cover the Ω and mass range of the protein of interest, as these would normally be of similar mobility to the analyte. To calibrate the TWIMS drift times, the relationship between mobility and drift time is determined by plotting the corrected drift time td′ as a function of Ω/z (Ω′, see Methods). Fitting the points with a power law then allows us to calculate the Ω of an ion from its drift time and charge state. However, soluble proteins and membrane proteins separate into two distinct Ω′ regimes, independent of the TWIMS wave height (Figure 2a–c). The membrane proteins are marked by both higher Ω′ values as well as much longer drift times than those of soluble proteins with a similar molecular mass or Ω. Therefore, they deviate substantially from the best calibration fit obtained when using soluble protein calibrants with similar Ω and mass, leading to an underestimation of their Ω in some cases by >20 % (Tables S3-S5).
Figure 2.
Charge-corrected Ω (Ω′) of a set of soluble (round data points) and membrane proteins (square data points) as a function of corrected drift time (td′). (a–c) At a fixed wave velocity of 350 m/s, membrane proteins and calibrant ions separate into distinct Ω′ regimes. This separation is not affected by wave height. The data from panel (a) are plotted as the ‘doubly-corrected’ drift time vs. Ω in Figure S2 to illustrate in this alternative representation the deviation of membrane proteins from soluble proteins. (d) The plot of Ω′ of native-like and unfolded membrane protein AmtB as a function of td′ shows that the species cannot be matched by a single calibration curve. (e) Charge reduction bridges the difference in mobility between ConA and the membrane protein AqpZ. (f) PKin is a suitable calibrant protein for approximating the Ω of AmtB by TWIMS. Selected charge states for each protein are listed in Table S6. WH: wave height; WV: wave velocity.
Mobility differences arise when the correlation between Ω and z (for native-like soluble proteins) no longer holds. This is the case when z deviates for the reduced charge of membrane proteins, but it is also possible when an analyte is in the same z range, but has a significantly different Ω. We therefore analysed the accuracy of Ω measurements in the context of collision-induced unfolding (CIU) experiments by TWIMS. CIU is widely used to study domain organization and relative gas-phase stabilities of protein complexes, and provides a means to distinguish complex biomolecules based on their unfolding “fingerprints”.22–25 Briefly, protein ions are subjected to increasing collisional activation inside the mass spectrometer, resulting in a quasi-thermal denaturation that can be monitored by using IMS.26 However, while the Ω of the ions increase, their charge states remain largely unaffected, and therefore the mobility decreases. We performed CIU experiments and compared TWIMS-derived and DTIMS-measured Ω of the different unfolded species. Plotting td′ as a function of Ω′ for the most unfolded states (maximum collision energy) illustrates that unfolding significantly reduces the mobility of the unfolded states relative to the initial native-like species in a charge-state dependent manner (Figure 2d). Therefore, in effect, a calibration strategy chosen on the basis of the native-like folded form will only approximate the Ω of the unfolded species. As such, rather than assigning partially folded conformations to these extended Ω values, stronger interpretations can currently be made by comparing the effect of ligand binding on the retention of folded structure,15 or by analyzing the changes in unfolding patterns.14
To probe the cause for the deviations between membrane proteins and soluble proteins, we compared the drift times of concanavalin A (ConA), a soluble protein, and the membrane protein AqpZ, which have similar molecular weights (103 and 99 kDa, respectively) and Ω (5,500 and 5,100 Å2, respectively). ConA has an average charge state of 20+ as opposed to 13+ for AqpZ solubilized in C8E4. To achieve a better charge-state match (and therefore more similar mobility), we reduced the charge of ConA by addition of 10 mM imidazole to the buffer,27 and additional exposure to acetonitrile vapor in the ion source,28 reducing the average charge to 15+. With decreasing charge, the Ω′ and td′ of ConA approach those observed for AqpZ (Figure 2e). Calibrating with charge-reduced ConA (charge states 13–17+; Ω values in Table S7) allows us to calculate an average Ω of 5,301 Å2 for AqpZ, an error of 1.2% that is within the anticipated accuracy of <5% for a suitable TWIMS calibration.11 Whilst the calibration with non-charge-reduced ConA (charge states 19–22+) can be extrapolated to yield a Ω for AqpZ, with reasonable error (3.9–13%), dependent on wave height, the increased accuracy using charge-reduced ConA suggests that the charge difference alone is sufficient to produce the observed discrepancies between soluble calibrants and membrane proteins.
