Abstract
Along with their cognate acyl-homoserine lactone signals, the quorum sensing regulators LasR and RhlR control the expression of hundreds of genes in the opportunistic human pathogen Pseudomonas aeruginosa. This extensive, overlapping regulatory network affords the opportunity to systematically investigate the sequence requirements and specificity determinants of large families of target promoters. Many of the P. aeruginosa quorum-controlled genes possess conserved palindromic promoter elements predicted to be binding sites for either one or both transcriptional regulators, but biochemical proof has not been reported. We have purified native LasR and characterized binding to various quorum-controlled promoters in vitro. Purified LasR was a dimer in solution that irreversibly bound two molecules of 3-oxo-C12-homoserine lactone. LasR bound several las-responsive promoters specifically and with high affinity, interacting cooperatively with some promoters and noncooperatively with others. LasR recognized some, but not all, of the predicted binding sites, and also bound to several unexpected sites. In contrast to predictions from genetic data, we found that the recognition sequences of las-specific promoters showed little overall sequence conservation and did not require dyad symmetry. We found distinct differences in sequence composition between las-specific noncooperative, las-specific cooperative, and rhl-responsive promoters. These results provide the basis for defining promoter specificity elements in P. aeruginosa quorum sensing. Insights into the molecular mechanism of LasR function have implications for the development of quorum-sensing targeted antivirulence compounds.
Keywords: bacterial signaling, cell communication, homoserine lactone, gene regulation
Intercellular communication by the exchange of chemical signals is a widespread phenomenon in bacteria. This process, termed quorum sensing, allows a bacterial population to coordinate gene expression in response to cell density (1, 2). In many Gram-negative bacteria, quorum sensing is accomplished by a signal transduction system comprised of a protein homologous to LuxI of Vibrio fischeri that generates a diffusable acylhomoserine lactone signal molecule and a protein homologous to LuxR that serves as a signal receptor and transcriptional activator (3, 4). The opportunistic human pathogen Pseudomonas aeruginosa provides one of the most intensely studied examples of quorum-controlled gene expression. This organism is known for its ability to cause persistent infections in immunocompromised patients, such as those with the genetic disorder cystic fibrosis. Quorum-sensing-regulated expression of an array of virulence factors, such as extracellular enzymes and secondary metabolites, plays an important role in this process (5, 6).
Overall, >300 P. aeruginosa genes are regulated by two acyl-homoserine lactone systems, LasR–LasI and RhlR–RhlI (7, 8). LasI and RhlI are synthases that produce the signal molecules 3-oxo-dodecanoyl-homoserine lactone (3OC12-HSL) and butanoyl (C4)-HSL, respectively. LasR and RhlR are the regulators that respond to their cognate signals and activate transcription. Both systems are connected in a hierachical fashion, because LasR, at least under some conditions, controls the expression of rhlI and rhlR (9–11). Target promoters respond to each system with varying degrees of specificity (8, 12), but the sequence determinants for this specificity are not known. Genetic evidence suggests that LasR and RhlR bind to conserved palindromic sequences of some quorum-controlled promoters (12–15), and more such sites have been located upstream of other quorum-controlled genes (7, 8). These so-called las–rhl box-like sequences show similarity to the lux box, which is the promoter element required for quorum control of the V. fischeri luminescence genes (16). In addition to LasR and RhlR, a third LuxR homolog, QscR, has been found to modulate the expression of individual quorum-controlled genes in P. aeruginosa (17). For QscR, the extent of regulation, the requirement, if any, for an acyl-HSL ligand, and the mechanism of action are not yet fully understood.
In a single previous study that attempted to biochemically characterize the DNA-binding activity of LasR, little or no active protein was purified (18). Quorum sensing regulators from other organisms, including V. fischeri, have been investigated biochemically in more detail (19–23), but the number of target promoters is either comparatively low or information about them is scarce. In P. aeruginosa, the large size of the quorum sensing regulon and its regulation by two, possibly three, acyl-HSL signaling systems poses additional questions: Which genes are regulated by quorum sensing directly, and what are the signal specificity determinants of quorum-controlled promoters? To begin to address these questions, we describe the in vitro characterization of purified LasR, including DNA binding, acyl-HSL binding, and oligomeric state.
