ABSTRACT
Thiamine (vitamin B1) is a precursor of thiamine pyrophosphate (TPP), an essential coenzyme in the central metabolism of all living organisms. Bacterial thiamine biosynthesis and salvage genes are controlled at the RNA level by TPP-responsive riboswitches. In Archaea, TPP riboswitches are restricted to the Thermoplasmatales order. Mechanisms of transcriptional control of thiamine genes in other archaeal lineages remain unknown. Using the comparative genomics approach, we identified a novel family of transcriptional regulators (named ThiR) controlling thiamine biosynthesis and transport genes in diverse lineages in the Crenarchaeota phylum as well as in the Halobacteria and Thermococci classes of the Euryarchaeota. ThiR regulators are composed of an N-terminal DNA-binding domain and a C-terminal ligand-binding domain, which is similar to the archaeal thiamine phosphate synthase ThiN. By using comparative genomics, we predicted ThiR-binding DNA motifs and reconstructed ThiR regulons in 67 genomes representing all above-mentioned lineages. The predicted ThiR-binding motifs are characterized by palindromic symmetry with several distinct lineage-specific consensus sequences. In addition to thiamine biosynthesis genes, the reconstructed ThiR regulons include various transporters for thiamine and its precursors. Bioinformatics predictions were experimentally validated by in vitro DNA-binding assays with the recombinant ThiR protein from the hyperthermophilic archaeon Metallosphaera yellowstonensis MK1. Thiamine phosphate and, to some extent, TPP and hydroxyethylthiazole phosphate were required for the binding of ThiR to its DNA targets, suggesting that ThiR is derepressed by limitation of thiamine phosphates. The thiamine phosphate-binding residues previously identified in ThiN are highly conserved in ThiR regulators, suggesting a conserved mechanism for effector recognition.
IMPORTANCE Thiamine pyrophosphate is a cofactor for many essential enzymes for glucose and energy metabolism. Thiamine or vitamin B1 biosynthesis and its transcriptional regulation in Archaea are poorly understood. We applied the comparative genomics approach to identify a novel family of regulators for the transcriptional control of thiamine metabolism genes in Archaea and reconstructed the respective regulons. The predicted ThiR regulons in archaeal genomes control the majority of thiamine biosynthesis genes. The reconstructed regulon content suggests that numerous uptake transporters for thiamine and/or its precursors are encoded in archaeal genomes. The ThiR regulon was experimentally validated by DNA-binding assays with Metallosphaera spp. These discoveries contribute to our understanding of metabolic and regulatory networks involved in vitamin homeostasis in diverse lineages of Archaea.
KEYWORDS: Archaea, comparative genomics, regulon reconstruction, thiamine metabolism, transcriptional regulation
INTRODUCTION
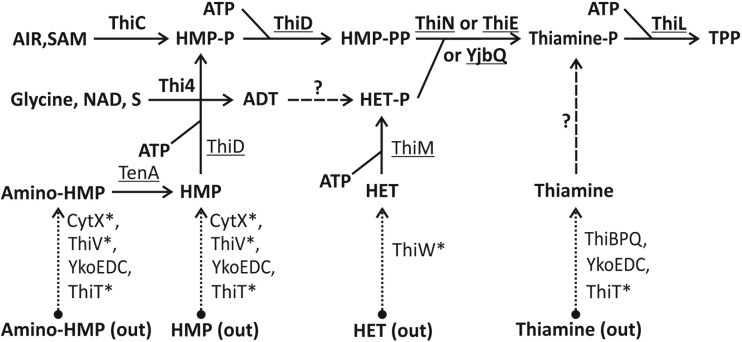
Vitamins of the B group are key precursors in the biosynthesis of indispensable enzyme cofactors driving numerous metabolic processes in all forms of life. Vitamin B1 (thiamine) in its active form, thiamine pyrophosphate (TPP), is an essential cofactor for several anabolic and catabolic reactions in the central metabolism (1). Archaeal vitamin biosynthesis pathways contain several unique enzymes that lack homology to those of Bacteria (Fig. 1 and Table 1). The thiamine biosynthesis pathway (TBP) utilizes a set of committed enzymes catalyzing the independent synthesis of 4-amino-5-hydroxy-2-methylpyrimidine (HMP) and 4-methyl-5-β-hydroxyethylthiazole (HET) moieties and their subsequent phosphorylation and coupling into a single molecule of thiamine phosphate, which is finally phosphorylated to TPP by the ThiL kinase (2). The pyrimidine moiety of thiamine, HMP pyrophosphate (HMP-PP), is synthesized from aminoimidazole ribotide (AIR), an intermediate of the purine biosynthesis pathway, by using HMP phosphate (HMP-P) synthase (ThiC) and HMP-P kinase (ThiD) (3). The HMP-PP synthesis pathway is conserved between Bacteria and Archaea; however, the synthesis of the thiazole moiety of thiamine, HET phosphate (HET-P), is catalyzed by two different biochemical routes (4). In Bacteria, HET-P is synthesized from 1-deoxy-xylulose phosphate, glycine (or tyrosine), and cysteine by using the thiS, thiF, thiH (or thiO), thiI, iscS, and thiG gene products (5). In contrast, the Thi4 family proteins catalyze the single-step synthesis of HET-P in eukaryotes and archaea (4, 6, 7). Thiamine phosphate synthase catalyzes the coupling of HMP-PP and HET-P moieties to yield thiamine phosphate and pyrophosphate. Bacteria and Archaea utilize analogous thiamine phosphate synthases from different protein families, ThiE and ThiN, respectively (4, 8–10). Thiamine phosphate is finally phosphorylated to TPP by the ThiL kinase in both Bacteria and Archaea (11, 12).
FIG 1.
Overview of thiamine biosynthesis and salvage pathways in Archaea. Biosynthetic and salvage enzymes (solid lines) are in boldface type and underlined, respectively. The dashed line with a question mark indicates as-yet-uncharacterized ADP-thiazole hydrolase and thiamine kinases activities. Uptake transporters (dotted lines) are listed along with their predicted substrates. The asterisk denotes transporters with predicted substrate specificity. Detailed functional roles, including Enzyme Commission (EC) numbers and transporter protein families, are listed in Table 1. All included enzymes and transporters (except ThiT) have representatives in both Archaea and Bacteria; however, Thi4 and ThiN are ubiquitous in Archaea, while ThiE is present mostly in Bacteria. Abbreviations: TPP, thiamine pyrophosphate; HMP, 4-amino-5-hydroxy-2-methylpyrimidine; HET, 4-methyl-5-β-hydroxyethylthiazole; AIR, aminoimidazole ribotide; ADT, ADP-thiazole; SAM, S-adenosylmethionine.
TABLE 1.