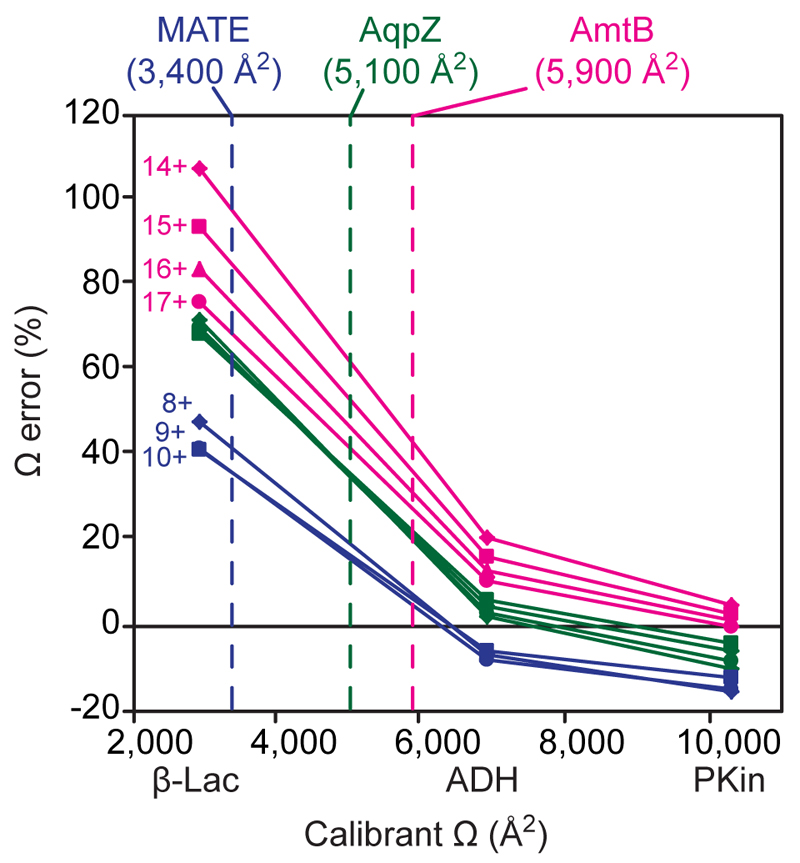
To investigate whether the use of calibrant ions in the same drift-time regime can alleviate the problems caused by differences in mobility, we determined the Ω error for each membrane protein when using individual calibrant proteins. Although the expected accuracy is low when using only one calibrant protein and its 3–4 main charge states, the results demonstrate that the accuracy of the calibration improves when the Ω of the calibrant ions are >30 % larger than the Ω of the membrane protein ions (Figure 3, Tables S3-S5). This is particularly evident for the membrane protein AmtB, for which we determined Ω values for all charge states within 1.4 % of the literature value when using only pyruvate kinase (PKin) as calibrant (Figure 2f and Table S4). Similarly, the Ω of the membrane proteins MATE and AqpZ can be approximated to 5–6 % of the literature values when calibrating with ConA and ADH, respectively (Figures 3 and S2, Table S4). Using a combination of ADH and PKin further reduced the error for AmtB to 1.3 %, and a combination of ConA and ADH allowed us to determine the Ω of AqpZ with 0.8 % error (Table S4).
Figure 3.
Effect of calibrant ion Ω on TWIMS calibration accuracy. The Ω of the main charge states of the membrane proteins AmtB (pink), AqpZ (green; charge states 11–14+) and MATE (blue) were determined using non-charge-reduced either only β-lactoglobulin (2,900 Å2), only alcohol dehydrogenase (6,900 Å2) or only pyruvate kinase (10,300 Å2) as calibrant ions. The smallest deviations from the literature values are achieved when the calibrant ions have a significantly larger Ω than the membrane proteins. Dashed lines indicate the literature Ω for the membrane proteins. Spectra were recorded using a wave velocity of 350 m/s and wave height of 13 V.