Materials and Methods
Overproduction and Purification of LasR. First, megapriming was used to introduce a silent mutation in the lasR coding region that eliminated an internal NdeI restriction site. A megaprimer was generated by PCR using pECP59 (pEX1.8 with ptac-lasR) (24) as template and 5′-NNNNNNNCATATGGCCTTGGTTGACGGTTTTC-3′ (NdeI restriction site is underlined) and 5′-GTGAACTTCCACATGGGAAATATTC-3′ as primers. With this megaprimer and primer 5′-NNNNNNCTCGAGTCAGAGAGTAATAAGACCCAAATT-3′ (XhoI restriction site is underlined), the lasR-coding region was amplified from pECP59 by PCR. The resulting PCR product was digested with NdeI and XhoI, and was cloned into pET17b (Novagen) to form pET17b.lasR. The integrity of the lasR gene was confirmed by sequencing.
For LasR overexpression, colonies of Escherichia coli Tuner (DE3) pLysS (Novagen) carrying pET17b.lasR were grown overnight on LB-ampicillin-chloramphenicol plates and were used to inoculate 1 liter of prewarmed LB containing 50 mM Mops, pH 7, 100 μg/ml ampicillin, 34 μg/ml chloramphenicol, and 5 μM 3OC12-HSL. The culture was incubated at 37°C. At an optical density of 0.5 at 600 nm, the culture was rapidly chilled to 20°C, induced with isopropyl-β-d-thiogalactoside (0.4 mM final concentration), and incubated at 17°C overnight, after which cells were harvested by centrifugation and frozen at –80°C. All subsequent steps were carried out at 0–4°C.
The cell pellets from 500 ml of culture were thawed and resuspended in 8.5 ml of LasR purification buffer (LRPB) (25 mM Tris·HCl, pH 7.8/150 mM NaCl/1 mM DTT/1 mM EDTA/10% glycerol/0.05% Tween 20). Cells were broken by sonication using a Branson Microtip Sonifier (7 × 20 s at an output of 0.4). Insoluble material was removed by ultracentrifugation (100,000 × g for 30 min). The supernatant fluid was precipitated with ammonium sulfate (40% saturation), and the precipitate was resuspended and dialyzed for 6 h in LRPB buffer. The dialysate was chromatographed on a tandemly arranged pair of columns comprising a 5-ml HiTrap Q HP anion exchange column and a 5-ml HiTrap Heparin HP affinity column (Amersham Pharmacia). Both columns had been equilibrated with LRPB. After the columns were washed with LRPB, the Q column was removed and the LasR protein bound to the heparin column was eluted with a 50-ml linear NaCl gradient of 150–500 mM. Fractions containing LasR were stored at –80°C. The resulting LasR preparation was >95% pure as judged by SDS/PAGE. Protein concentrations were determined by using a Bradford assay (Bio-Rad).
Electrophoretic Mobility-Shift Assays. Gel-shift experiments were based on a protocol by Urbanowski et al. (25). Each gel-shift reaction contained both specific and nonspecific DNA probes. Specific DNA probes ≈280 bp in length were generated by PCR-amplification of the regulatory regions upstream of hcnA, lasB, rhlA, rsaL, PA0572, PA1897, PA3904, PA4078, PA4131, and PA4677. A nonspecific DNA probe of 176 bp was generated by PCR amplification of DNA encompassing most of the rsaL-coding region. The resulting PCR products were labeled at both ends by using [γ-32P]ATP and T4 nucleotide kinase. Binding reactions contained 0.5–1 pM each of specific and nonspecific DNA in a final volume of 20 μl of DNA-binding buffer [20 mM Tris·HCl, pH 7.5/50 mM KCl/1 mM EDTA/1 mM DTT/100 μg/ml BSA/10 μg/ml poly(dIdC)/5% glycerol]. Purified LasR and 3OC12-HSL were added at the indicated concentrations, and the reaction mixtures were incubated at room temperature for 20 min. After the addition of 1 μl of loading dye, the reaction mixtures were electrophoresed on native 5% Tris-glycine-EDTA polyacrylamide gels (1:30 bis-acrylamide to acrylamide ratio) at 10 V/cm at room temperature. Gels were dried, and probes were detected by using PhosphorImager technology.