Thiamine and TPP biosynthesis enzymes and transporters identified in archaeal genomes
| Protein | Functional role (EC no.)a | No. of genes in the ThiR regulonb (no. of respective genes in 67 studied genomes) | Reference(s) |
|---|---|---|---|
| ThiC | Phosphomethylpyrimidine synthase (EC 4.1.99.17) | 37 (44) | 3 |
| Thi4 | Thiamine thiazole synthase, archaeal/eukaryotic | 55 (59) | 6, 7 |
| ThiM | Hydroxyethylthiazole kinase (EC 2.7.1.50) | 12 (23) | 51 |
| ThiE | Thiamine phosphate synthase (EC 2.5.1.3), bacterial | 12 (27) | 10 |
| ThiD | HMP/phosphomethylpyrimidine kinase (EC 2.7.1.49, EC 2.7.4.7) | 1 (6) | 52, 53 |
| ThiN | Bifunctional thiamine phosphate synthase ThiN (EC 2.5.1.3)/HMP/phosphomethylpyrimidine kinase (EC 2.7.1.49, EC 2.7.4.7), archaeal | 26 (55) | 8, 9 |
| YjbQ | Low-activity thiamine phosphate synthase YjbQ (EC 2.5.1.3), bacterial | 7 (15) | 8, 9, 11, 26 |
| TenA | Aminopyrimidine aminohydrolase (EC 3.5.99.2) | 46 (72) | 54, 55 |
| ThiL | Thiamine monophosphate kinase (EC 2.7.4.16) | 0 (67) | 12, 56 |
| ThiR | Transcriptional regulator of thiamine metabolism | 12 (67) | This work |
| ThiBPQ | Thiamine transporter, ABC family, bacterial | 26 (30) | 57 |
| YkoEDC | Predicted thiamine/HMP moiety transporter, ECF family | 12 (12) | 58, 59 |
| ThiT | Predicted thiamine/HMP transporter, MFS | 18 (23) | |
| ThiW | Predicted thiazole moiety transporter, ECF family | 2 (2) | |
| CytX | Predicted HMP moiety transporter, NCS1 family | 10 (10) | |
| ThiV | Predicted HMP moiety transporter, SSS family | 6 (7) |
Predicted functional roles of thiamine-related transporters determined by using comparative genomics analysis of TPP riboswitch regulons were previously proposed (4). NCS1, nucleobase:cation symporter 1; SSS, solute:sodium symporter.
Numbers of thiamine metabolism genes that are preceded by a candidate ThiR-binding site. The total numbers of the respective genes (including paralogs) found in 67 studied genomes possessing the ThiR regulon are given in parentheses.
The TBP genes in bacteria are controlled by cis-regulatory RNAs (the so-called THI elements) that were first discovered by comparative genomics (4) and then attributed to a class of TPP-sensing riboswitches that control gene expression through the formation of alternative leader mRNA structures (13, 14). TPP riboswitches provide a negative-feedback loop for the control of TBP genes that are often clustered into one or several thi operons in bacterial genomes. In addition to the TBP genes, the reconstructed TPP riboswitch regulons in many proteobacteria and several bacteria from other phyla include the thiBPQ operon, encoding the known ABC transporter for thiamine, thiamine phosphate, and TPP (4, 15). Analysis of the distribution of TPP riboswitches in bacterial genomes combined with subsystem-based metabolic reconstruction revealed other families of putative uptake transporters for thiamine, HET, and HMP (4). The predicted functions of some of these transporters, including the energy-coupling factor (ECF) family transporters YuaJ/ThiT, YkoEDC, and ThiW and the ABC family transporter ThiXYZ, have been experimentally validated (16–18). Interestingly, the TPP riboswitches were also identified in a single taxonomic group of Archaea, the Thermoplasmatales, including Thermoplasma, Ferroplasma, and Picrophilus spp., where they coregulate two predicted thiamine transporters, ThiT1 and ThiT2, of the major facilitator superfamily (MFS) (4). However, TPP riboswitches were not found in all other lineages of Archaea, and the mechanism of transcriptional regulation of TBP genes in Archaea was not known before this study.
Comparative analysis of multiple closely related genomes is a powerful approach for the identification and reconstruction of novel transcriptional regulons in Bacteria (19), and it is also applicable to archaeal genomes (20, 21). In this study, we combined comparative genomics to map genes for the thiamine biosynthesis and uptake pathways and discovered putative thiamine regulons in archaeal genomes. We found a novel transcriptional regulator for the thiamine metabolism genes, termed ThiR, which represents a fusion of an N-terminal DNA-binding domain and a C-terminal domain that is similar to the archaeal thiamine phosphate synthase ThiN. Evidence from comparative genomics suggests that ThiR is a thiamine phosphate-responsive repressor, which is conserved in diverse lineages of Archaea. The reconstructed ThiR regulons in 67 archaeal genomes include the TBP genes and/or various genes encoding transporters for thiamine and its metabolic precursors. To evaluate the accuracy of the genomic reconstruction, we performed experimental testing of the ThiR regulon in Metallosphaera yellowstonensis MK-1, a thermoacidophilic species of the Crenarchaeota phylum that was isolated in Yellowstone National Park (22). In vitro-binding assays with the purified ThiR regulator confirmed its predicted DNA sites and revealed thiamine phosphates as molecular effectors that are required for ThiR-DNA binding. A putative mechanism for the ThiR-dependent negative regulation of the TBP and/or thiamine uptake genes was proposed.
RESULTS AND DISCUSSION
Genomic identification and reconstruction of thiamine regulons.
We began this study by performing a bioinformatics survey of candidate genes for transcriptional regulators of thiamine metabolism in 41 available archaeal genomes of the Crenarchaeota phylum (the Thermoprotei class) that do not contain TPP riboswitches. Prioritization of candidate thiamine regulatory genes was performed based on an analysis of their domain structure and genome context evidence, including chromosomal gene clustering with thiamine metabolism genes. As a primary candidate for the thiamine regulator, we identified a family of archaeal thiamine phosphate synthase (ThiN) paralogs fused to an N-terminal DNA-binding domain resembling a helix-turn-helix (HTH) motif, which is characteristic of many prokaryotic transcription factors (TFs). Below we summarize major observations that support the proposed functional role for members of the predicted family of thiamine regulators, which was named ThiR.
Orthologs of the thiR genes were identified in 35 out of 41 analyzed genomes of the Crenarchaeota phylum (see Fig. S1 in the supplemental material). The only species that apparently had lost thiR genes among the Crenarchaeota are two Aeropyrum spp., Ignicoccus hospitalis, Pyrolobus fumarii, Acidilobus saccharovorans, and Caldisphaera lagunensis. In addition, thiR orthologs are present in all available genomes of the Halobacteria (17 genomes) and Thermococci (15 genomes) classes of the Euryarchaeota phylum. Overall, the putative thiR thiamine regulatory genes were found in 67 genomes that belong to three major classes of Archaea. Other classes of Archaea do not have thiR orthologs.
The thiR genes are present in a single copy in most genomes, with the exception of Halobacteria (10 genomes), where we have identified two thiR paralogs. The phylogenetic tree of the ThiR proteins largely coincides with the taxonomic species tree of Archaea, with the single exception of the Thermofilum branch, which is not clustered with other Thermoproteales (see Fig. S2 in the supplemental material). ThiR1 and ThiR2 paralogs in Halobacteria form two distinct branches (average sequence identity, 40%), suggesting their possible appearance by an ancient lineage-specific duplication.