We then explored the use of larger calibrants for CIU. We found that when PKin is used as a calibrant for unfolding of AmtB, TWIMS reports an increase from 6,200 Å2 to 7,800 Å2 for the 16+ charge state, which unfolds readily above 100 V. For the same charge state and energy range, DTIMS shows a Ω increase from 5,950 Å2 to 8,300 Å2, revealing an error of approximately 5 % for the TWIMS measurements (Figure S4). We conclude that the use of larger calibrants also reduces the errors that arise in Ω during CIU for the resulting unfolded states of membrane protein complexes. The relationship between Ω′ and td′ also appears, particularly for charge-reduced membrane proteins, to differ on a protein-to-protein basis, as there is no general fit that would encompass all charges states of all proteins (Figure 2a-c). Although this effect is relatively small for soluble proteins, it will manifest itself in calibration-dependent deviations from expected Ω that vary for each charge state, potentially more so than the variations observed for DTIMS measurements. This further reinforces that a good calibration should be consistent across all folded native-like charge states and requires the calibrant and analyte ions to be matched individually. For example, a protein with an extremely broad charge state distribution would require a moving calibration window with calibrant ions of increasing Ω as analyte charge decreases.
Conclusions
Taken together, our results reveal important pitfalls in TWIMS calibrations for native MS Ω determination of membrane proteins, and gas-phase unfolding studies. In both cases, the difference in Ω′ of the calibrant and analyte ions renders the common practice of calibrant ion selection based on mass ineffective. In case of native-like membrane proteins, the use of membrane protein standards in the same detergent and matching the mass of the analyte may be a viable strategy, but the lack of widely available membrane protein standards with consistent quality and a broad size range hamper this approach. In addition, the low charges create significant mobility differences between the individual charge states, causing additional error. A simple way to avoid error-prone long-range extrapolations of the calibration function is to use much larger calibrants, which we show can provide reasonably accurate TWIMS calibrations for charge-reduced, native-like membrane protein complexes.
Supplementary Material
Acknowledgements
ML holds a Marie Curie Early Career Development grant and a Junior Research Fellowship from St Cross College, University of Oxford. JLPB is a Royal Society University Research Fellow. CVR is supported by an ERC Advanced Investigator Award (26851) (IMPRESS) and a Royal Society Research Professorship. The authors thank Idlir Liko, Joseph Gault, Stephen Ambrose, and Shahid Mehmood for membrane protein samples and illuminating discussions.
Abbreviations
- ADH
alcohol dehydrogenase
- AmtB
ammonia channel
- AqpZ
aquaporin channel Z
- β-lactoglobulin
- CIU
collision-induced unfolding
- ConA
concanavalin A
- DTIMS
drift tube ion mobility spectrometry
- FocA
formate channel
- MATE
multiantimicrobial transporter
- MscL
mechanosensitive channel of large conductance
- nESI-MS
nano-electrospray ionization mass spectrometry
- PKin
pyruvate kinase
- TWIMS
travelling-wave ion mobility spectrometry
Footnotes
ASSOCIATED CONTENT
Supporting Information available: Ω and mobility values, calibration errors with different calibrant combinations, charge states used for analysis, illustrations of calibration accuracy, calibrated unfolding plot. This material is available free of charge via the internet at http://pubs.acs.org
Notes
The authors declare no competing financial interest.
Author Contributions
TMA, ML and CVR designed the study. TMA and ML performed experiments. TMA and ML wrote the manuscript with contributions from all authors. All authors have given approval to the final version of the manuscript.