DNase I Footprinting. DNase I protection assays were based on described methods (25–27). For the gel-shift experiments described above, the lasB, hcnA, rsaL, PA0572, PA3904, and PA4677 promoter fragments were amplified with primers containing restriction sites resulting in fragments with a 5′ BamHI site and a 3′ EcoRI site, and were labeled at both ends. To generate probes that were labeled at only one end, the DNA fragments were digested with either BamHI or EcoRI as indicated. The end-labeled ≈280-bp fragments were then purified by sizing on 5% PAGE. Approximately 0.1 nM each labeled probe, 0.8 nM purified LasR, and 5 μM 3OC12-HSL were incubated in 120 μl of the gel-shift DNA binding buffer for 20 min at room temperature. The reaction mixtures were then treated with 6 μl of RQ1 DNase (0.24 units in 25 mM Tris·HCl, pH 7.4/30 mM MgCl2/30 mM CaCl2; Promega) for an additional 5 min. The reactions were stopped by the addition of 25 μl of footprint stop solution (3 M ammonium acetate/0.25 M EDTA/15 μg/ml sheared calf thymus DNA), and DNA was precipitated with 370 μl of ethanol. DNase I digestion products were separated on 5% denaturing polyacrylamide gels and were visualized by using PhosphorImager technology. For the detection of low-affinity binding of LasR to lasB operator 1 (OP1), we modified the footprinting assay as follows: The DNA binding reaction contained ≈5 pM labeled probe and 0, 10, 30, 90, 270, or 800 pM purified LasR. Because of the low concentration of radiolabeled probe, footprinting gels were exposed for several weeks before PhosphorImager analysis.
Analysis of Oligomeric State of LasR. For gel filtration, 0.2 ml of LasR (0.4 mg/ml) purified in the presence of 3OC12-HSL was chromatographed on a Superose 12 10/300 GL column (Amersham Pharmacia) at 4°C by using a Bio-Rad DuoFlow fast performance liquid chromatography apparatus. LasR was eluted with LRPB buffer containing 5 μM 3OC12-HSL at a flow-rate of 0.4 ml/min. The column was calibrated with protein standards from a low molecular weight gel filtration kit (Sigma). Dynamic light scattering of LasR purified in the presence of 3OC12-HSL was performed at 19°C by using a DynaPro-MS spectrophotometer (Protein Solutions, Charlottesville, VA) at the University of Iowa Protein Crystallography Facility.
Analysis of 3OC12-HSL Content of LasR. LasR was purified as described above except that 3OC12-HSL was omitted from the purification buffer. A 20-μl sample of each heparin column fraction was digested with 0.2 mg/ml proteinase K in 200 μl for 1 h at 37°C, extracted twice with ethyl acetate, evaporated to dryness, and resuspended in ethyl acetate. The amount of 3OC12-HSL in the ethyl acetate extract was determined by using a 3OC12-HSL bioassay (28). To estimate the LasR/3OC12-HSL molar ratio, LasR purified in the absence of 3OC12-HSL was dialyzed against phosphate buffer (50 mM potassium phosphate, pH 7.8/150 mM NaCl) without 3OC12-HSL. This LasR preparation and a synthetic 3OC12-HSL standard were subjected to amino acid analysis at the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University (New Haven, CT). In attempts to remove bound 3OC12-HSL from LasR, the purified protein was dialyzed for up to 6 days against buffers containing detergents (up to 3% Tween 20 or Triton X-100) or denaturant (up to 3 M urea).