In order to reconstruct putative ThiR regulons, we first identified the repertoire of known and putative genes involved in thiamine biosynthesis, salvage, and uptake in 73 genomes of Archaea from the above-mentioned three taxonomic ranks (Table 1; see also Table S1 in the supplemental material). In the group of 4 Thermofilum spp., the thiR gene is colocalized and divergently transcribed with the predicted thiamine transporter operon ykoEDC. In Fervidicoccus fontis, thiR is also colocalized with the ykoDCE transport locus, but their relative orientation is convergent (Fig. 2). In contrast, thiR orthologs in other analyzed genomes are not clustered on the chromosome with TBP or thiamine transport genes. We have analyzed the predicted prototrophy/auxotrophy phenotypes and the distribution of ThiR regulators across three studied classes of Archaea (see Fig. S1 in the supplemental material). Among 67 ThiR-encoding archaea, 43 species possess the complete sets of TBP genes (B1 prototrophs), while the remaining 24 species lack at least one essential TBP gene (B1 or HMP auxotrophs). HMP auxotrophs (14 species) are not able to synthesize the HMP precursor of thiamine due to a lack of ThiC, while the HET biosynthesis pathway is intact in these species. The remaining B1 auxotrophs could be further classified into two subgroups. Three species lack the HMP and HET synthesis pathways but possess the salvage pathways for both precursors (ThiM, ThiD, and ThiE/N), whereas seven species lack both de novo biosynthesis and salvage pathways for B1 precursors.
FIG 2.
Distribution of thiamine biosynthesis and uptake genes and reconstructed ThiR regulons in representative genomes of Thermoprotei (A), Thermococcales (B), and Halobacteria (C). Detailed information for each analyzed genome is provided in Table S1 in the supplemental material. Total numbers of analyzed species in selected genera of Thermoprotei are shown in parentheses. Genes are shown by arrows, and the same functional roles are marked by matching colors. Candidate ThiR-binding sites are shown as pink or red circles. Five different types of DNA motifs identified in Thermoprotei are marked with roman numerals inside the circle. Sequence logos representing the consensus binding site motifs were built by using all candidate sites in the respective groups of genomes.
To facilitate the comparative genomics analysis, all analyzed genomes were subdivided into individual taxonomic groups according to the phylogenetic tree of ThiR proteins. By applying the DNA motif recognition procedure to a training set of upstream regions of TBP and/or thiamine transporter genes, we identified putative ThiR-binding site motifs in each taxonomic group. The identified putative ThiR-binding site motifs were refined by the construction of multiple alignments of orthologous upstream regions (see Fig. S3 in the supplemental material). Based on the obtained lineage-specific profiles, we searched for additional sites in archaeal genomes and identified additional candidate members of ThiR regulons such as the putative thiamine transporter thiT in the Sulfolobales and Thermoproteales (see Table S2 in the supplemental material).
The predicted ThiR motifs and the respective sets of coregulated operons in all studied lineages are illustrated in Fig. 2. ThiR motifs in the Thermoprotei class share a 13-bp palindrome symmetry and a conserved core with a consensus ATAA-N5-TTAT sequence; however, the Sulfolobales-specific motif has a 6-bp spacer between the half-sites. ThiR motifs in the Thermococci and Halobacteria classes as well as in Thermofilum species share a consensus sequence (ANNAC-N3-GTNNT), which is partially similar to the above-mentioned consensus sequence in the Thermoprotei. Despite these lineage-specific variations in the consensus sequences of palindromic half-sites and the central spacer lengths, the common consensus signals of the predicted ThiR-binding half-sites can be deduced as ATAA and TTAT, respectively (see Fig. S2 in the supplemental material).
Interestingly, candidate ThiR boxes occur in tandem in 63 and 76% of regulatory gene regions in the Thermococci and Halobacteria genomes, as the distance between the centers of palindromes equals an integer number of DNA turns (21 bp and 12 bp, respectively) (see Table S2 in the supplemental material). The presence of multiple regulatory sites at a specific distance ensures cooperative binding of ThiR dimers to DNA. Within the Thermoprotei class, the majority of predicted ThiR-regulated genes in Fervidicoccus, Staphylothermus, Thermofilum, and Thermogladius spp. and ∼30% of the candidate ThiR targets in Pyrobaculum spp. are preceded by two or three palindromic ThiR sites, suggesting cooperative binding of two or more ThiR dimers to DNA. In contrast, the candidate ThiR operators in the Sulfolobales order contain a single palindromic ThiR site.
We analyzed the relative positions of the identified ThiR-binding sites and known transcription start sites (TSSs) from previously reported transcriptomes of Sulfolobus solfataricus, Pyrococcus furiosus, and Haloferax volcanii (23, 24) (see Table S2 in the supplemental material). The transcription start sites coincide with the translation start sites of the thi4, thiT, and tenA genes in S. solfataricus (23) and the thi4 and thiC genes in H. volcanii (25), as the predicted ThiR-binding sites overlap TATA-like conserved core promoter motifs across all related genomes (see Fig. S3 in the supplemental material). The TSS of thi4 in P. furiosus is located ∼7 nucleotides (nt) upstream of the translation start site, and its respective TATA-like core promoter motif is inserted between tandem ThiR sites. The TSS of thiB in P. furiosus is located in the middle of the proximal site of the tandem ThiR operator. Thus, all of the known promoters of ThiR-regulated genes overlapped predicted ThiR sites, making it possible to predict that ThiR acts as a repressor of transcription.
The content of reconstructed ThiR regulons is quite uniform in the representatives of all three classes of Archaea, although individual genomes may have between one and five target operons (Fig. 2). The core regulon group includes the de novo TBP genes thi4, thiC, and thiDN as well as the thiEM and tenA genes involved in the salvage of thiamine precursors (Table 1). The thiR genes are preceded by candidate ThiR boxes in 4 Thermofilum and 7 Thermococcus species as well as in F. fontis, suggesting potential autoregulation of ThiR. The predicted ThiR regulons in Thermococcus spp. also include the YjbQ family proteins (encoded by a putative operon with thiR) that were previously characterized for their low thiamine phosphate synthase activity (26).
Among the taxon-specific members of predicted ThiR regulons are various genes presumably involved in the transport of thiamine and/or its metabolic precursors HET and HMP (Table 1). These members include a novel group of MFS-type transporters in the Sulfolobales and Thermoproteales that are orthologous to the TPP riboswitch-regulated transporter ThiT from the Thermoplasmatales order of Archaea (4). Other predicted ThiR-regulated transporters in Archaea (thiBPQ, ykoEDC, thiV, cytX, and thiW) are shared with Bacteria, where they are commonly regulated by TPP riboswitches (18). An integrated genomic approach combining the identification of ThiR regulons with subsystem-based metabolic reconstruction allowed us to predict likely specificities for thiamine-regulated transporters (Fig. 1). The CytX transporters of the nucleoside transporter family of the Thermococci and two Desulfurococcales species as well as the sodium-dependent ThiV transporters in the Halobacteria are presumably involved in the uptake of pyrimidine precursors of thiamine (HMP or amino-HMP). The ECF family transporter ThiW found in two Thermococcus spp. is likely involved in HET salvage. The novel group of MFS ThiT transporters in the Sulfolobales and Thermoproteales as well as the ECF family YkoEDC system in the Desulfurococcales, Thermofilum, and Fervidicoccus are likely involved in the uptake of thiamine and/or the HMP precursors, as they are present in both B1 prototrophs and auxotrophs. The ABC family transporter ThiBPQ found in the reconstructed ThiR regulons of the Thermococci and Halobacteria is presumably involved in the salvage of thiamine.