References
- (1).Uetrecht C, Rose RJ, van Duijn E, Lorenzen K, Heck AJR. Chem Soc Rev. 2010;39:1633–1655. doi: 10.1039/b914002f. [DOI] [PubMed] [Google Scholar]
- (2).Lanucara F, Holman SW, Gray CJ, Eyers CE. Nat Chem. 2014;6:281–294. doi: 10.1038/nchem.1889. [DOI] [PubMed] [Google Scholar]
- (3).Mehmood S, Allison TM, Robinson CV. Annu Rev Phys Chem. 2015;66:453–474. doi: 10.1146/annurev-physchem-040214-121732. [DOI] [PubMed] [Google Scholar]
- (4).Laganowsky A, Reading E, Hopper JT, Robinson CV. Nat Protoc. 2013;8:639–651. doi: 10.1038/nprot.2013.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, Baldwin AJ, Robinson CV. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Konijnenberg A, Yilmaz D, Ingolfsson HI, Dimitrova A, Marrink SJ, Li Z, Venien-Bryan C, Sobott F, Kocer A. Proc Natl Acad Sci. 2014;111:17170–17175. doi: 10.1073/pnas.1413118111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Mc Daniel EW, Mason EA. Transport properties of ions in gases. Wiley; New York: 1988. [Google Scholar]
- (8).Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. Int J Mass Spectrom. 2007;261:1–12. [Google Scholar]
- (9).Shvartsburg AA, Smith RD. Anal Chem. 2008;80:9689–9699. doi: 10.1021/ac8016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Thalassinos K, Grabenauer M, Slade SE, Hilton GR, Bowers MT, Scrivens JH. Anal Chem. 2009;81:248–254. doi: 10.1021/ac801916h. [DOI] [PubMed] [Google Scholar]
- (11).Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Anal Chem. 2010;82:9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- (12).Sun Y, Vahidi S, Sowole MA, Konermann L. J Am Soc Mass Spectrom. 2016;27:31–40. doi: 10.1007/s13361-015-1244-5. [DOI] [PubMed] [Google Scholar]
- (13).Clemmer DE. [(Accessed Jan. 6, 2016)];Cross section database. http://www.indiana.edu/~clemmer/Research/Cross%20Section%20Database/cs_database.php.
- (14).Ruotolo BT, Benesch JLP, Sandercock AM, Hyung SJ, Robinson CV. Nat Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- (15).Allison TM, Reading E, Liko I, Baldwin AJ, Laganowsky A, Robinson CV. Nat Commun. 2015;6:8551. doi: 10.1038/ncomms9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ruotolo BT, Benesch JL, Sandercock AM, Hyung SJ, Robinson CV. Nat Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- (17).Robinson EW, Shvartsburg AA, Tang K, Smith RD. Anal Chem. 2008;80:7508–7515. doi: 10.1021/ac800655d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Morgner N, Montenegro F, Barrera NP, Robinson CV. J Mol Biol. 2012;423:1–13. doi: 10.1016/j.jmb.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Mehmood S, Marcoux J, Hopper JTS, Allison TM, Liko I, Borysik AJ, Robinson CV. J Am Chem Soc. 2014;136:17010–17012. doi: 10.1021/ja510283g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Reading E, Liko I, Allison TM, Benesch JL, Laganowsky A, Robinson CV. Angew Chem Int Ed. 2015;54:4577–4581. doi: 10.1002/anie.201411622. [DOI] [PubMed] [Google Scholar]
- (21).Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Anal Chem. 2010;82:9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- (22).Hopper JTS, Oldham NJ. J Am Soc Mass Spectr. 2009;20:1851–1858. doi: 10.1016/j.jasms.2009.06.010. [DOI] [PubMed] [Google Scholar]
- (23).Nabuchi Y, Hirose K, Takayama M. Anal Chem. 2010;82:8890–8896. doi: 10.1021/ac101482b. [DOI] [PubMed] [Google Scholar]
- (24).Ninonuevo MR, Leary JA. Anal Chem. 2012;84:3208–3214. doi: 10.1021/ac2030249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Tian YW, Han LJ, Buckner AC, Ruotolo BT. Anal Chem. 2015;87:11509–11515. doi: 10.1021/acs.analchem.5b03291. [DOI] [PubMed] [Google Scholar]
- (26).Benesch JLP. J Am Soc Mass Spectrom. 2009;20:341. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- (27).Liu L, Bagal D, Kitova EN, Schnier PD, Klassen JS. J Am Chem Soc. 2009;131:15980–15981. doi: 10.1021/ja9060454. [DOI] [PubMed] [Google Scholar]
- (28).Hopper JTS, Sokratous K, Oldham NJ. Anal Biochem. 2012;421:788–790. doi: 10.1016/j.ab.2011.10.034. [DOI] [PubMed] [Google Scholar]
- (29).Hall Z, Politis A, Bush MF, Smith LJ, Robinson CV. J Am Chem Soc. 2012;134:3429–3438. doi: 10.1021/ja2096859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.