Results
Purified LasR Binds Specifically and with High Affinity to DNA Containing a las–rhl Box. The lasR gene product was overproduced in E. coli in its native form. In the absence of 3OC12-HSL or at 37°C, LasR formed insoluble inclusion bodies. However, in the presence of 5 μM 3OC12-HSL and at low temperatures, ≈70% of the total LasR protein was soluble. Based on purification schemes used for the LasR homologues TraR from Agrobacterium tumefaciens (21) and LuxR from V. fischeri (25), LasR was purified to >95% homogeneity (see Materials and Methods).
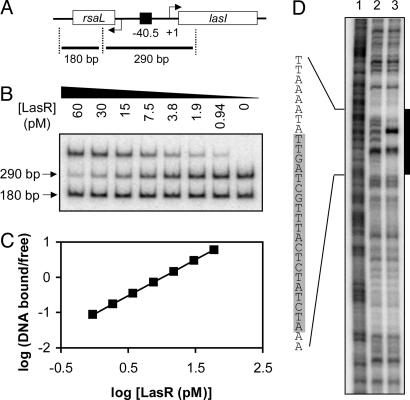
To begin to investigate the DNA binding properties of LasR, we chose the lasI–rsaL intergenic region as a specific probe for gel-shift analysis. This region contains a las-rhl box-like element that is centered nearly equidistant from the lasI and rsaL ORFs (Fig. 1A), and this element serves in the bidirectional LasR-3OC12HSL-dependent activation of both genes (12, 29). Moreover, the rsaL gene is among the genes most strongly activated by LasR-3OC12-HSL (8). Our gel-shift assay reaction also contained a nonspecific probe encompassing most of the rsaL coding region. In the presence of 5 μM 3OC12-HSL, LasR bound specifically to the las–rhl box probe with an affinity (Kd) of 11 pM (Fig. 1B) and a Hill coefficient of 1.0 (Fig. 1C), indicating that LasR bound this promoter probe with high affinity and without any cooperativity.
Fig. 1.
Binding of LasR to the rsaL–lasI intergenic region. (A) Map of the rsaL–lasI intergenic region. The black square indicates the las–rhl box sequence centered 40.5 bp upstream of the transcriptional start of the lasI gene. Specific and nonspecific fragments (290 and 180 bp in length, respectively) are shown. (B) Gel shift assay. All lanes contained ≈0.01 fmol of an equimolar mixture of the two probes and the indicated concentration of LuxR in the presence of 5 μM 3OC12-HSL. (C) Hill plot of the data obtained in A.(D) DNase I protection assay. The 290-bp specific fragment, end-labeled on the rsaL coding strand, was incubated without LasR (lane 2) or with LasR (lane 3) in the presence of 5 μM 3OC12-HSL. Lane 1 shows a Maxam–Gilbert A+G sequencing ladder of the probe. The black bar indicates the approximate region protected by LasR; the shaded area within the protected sequence indicates the predicted 20-bp las–rhl box.
DNase I footprinting showed that LasR protected a region of ≈30 bp that included the predicted 20-bp las–rhl box (Fig. 1D). The presence of two unprotected nucleotides within this area suggested that, like TraR and LuxR (21, 25), LasR may only bind to a single face of this DNA.
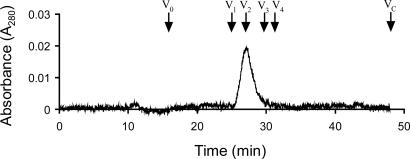
LasR Is a Dimer in Solution. In size exclusion chromatography, purified LasR eluted with a single peak corresponding to a molecular mass of 42 kDa (Fig. 2). This observation is below the expected molecular mass of 53 kDa for a dimer, although the purified LasR homologs TraR and CarR have been shown to exist as dimers in solution (20, 22, 23). Therefore, we used dynamic light scattering as an alternative method to determine the oligomeric state of LasR. We observed a single peak with a radius of 3.15 nm (data not shown) corresponding to a molecular mass of 49.3 kDa. This finding suggested that purified LasR-3OC12-HSL exists as a dimer in solution, consistent with a genetic study showing that LasR functions as a multimer in vivo (30).