Experimental validation of the ThiR regulon in M. yellowstonensis.
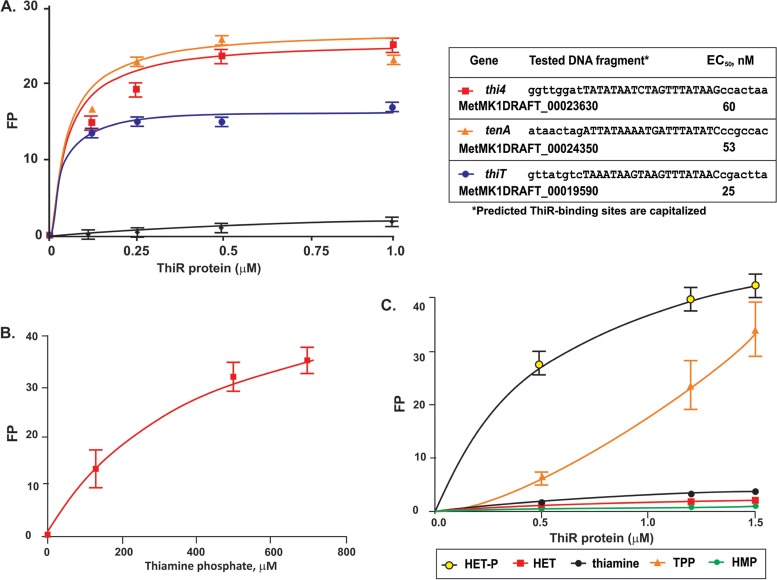
In order to validate the computationally predicted DNA-binding motif of ThiR in the Sulfolobales, we heterologously expressed and purified the ThiR protein from M. yellowstonensis MK1. This extremophile is a dominant community member in acidic thermophilic Fe(II)-oxidizing microbial mats of Yellowstone National Park (22). Our genomic reconstruction suggests that M. yellowstonensis is the only B1 auxotroph within the Sulfolobales order, which is unable to synthesize the HMP precursor of thiamine (see Fig. S1 in the supplemental material). Three synthetic fluorescence-labeled 46-bp DNA fragments containing the predicted ThiR-binding sites upstream of the thiT, thi4, and tenA genes in M. yellowstonensis were assessed for their specific interaction with the purified ThiR protein. Each synthetic DNA fragment contained a 20-bp ThiR site located in the middle of the 35-bp natural DNA fragment of the promoter region (see Fig. S3 in the supplemental material) and flanked by polycytosine stretches on each side. In the presence of excess thiamine phosphate (0.5 mM), ThiR specifically binds to the synthetic DNA fragments containing ThiR-binding sites at all three predicted target genes in M. yellowstonensis, as determine by a fluorescence polarization assay (Fig. 3A). The apparent affinity, estimated as 50% effective concentrations (EC50), for the ThiR protein interacting with the tested DNA fragments was in the range of 25 to 60 nM. The exclusion of thiamine phosphate from the incubation mixture led to the disruption of ThiR-DNA binding. Titration of the thiamine phosphate effect at a fixed concentration of the ThiR protein (600 nM) and the thi4 DNA fragment determined an EC50 for the effector in the range of 0.3 to 0.4 mM (Fig. 3B). TPP and HET-P (0.5 mM) also stimulated the binding of ThiR with the thi4 DNA fragment, although the effect of these metabolites was weaker than the effect of thiamine phosphate (Fig. 3C). In contrast, thiamine, HET, and HMP (up to 1 mM) had no effect on the binding of ThiR to the target DNA. Physiological concentrations of thiamine phosphates in the cell and the EC50 for TPP of TPP-sensing riboswitches in bacteria are at least 2 orders of magnitude lower than the determined EC50 for thiamine phosphates in the ThiR-DNA binding assays (11, 12, 27). Nonphysiologically high concentrations of thiamine phosphates required for ThiR-DNA binding could be explained by nonoptimized conditions of the fluorescence polarization assay, such as moderate temperatures and neutral pH, while physiological conditions for M. yellowstonensis include extremely low pH and high temperatures (28). Overall, this analysis of the M. yellowstonensis ThiR protein provided sufficient experimental confirmation of the target ThiR-binding sites and ThiR-regulated genes tentatively identified in the Sulfolobales lineage by comparative genomics, while many aspects of the proposed mechanism are yet to be investigated. Experimental validation of candidate DNA-binding motifs for ThiR orthologs in other lineages of Archaea was beyond the scope of this mostly bioinformatics-based study.
FIG 3.
Experimental validation of the ThiR regulon in M. yellowstonensis via a fluorescence polarization (FP) DNA-binding assay. (A) Increasing concentrations of the recombinant ThiR protein were mixed with 46-bp fluorescence-labeled DNA fragments containing predicted ThiR sites upstream of the thiT, thi4, and tenA genes of M. yellowstonensis in the presence of thiamine phosphate (0.5 mM). The tested DNA fragments containing candidate ThiR sites (capitalized) and their apparent EC50 for the ThiR protein are listed. The locations of tested DNA fragments within the promoter gene regions are shown in Fig. S3 in the supplemental material. As a negative control (black line), a 28-bp DNA fragment of the M. yellowstonensis trxA gene from the autotrophic regulon HhcR was used (21). The values are means from three measurements, and the standard deviations are shown. (B) Titration of thiamine phosphate that has a positive effect on the interaction between ThiR (600 nM) and the thi4 DNA fragment. (C) Testing of the influence of other potential effectors, including HET, HET phosphate, HMP, thiamine, and TPP (0.5 mM), on protein-DNA binding. Increasing concentrations of ThiR and the thi4 DNA fragment were used in a fluorescence polarization assay.
Proposed ThiR regulatory mechanism.
Based on the in vitro effector screening results and the positional analysis of candidate ThiR sites relative to experimentally determined TSSs/promoter elements (see above), we propose the following regulatory mechanism for ThiR. In the presence of an excessive amount of thiamine phosphates in the cell, ThiR binds its DNA target sites and represses the expression of genes involved in the biosynthesis and/or salvage of vitamin B1. When levels of thiamine phosphates become limiting in the cell, ThiR dissociates from DNA, resulting in the derepression of ThiR-regulated genes. Thus, thiamine phosphates act as a corepressor of ThiR. The proposed regulatory mechanism for ThiR in Archaea partially resembles the known mechanism for the transcriptional/translational control of thiamine metabolism genes in Bacteria, involving a metabolite-sensing riboswitch, which binds TPP as a corepressor and thus promotes the repressive conformation of 5′ untranslated leader mRNA (14).
The concentration of thiamine phosphates in the cell is linked to the activity of the de novo biosynthesis pathway and/or the uptake and phosphorylation of exogenous vitamin B1. Despite genomic identification of candidate thiamine-related transporters, kinases responsible for the further phosphorylation of thiamine for TPP production are largely unknown in Archaea. Recent biochemical studies with Pyrobaculum caldifontis cell extracts suggest that the phosphorylation of thiamine does not take place in this hyperthermophilic archaeon (11). The ThiL kinase in this species is strictly specific for thiamine phosphate and cannot phosphorylate thiamine (11). P. calidifontis is an HMP auxotroph possessing the thiDN and thi4 biosynthetic genes and the predicted HMP/thiamine transporter thiT. Since the salvaged thiamine is readily hydrolyzed into HMP and HET in hyperthermophilic environments, it was suggested that P. calidifontis can reutilize the HMP moiety. The thiL kinases are ubiquitous in Archaea, but they do not belong to the ThiR regulons. Thus, the above-described hypothesis for P. calidifontis is not applicable to several B1-auxotrophic Thermofilum and Thermococcus species possessing only a candidate thiamine transporter (ykoEDC or thiBPQ) and the thiL kinase. It is plausible that either some ThiL orthologs may have a broader substrate specificity toward thiamine or there are alternative, as-yet-unknown thiamine kinases. Further investigation of thiamine salvage pathways in diverse archaeal lineages may help to resolve these puzzles in the future.
Domain analysis of ThiR proteins and comparison with ThiN enzymes.
The archaeal ThiR proteins represent a fusion of the N-terminal HTH DNA-binding domain (COG2522 [XRE family]) and the C-terminal ThiN-like domain (COG1992, arCOG0002, or Pfam10120). Multiple alignments of ThiR proteins showed a significant conservation of both domains across various archaeal lineages (see Fig. S4 in the supplemental material). We then compared the sequences of ThiN-like domains in regulators with the thiamine phosphate synthase ThiN domain of the bifunctional ThiDN protein from P. furiosus with an available tertiary structure (PDB accession no. 2PB9). The structure reported under PDB accession no. 2PB9 contains a phosphoric acid molecule bound to Asn298, Arg320, and residues 339 to 341 (Ser-Asp-His-Ile). The amino acid sequence alignment showed a high level of conservation of the corresponding residues in ThiR proteins (see Fig. S4 in the supplemental material). Asn298 and Ser339 are absolutely invariant in all ThiR proteins. Arg320 is conserved in all ThiR proteins except for a single protein in Hyperthermus butylicus. His341 is present in ThiR orthologs from most archaeal lineages except for the Thermoproteus, Pyrobaculum, and Thermococcus genera, where it is replaced with phenylalanine. Thus, we propose that the above-mentioned four residues are involved in the interaction with thiamine phosphate in ThiR regulators. A previously proposed structure model of the enzyme-product complex of ThiN of Pyrobaculum calidifontis followed by site-directed mutagenesis confirmed the importance of the Arg320 and His341 residues for thiamine phosphate synthase activity (8). Thus, we propose that the ThiN-like domains in regulators are involved in the binding of thiamine phosphate as a corepressor.
It is yet to be assessed experimentally whether ThiR regulators are able to catalyze the thiamine phosphate synthase reaction, although this possibility seems to us rather unlikely. The genomic distribution of the thiDN genes, encoding a fusion between the HMP/HMP-P kinase (ThiD) and thiamine phosphate synthase (ThiN) domains, suggests that this essential thiamine biosynthetic enzyme is present in the majority of B1 prototrophs, with only a few exceptions in the Halobacteria genomes that possess the ThiE-type thiamine phosphate synthase (thiE) and stand-alone HMP/HMP-P kinase (thiD) genes. The thiDN and/or thiE gene was also found in B1 auxotrophs, where it is presumably involved in the salvage of the thiamine precursors HMP and HET. Genomes of the Thermofilum genus and two Thermococcus species are the only archaeal genomes that lack both types of thiamine phosphate synthases; these genomes lack any thiamine biosynthesis gene except for the thiL kinase gene. The predicted ThiR regulons in these B1-auxotrophic species contain only putative vitamin B1 transporters. Thus, the genomics-based reconstruction of B1 synthesis pathways in archaeal genomes suggests that ThiR regulators may have lost the thiamine phosphate synthase activity and use the ThiN-like domains exclusively for effector sensing.
Conclusions.
The identification of a novel family of DNA-binding regulators for thiamine metabolism allowed us to fill a substantial gap in the knowledge of the transcriptional regulation of key metabolic pathways in Archaea. The ThiR regulator represents a unique type of metabolite-sensing TF emerging via fusion between DNA-binding and enzymatic domains. A few other prokaryotic TFs with similar domain architecture principles include the bifunctional biotin repressor/ligase BirA (29), the trifunctional NAD repressor/nicotinamide ribose kinase/adenylyltransferase NadR (30), and the proline utilization repressor/dehydrogenase PutA (31). Using the comparative genomics approach, we identified palindromic ThiR-binding DNA motifs and reconstructed the respective transcriptional regulons in available ThiR-encoding genomes. The core of the reconstructed ThiR regulons includes the thiamine biosynthesis and salvage enzymes as well as known and predicted uptake transporters for thiamine and its precursors. We chose one representative ThiR regulator to experimentally assess its specific DNA-binding properties and to test possible small-molecule effectors. Thiamine phosphate was most effective in stimulating the in vitro binding of ThiR to its DNA fragments. The obtained genomics and experimental data are generally supportive of a role of ThiR-effector interactions in the proposed mechanism of repression of ThiR regulon genes.
MATERIALS AND METHODS
Bioinformatics tools and databases.
The 73 analyzed archaeal genomes were downloaded from GenBank (32). These genomes include 41 complete genomes from the Thermoprotei class (the Crenarchaeota phylum) that were available in GenBank at the time of analysis (July 2015). In addition, ThiR orthologs were identified in all available genomes of the Halobacteria and Thermococci classes in the Euryarchaeota phylum, which included 17 and 15 genomes, respectively. The phylogenetic tree of the 73 analyzed archaeal species was constructed with PhyML, using the multiple alignments of concatenated sequences of universal ribosomal proteins extracted from each genome (33). Orthologs were identified as bidirectional best hits by using protein BLAST and the ortholog search tool in Genome Explorer software (34) and additionally confirmed via the construction of phylogenetic trees by using PhyML with 100 bootstrap replicates. To perform reconstruction of the thiamine biosynthesis pathways in archaeal genomes, we utilized the subsystem-based comparative genomics approach (35) and a set of experimentally characterized and previously predicted enzymes and transporters of Bacteria and Archaea (Table 1). A gene neighborhood analysis was performed by using the IMG ER, PubSEED, and MicrobesOnline databases (36–38). Closely related gene orthologs with experimentally determined function were identified via BLAST searches of the UniProtKB/Swiss-Prot database (39). Protein domain architecture was determined by using the COG (40), arCOG (41), and Pfam (42) databases. Transporters were classified according to the TCDB database (43). Computational searches of TPP riboswitches were performed by using covariance models uploaded from the Rfam database (44; recently reviewed in reference 45). Sequence logos for DNA motifs were drawn by using the WebLogo package (46).