Fig. 2.
Gel-filtration chromatography of LasR. Arrows indicate the elution times for a set of standard proteins (Amersham Pharmacia) including V1, BSA (Mr = 67,000); V2, ovalbumin (Mr = 45,000); V3, chymotrypsinogen A (Mr = 25,000); V4, ribonuclease A (Mr = 13,700). V0 marks the elution time of blue dextran (Mr =≈2,000,000), and VC marks the total bed volume of the column.
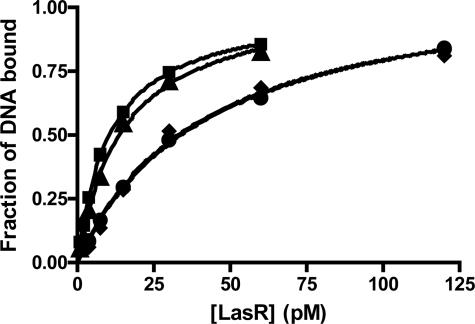
LasR Binds 3OC12-HSL Tightly. In the above experiments, LasR activity was studied in the presence of 3OC12-HSL. In an attempt to characterize the apo, 3OC12-HSL-free form of the protein, purified LasR was diluted in buffer lacking 3OC12-HSL. The DNA-binding affinity of this LasR preparation (Kd = 43 pM) was only ≈3-fold lower than that of LasR in the presence of 3OC12-HSL (Kd = 15 pM) (Fig. 3). Extensive dialysis of LasR in the absence of 3OC12-HSL did not result in a further reduction of gel-shift activity. Addition of signal restored DNA binding to levels observed before dialysis. As mentioned above, attempts to purify apo-LasR from overexpressing E. coli in the absence of exogenous 3OC12-HSL resulted in insoluble protein. Interestingly, when expressed at native levels in a P. aeruginosa lasI signal generation mutant, soluble LasR was detectable in cell lysates by Western immunoblotting, but these cell lysates showed no gel shift activity even after addition of exogenous signal. As expected, cell lysates from the same strain grown in the presence of 3OC12-HSL did exhibit gel-shift activity (data not shown).
Fig. 3.
Binding of various LasR preparations to the rsaL-lasI intergenic region in the presence and in the absence of 3OC12-HSL. Results from gel-shift assays (as described in Fig. 1) were quantified by plotting the fraction of shifted specific DNA versus LasR concentration. Circles, LasR diluted in the absence of 3OC12-HSL; diamonds, dialyzed LasR in the absence of 3OC12-HSL; triangles, dialyzed LasR in the presence of 5 μM 3OC12-HSL; squares, LasR before dialysis in the presence of 5 μM 3OC12-HSL.