For the identification of candidate ThiR-binding site motifs and regulon reconstruction, we used a previously established comparative genomics approach (19) implemented in GenomeExplorer software (34) and the RegPredict Web server (47). We started regulon reconstruction from a collection of initial training sets of known genes involved in the TBP (thiC, thi4, thiMED, and thiDN) and/or thiamine salvage and transport (tenA, thiBPQ, ykoEDC, thiV, cytX, and thiW) in each archaeal lineage containing a ThiR ortholog (see Table S1 in the supplemental material). For each training set of genes, we determined orthologs in related genomes, analyzed the potential operon structure of their loci, and collected their upstream DNA regions up to 200 nucleotides upstream of the translation start site (excluding the coding regions of any upstream gene if the intergenic region was less than 200 nt). The prepared training sets for each ThiR-containing archaeal lineage, including the Sulfolobales, Thermoproteales, Desulfurococcales, Fervidicoccales, Thermococcales, and Halobacteria, were used for a search of lineage-specific conserved DNA motifs.
The Discover Profile tool in RegPredict uses expectation maximization methods and the palindromic or direct-repeat symmetry of motifs, a feature that is typical of many prokaryotic TFs that interact with their TF-binding sites (TFBSs) as symmetric dimers (19). The algorithm identifies all weak palindromes (direct repeats) in an input group of sequences, iteratively clusters them to convergence, and outputs several putative motifs ranked by their average positional information content. A search for palindromic DNA motifs of 13 to 20 bp of both odd and even lengths was carried out by using the following parameters: minimum number of GC pairs of 0, number of palindromic positions of 4, and size of the training set of 50. Motifs were further validated by the construction of multiple alignments of orthologous DNA fragments using MUSCLE (48). Conservative palindromic sites were selected as potential binding sites for ThiR and used for the construction of positional weight matrices (PWMs), which describe the probability of each possible letter at each position in the pattern. Positional nucleotide weights in PWMs and site scores were defined as previously described (49).
Further PWM-based searches for additional sites identified a second copy of the putative ThiR site in the majority of DNA upstream regions from the initial training set in the Thermococcales, Halobacteria, and Fervidicoccales lineages as well as in Thermofilum and Staphylothermus spp. The tandem ThiR sites were added to the final training sets used for PWM construction.
The constructed PWMs were used to search for additional TFBSs and regulon members in the analyzed genomes by using GenomeExplorer. The genome scan parameters were set up to reduce the chance of nonfunctioning sequences being detected. Specifically, positions of candidate regulatory sites were set between 200 nt upstream and 25 nt downstream of a gene start codon. The score thresholds for site searches were selected as the lowest site score in the training set (see Table S2 in the supplemental material). Weaker sites (with scores 15% lower than the threshold) were also taken into account if their positions were similar to the positions of stronger sites upstream of orthologous genes. False-positive TFBS predictions were eliminated by using the consistency check approach (19). New candidate members were attributed to the regulon if they were preceded by candidate TFBSs in more than 50% of genomes in each taxonomic group. The reconstructed regulons were extended to include all genes in putative operons. Genes were considered to belong to an operon if they were transcribed in the same direction and had intergenic distances not exceeding 50 nt. All reconstructed regulons, including predicted sites, regulators, and regulated genes, are available in Tables S1 and S2 in the supplemental material.
Gene cloning and protein purification.
The thiR gene (GenBank accession no. EHP68530.1; locus tag MetMK1_29690) from M. yellowstonensis MK1 was synthesized by GenScript Inc. with optimized codons for expression in Escherichia coli. The synthesized gene fragments were cloned into the pSMT3 expression vector under the control of the T7 promoter (50). The obtained vector encodes a fusion between the target protein and an N-terminally hexahistidine-tagged Smt3 polypeptide (a yeast SUMO ortholog), which enhances protein solubility. The resulting constructs were transformed into E. coli BL21(DE3) cells. Cells were grown in LB medium (1 liter) at 37°C to an optical density at 600 nm (OD600) of ∼0.8, protein expression was induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and cells were harvested after shaking at 20°C overnight or at 37°C during 4 h. The recombinant ThiR protein was purified to homogeneity by using Ni2+ chelation chromatography. The harvested cells were resuspended in 20 mM HEPES buffer (pH 7) containing 100 mM NaCl, 0.03% Brij 35, 2 mM β-mercaptoethanol, and 2 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich). Cells were lysed by incubation with lysozyme (1 mg/ml) for 30 min at 4°C, followed by a freeze-thaw cycle and sonication. For purification of the soluble fraction after centrifugation, Tris-HCl buffer (pH 8) was added to the supernatant (50 mM), which was loaded onto a Ni-nitrilotriacetic acid-agarose minicolumn (0.3 ml) (Qiagen Inc.). After washing with starting buffer containing 1 M NaCl and 0.3% Brij 35, the bound proteins were eluted with 0.3 ml of the same buffer supplemented with 250 mM imidazole. The purified protein was controlled by gel filtration with a fast protein liquid chromatography system. The protein concentration was determined by using a Quick Start Bradford protein assay kit (Bio-Rad). The ThiR protein was obtained with high purity (>90%, as determined by SDS-PAGE).
DNA-binding assays.
The interaction between the purified recombinant regulatory proteins and their cognate DNA binding sites in M. yellowstonensis MK1 was assessed by using a fluorescence polarization DNA-binding assay. The His6-Smt3 tag was cleaved from the purified ThiR protein by digestion with Ulp1 protease at 4°C overnight. The synthetic DNA oligonucleotides containing the predicted binding sites were synthesized by Integrated DNA Technology. One strand of the oligonucleotide was 3′ labeled by a fluorescence label, 6-carboxyfluorescein. The double-stranded labeled DNA fragments were obtained by annealing the labeled oligonucleotides with unlabeled complementary oligonucleotides at a 1:10 ratio. ThiR was tested with three 46-bp DNA fragments containing binding sites upstream of the thiT (MetMK1_19590), thi4 (MetMK1_23630), and tenA (MetMK1_24350) genes (see Table S3 in the supplemental material). Another 28-bp DNA fragment containing a binding site for HhcR at the trxA (MetMK1_12900) gene in M. yellowstonensis (21) was used as a negative control. The obtained fluorescence-labeled double-stranded DNA fragments (10 nM) were incubated with increasing concentrations of the ThiR protein (0.12 to 1 μM) in a 100-μl reaction mixture in 96-well black plates (VWR, Radnor, PA) for 20 min. The binding buffer contained 0.2× phosphate-buffered saline (PBS) (pH 6.5), 0.5 mM EDTA, 7 mM MgCl2, 100 mM KCl, and 5 mM phosphate, and the incubation temperature was 50°C. Herring sperm DNA (1 μg) was added to the reaction mixture as a nonspecific competitor DNA to suppress nonspecific binding. The fluorescence-labeled DNA was detected with the FLA-5100 fluorescence image analyzer. The effects of thiamine, thiamine phosphate, TPP, HET, HMP, and HET-P (0 to 500 μM) on ThiR binding to DNA were tested by adding them to the incubation mixture.
ADDENDUM
Since this work was submitted, an independent study and manuscript kindly shared with us by J. Maupin-Furlow from the University of Florida identified and characterized the ThiR regulator and its regulon in Haloferax volcanii. Based on the detailed characterization of ThiR using in vivo genetic techniques, the researchers came to the conclusion that ThiR is a repressor of the thiamine biosynthesis genes in the presence of thiamine or thiazole that lacks thiamine phosphate synthase activity in vivo. We note that the ThiR-binding DNA motif proposed in our work coincides with the ThiR operator that was identified in H. volcanii by using genetic techniques and reporter assays.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Russian Science Foundation (grant number 14-14-00289). Additional funding for experimental validation of ThiR function was provided by the Genomic Science Program (GSP), Office of Biological and Environmental Research (OBER), U.S. Department of Energy (DOE).
We thank Margie F. Romine for providing tables of orthologous genes for the Sulfolobales and Thermoproteales genomes and Andrei L. Osterman for useful discussions on experimental assessment of the ThiR regulator.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00743-16.
REFERENCES
- 1.Begley TP, Ealick SE. 2010. Thiamin biosynthesis, p 547–559. In Mander LN, Liu H (ed), Comprehensive natural products II: chemistry and biology, vol 7 Elsevier Ltd., Kidlington, United Kingdom. [Google Scholar]
- 2.Begley TP, Downs DM, Ealick SE, McLafferty FW, Van Loon AP, Taylor S, Campobasso N, Chiu HJ, Kinsland C, Reddick JJ, Xi J. 1999. Thiamin biosynthesis in prokaryotes. Arch Microbiol 171:293–300. doi: 10.1007/s002030050713. [DOI] [PubMed] [Google Scholar]
- 3.Palmer LD, Downs DM. 2013. The thiamine biosynthetic enzyme ThiC catalyzes multiple turnovers and is inhibited by S-adenosylmethionine (AdoMet) metabolites. J Biol Chem 288:30693–30699. doi: 10.1074/jbc.M113.500280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. 2002. Comparative genomics of thiamin biosynthesis in prokaryotes. New genes and regulatory mechanisms. J Biol Chem 277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- 5.Jurgenson CT, Begley TP, Ealick SE. 2009. The structural and biochemical foundations of thiamin biosynthesis. Annu Rev Biochem 78:569–603. doi: 10.1146/annurev.biochem.78.072407.102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee A, Abeydeera ND, Bale S, Pai PJ, Dorrestein PC, Russell DH, Ealick SE, Begley TP. 2011. Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase. Nature 478:542–546. doi: 10.1038/nature10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang S, Cordova B, Chavarria N, Elbanna D, McHugh S, Rojas J, Pfeiffer F, Maupin-Furlow JA. 2014. Conserved active site cysteine residue of archaeal THI4 homolog is essential for thiamine biosynthesis in Haloferax volcanii. BMC Microbiol 14:260. doi: 10.1186/s12866-014-0260-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi M, Kobayashi K, Esaki H, Konno H, Akaji K, Tazuya K, Yamada K, Nakabayashi T, Nosaka K. 2014. Enzymatic and structural characterization of an archaeal thiamin phosphate synthase. Biochim Biophys Acta 1844:803–809. doi: 10.1016/j.bbapap.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 9.Morett E, Korbel JO, Rajan E, Saab-Rincon G, Olvera L, Olvera M, Schmidt S, Snel B, Bork P. 2003. Systematic discovery of analogous enzymes in thiamin biosynthesis. Nat Biotechnol 21:790–795. doi: 10.1038/nbt834. [DOI] [PubMed] [Google Scholar]
- 10.Reddick JJ, Nicewonger R, Begley TP. 2001. Mechanistic studies on thiamin phosphate synthase: evidence for a dissociative mechanism. Biochemistry 40:10095–10102. doi: 10.1021/bi010267q. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi M, Nosaka K. 2015. Characterization of thiamin phosphate kinase in the hyperthermophilic archaeon Pyrobaculum calidifontis. J Nutr Sci Vitaminol (Tokyo) 61:369–374. doi: 10.3177/jnsv.61.369. [DOI] [PubMed] [Google Scholar]
- 12.Webb E, Downs D. 1997. Characterization of thiL, encoding thiamin-monophosphate kinase, in Salmonella typhimurium. J Biol Chem 272:15702–15707. doi: 10.1074/jbc.272.25.15702. [DOI] [PubMed] [Google Scholar]
- 13.Breaker RR. 2011. Prospects for riboswitch discovery and analysis. Mol Cell 43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winkler W, Nahvi A, Breaker RR. 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 15.Sun EI, Leyn SA, Kazanov MD, Saier MH Jr, Novichkov PS, Rodionov DA. 2013. Comparative genomics of metabolic capacities of regulons controlled by cis-regulatory RNA motifs in bacteria. BMC Genomics 14:597. doi: 10.1186/1471-2164-14-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodionov DA, Hebbeln P, Eudes A, ter Beek J, Rodionova IA, Erkens GB, Slotboom DJ, Gelfand MS, Osterman AL, Hanson AD, Eitinger T. 2009. A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol 191:42–51. doi: 10.1128/JB.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodionova IA, Li X, Plymale AE, Motamedchaboki K, Konopka AE, Romine MF, Fredrickson JK, Osterman AL, Rodionov DA. 2015. Genomic distribution of B-vitamin auxotrophy and uptake transporters in environmental bacteria from the Chloroflexi phylum. Environ Microbiol Rep 7:204–210. doi: 10.1111/1758-2229.12227. [DOI] [PubMed] [Google Scholar]
- 18.Jaehme M, Slotboom DJ. 2015. Diversity of membrane transport proteins for vitamins in bacteria and archaea. Biochim Biophys Acta 1850:565–576. doi: 10.1016/j.bbagen.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Rodionov DA. 2007. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem Rev 107:3467–3497. doi: 10.1021/cr068309+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leyn SA, Rodionov DA. 2015. Comparative genomics of DtxR family regulons for metal homeostasis in Archaea. J Bacteriol 197:451–458. doi: 10.1128/JB.02386-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leyn SA, Rodionova IA, Li X, Rodionov DA. 2015. Novel transcriptional regulons for autotrophic cycle genes in Crenarchaeota. J Bacteriol 197:2383–2391. doi: 10.1128/JB.00249-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozubal MA, Dlakic M, Macur RE, Inskeep WP. 2011. Terminal oxidase diversity and function in “Metallosphaera yellowstonensis”: gene expression and protein modeling suggest mechanisms of Fe(II) oxidation in the Sulfolobales. Appl Environ Microbiol 77:1844–1853. doi: 10.1128/AEM.01646-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wurtzel O, Sapra R, Chen F, Zhu Y, Simmons BA, Sorek R. 2010. A single-base resolution map of an archaeal transcriptome. Genome Res 20:133–141. doi: 10.1101/gr.100396.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon SH, Reiss DJ, Bare JC, Tenenbaum D, Pan M, Slagel J, Moritz RL, Lim S, Hackett M, Menon AL, Adams MW, Barnebey A, Yannone SM, Leigh JA, Baliga NS. 2011. Parallel evolution of transcriptome architecture during genome reorganization. Genome Res 21:1892–1904. doi: 10.1101/gr.122218.