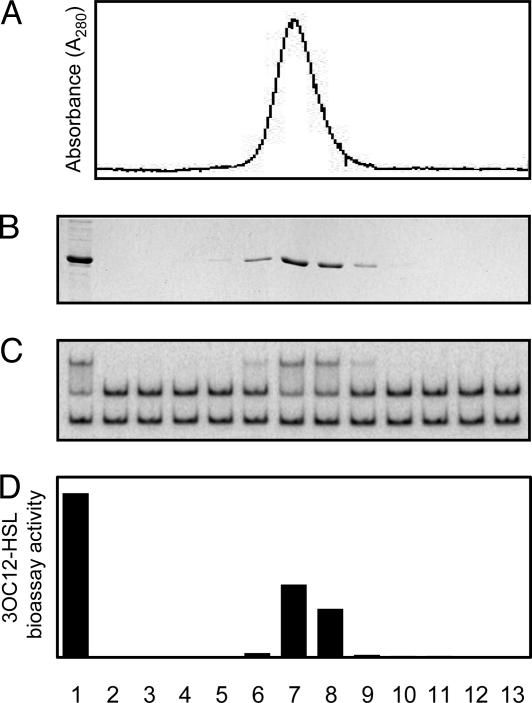
We considered the possibility that 3OC12-HSL remained tightly associated with a fraction of the diluted or dialyzed protein, similar to the tight binding of 3OC8-HSL to the LasR homolog TraR (21, 23). A bioassay revealed 3OC12-HSL activity in the dialyzed LasR preparation (data not shown). A bioassay also demonstrated coelution of 3OC12-HSL with LasR, which had originally been overexpressed in cultures containing 3OC12-HSL but was subsequently purified by using buffers that did not contain any exogenous signal (Fig. 4). To estimate the number of 3OC12-HSL molecules bound per LasR monomer, we subjected a dialyzed LasR preparation to amino acid analysis. Because this procedure causes the conversion of synthetic acyl-HSL to homoserine (21), we were able to determine the LasR-3OC12-HSL molar ratio based on the quantitation of each amino acid and homoserine. We calculated a ratio of 1.03, implying that each monomer contains one molecule of 3OC12-HSL. This result further suggested that 3OC12-HSL remained tightly bound to LasR. Why, then, did LasR exhibit decreased gel-retardation activity in the absence of exogenous 3OC12-HSL? It appeared that during the gel-shift reaction LasR slowly released some 3OC12-HSL. The gel-shift assay was performed at room temperature, whereas the dialysis attempts were at 4°C. However, when dialyzed at room temperature (as short as 1 h, with or without 3OC12-HSL), the protein completely lost activity. This finding suggests that the presence of a DNA template (as was the case in the DNA-binding reaction) is necessary for LasR-3OC12-HSL complex stability at elevated temperatures.
Fig. 4.
Copurification of LasR and 3OC12-HSL. (A) Absorption profile of a heparin column eluate containing LasR. (B) SDS/PAGE analysis of each eluted fraction. Nonfractionated proteins are shown in lane 1. (C) Gel-shift activity of each fraction shown in B.(D) 3OC12-HSL content of each fraction shown in B. 3OC12-HSL content in proteolyzed and ethyl acetate-extracted fractions was measured with a bioassay (28).
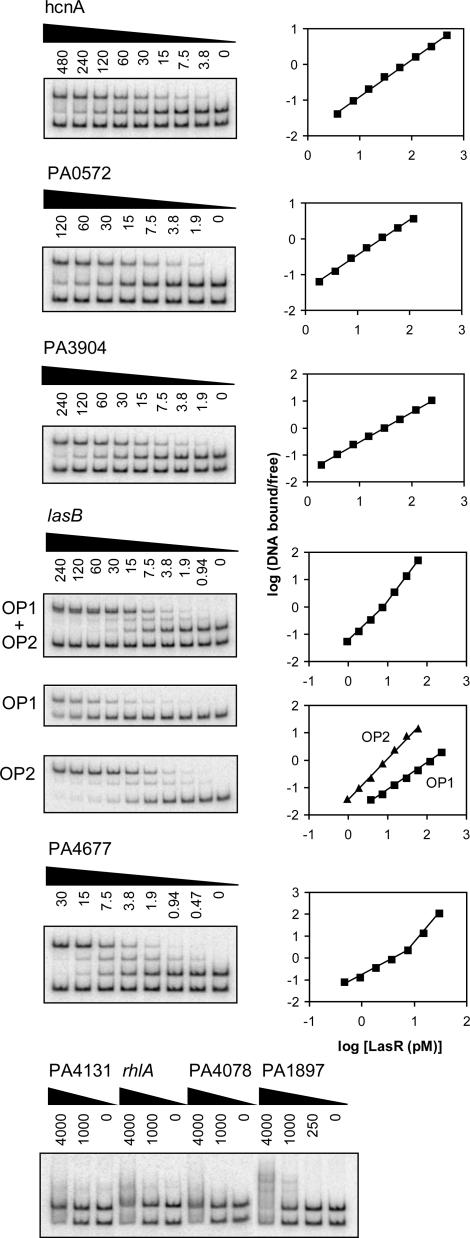
In Vitro LasR Binding to Different Quorum-Controlled Promoters Reveals Distinct Binding Modes and Sequence Determinants. To gain insights into the direct and indirect regulation of quorum-controlled genes, and to begin to understand the nature of las-specific regulatory DNA, we studied several different quorum-controlled promoters. According to our previous transcriptome analysis (8), the chosen promoters exhibited a broad range of signal specificities and induction strengths. Some were predicted to contain a las–rhl box-like sequence, whereas others were not (Table 1). LasR bound specifically to 6 of the 10 promoters tested (Fig. 5). Binding affinities varied, but all were in the picomolar