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brenneis M, Hering O, Lange C, Soppa J. 2007. Experimental characterization of cis-acting elements important for translation and transcription in halophilic archaea. PLoS Genet 3:e229. doi: 10.1371/journal.pgen.0030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morett E, Saab-Rincon G, Olvera L, Olvera M, Flores H, Grande R. 2008. Sensitive genome-wide screen for low secondary enzymatic activities: the YjbQ family shows thiamin phosphate synthase activity. J Mol Biol 376:839–853. doi: 10.1016/j.jmb.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 27.Winkler WC, Cohen-Chalamish S, Breaker RR. 2002. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci U S A 99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jennings RM, Whitmore LM, Moran JJ, Kreuzer HW, Inskeep WP. 2014. Carbon dioxide fixation by Metallosphaera yellowstonensis and acidothermophilic iron-oxidizing microbial communities from Yellowstone National Park. Appl Environ Microbiol 80:2665–2671. doi: 10.1128/AEM.03416-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood ZA, Weaver LH, Brown PH, Beckett D, Matthews BW. 2006. Co-repressor induced order and biotin repressor dimerization: a case for divergent followed by convergent evolution. J Mol Biol 357:509–523. doi: 10.1016/j.jmb.2005.12.066. [DOI] [PubMed] [Google Scholar]
- 30.Grose JH, Bergthorsson U, Roth JR. 2005. Regulation of NAD synthesis by the trifunctional NadR protein of Salmonella enterica. J Bacteriol 187:2774–2782. doi: 10.1128/JB.187.8.2774-2782.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, Zhu W, Bellur PS, Rewinkel D, Becker DF. 2008. Direct linking of metabolism and gene expression in the proline utilization A protein from Escherichia coli. Amino Acids 35:711–718. doi: 10.1007/s00726-008-0053-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2015. GenBank. Nucleic Acids Res 43:D30–D35. doi: 10.1093/nar/gku1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 34.Mironov AA, Vinokurova NP, Gel'fand MS. 2000. Software for analyzing bacterial genomes. Mol Biol (Mosk) 34:253–262. [PubMed] [Google Scholar]
- 35.Osterman AL, Overbeek R, Rodionov DA. 2010. The use of subsystems to encode biosynthesis of vitamins and cofactors, p 141–159. In Mander LN, Liu H (ed), Comprehensive natural products II: chemistry and biology, vol 7 Elsevier Ltd., Kidlington, United Kingdom. [Google Scholar]
- 36.Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS, Dubchak IL, Alm EJ, Arkin AP. 2010. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res 38:D396–D400. doi: 10.1093/nar/gkp919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. 2009. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25:2271–2278. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- 38.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.UniProt Consortium. 2015. UniProt: a hub for protein information. Nucleic Acids Res 43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galperin MY, Makarova KS, Wolf YI, Koonin EV. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolf YI, Makarova KS, Yutin N, Koonin EV. 2012. Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer. Biol Direct 7:46. doi: 10.1186/1745-6150-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saier MH Jr, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. 2016. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res 44:D372–D379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nawrocki EP, Burge SW, Bateman A, Daub J, Eberhardt RY, Eddy SR, Floden EW, Gardner PP, Jones TA, Tate J, Finn RD. 2015. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res 43:D130–D137. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun EI, Rodionov DA. 2014. Computational analysis of riboswitch-based regulation. Biochim Biophys Acta 1839:900–907. doi: 10.1016/j.bbagrm.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novichkov PS, Rodionov DA, Stavrovskaya ED, Novichkova ES, Kazakov AE, Gelfand MS, Arkin AP, Mironov AA, Dubchak I. 2010. RegPredict: an integrated system for regulon inference in prokaryotes by comparative genomics approach. Nucleic Acids Res 38:W299–W307. doi: 10.1093/nar/gkq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mironov AA, Koonin EV, Roytberg MA, Gelfand MS. 1999. Computer analysis of transcription regulatory patterns in completely sequenced bacterial genomes. Nucleic Acids Res 27:2981–2989. doi: 10.1093/nar/27.14.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mossessova E, Lima CD. 2000. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell 5:865–876. doi: 10.1016/S1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 51.Yazdani M, Zallot R, Tunc-Ozdemir M, de Crecy-Lagard V, Shintani DK, Hanson AD. 2013. Identification of the thiamin salvage enzyme thiazole kinase in Arabidopsis and maize. Phytochemistry 94:68–73. doi: 10.1016/j.phytochem.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 52.French JB, Begley TP, Ealick SE. 2011. Structure of trifunctional THI20 from yeast. Acta Crystallogr D Biol Crystallogr 67:784–791. doi: 10.1107/S0907444911024814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mizote T, Tsuda M, Smith DD, Nakayama H, Nakazawa T. 1999. Cloning and characterization of the thiD/J gene of Escherichia coli encoding a thiamin-synthesizing bifunctional enzyme, hydroxymethylpyrimidine kinase/phosphomethylpyrimidine kinase. Microbiology 145(Part 2):495–501. [DOI] [PubMed] [Google Scholar]
- 54.Benach J, Edstrom WC, Lee I, Das K, Cooper B, Xiao R, Liu J, Rost B, Acton TB, Montelione GT, Hunt JF. 2005. The 2.35 A structure of the TenA homolog from Pyrococcus furiosus supports an enzymatic function in thiamine metabolism. Acta Crystallogr D Biol Crystallogr 61:589–598. doi: 10.1107/S0907444905005147. [DOI] [PubMed] [Google Scholar]
- 55.Zallot R, Yazdani M, Goyer A, Ziemak MJ, Guan JC, McCarty DR, de Crecy-Lagard V, Gerdes S, Garrett TJ, Benach J, Hunt JF, Shintani DK, Hanson AD. 2014. Salvage of the thiamin pyrimidine moiety by plant TenA proteins lacking an active-site cysteine. Biochem J 463:145–155. doi: 10.1042/BJ20140522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayashi M, Kijima Y, Tazuya-Murayama K, Yamada K. 2015. The biosynthesis of the thiazole moiety of thiamin in the archaeon Halobacterium salinarum. J Nutr Sci Vitaminol (Tokyo) 61:270–274. doi: 10.3177/jnsv.61.270. [DOI] [PubMed] [Google Scholar]
- 57.Webb E, Claas K, Downs D. 1998. thiBPQ encodes an ABC transporter required for transport of thiamine and thiamine pyrophosphate in Salmonella typhimurium. J Biol Chem 273:8946–8950. doi: 10.1074/jbc.273.15.8946. [DOI] [PubMed] [Google Scholar]
- 58.Josts I, Almeida Hernandez Y, Andreeva A, Tidow H. 2016. Crystal structure of a group I energy coupling factor vitamin transporter S component in complex with its cognate substrate. Cell Chem Biol 23:827–836. doi: 10.1016/j.chembiol.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schyns G, Potot S, Geng Y, Barbosa TM, Henriques A, Perkins JB. 2005. Isolation and characterization of new thiamine-deregulated mutants of Bacillus subtilis. J Bacteriol 187:8127–8136. doi: 10.1128/JB.187.23.8127-8136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.