range. Consistent with previous genetic data (13–15, 24), LasR bound to the promoters of las-responsive genes lasB and hcnA, but did not bind to the promoter of the rhl-specific gene rhlA (Fig. 5). Overall, there was no correlation between specificity, the presence of a las–rhl box-like sequence, and observed binding. For instance, LasR did not bind to the las-responsive promoter PA1897 (qsc102), which was predicted to contain a las–rhl box (8, 12, 31), but LasR did bind to the las-responsive promoter PA4677, which was not predicted to contain a las–rhl box (Table 1).
Table 1. Characteristics of quorum-controlled promoters analyzed in this study.
| Gene/promoter | Gene product | Specificity | Predicted las-rhl box | Timing of induction* | Fold induction† | Binding affinity, pM | Hill coefficient |
|---|---|---|---|---|---|---|---|
| hcnABC | Hydrogen cyanide synthase | las/rhl | Yes | Mid-log | 140 (190) | 72 | 1.0 |
| lasB | LasB elastase | las/rhl | Yes | Log-stat | 110 (180) | 1.4, 1.9 | |
| OP1 | 160 | 1.0 | |||||
| OP2 | 8.9 | 1.5 | |||||
| rhlAB | Rhamnosyl transferase | rhl | Yes | Log-stat | 10 (120) | No binding | - |
| rsaL | lasI repressor | las | Yes | Mid-log | 350 (340) | 11 | 1.0 |
| PA0572 | Hypothetical protein | las | No | Log-stat | 19 (22) | 25 | 1.0 |
| PA1897 | Hypothetical protein | las | Yes | Log-stat | 130 (130) | No binding | - |
| PA3904 | Hypothetical protein | las | No | Early log | 49 (42) | 30 | 1.1 |
| PA4078 | Nonribosomal peptide synthetase | las/rhl | Yes | Stat | 3.2 (4.6) | No binding | - |
| PA4131 | Fe-S protein | las/rhl | No | Mid-log | 24 (30) | No binding | - |
| PA4677 | Hypothetical protein | las | No | Log-stat | 16 (13) | 5.1 | 1.3, 2.8 |
Timing of induction during batch culture growth of the P. aeruginosa lasI, rhlI signal generation mutant in the presence of 3OC12-HSL and C4-HSL versus no signal, and during batch culture growth of the parent strain versus the lasR, rhlR signal receptor mutant (taken from ref. 8). Early logarithmic phase (Early log) < mid-logarithmic phase (Mid-log) < transition from logarithmic to stationary phase (Log-stat) < stationary phase (Stat). PA0572 and PA4677 showed some low-level induction early in growth.
Changes in gene expression in the signal generation mutant in the presence of 3OC12-HSL (values in the presence of 3OC12-HSL and C4-HSL are shown in parentheses) versus the absence of signal (taken from ref. 8).
Fig. 5.
Gel-shift analysis of various quorum-controlled promoters. Gel-shift assays are shown in Left, and the corresponding Hill plots are shown in Right. All lanes contained ≈0.01 fmol of an equimolar mixture of the two probes and the indicated concentration of LasR (in pM) in the presence of 5 μM 3OC12-HSL. In lasB gel shift reactions with OP1 only and OP2 only, a nonspecific probe was omitted.
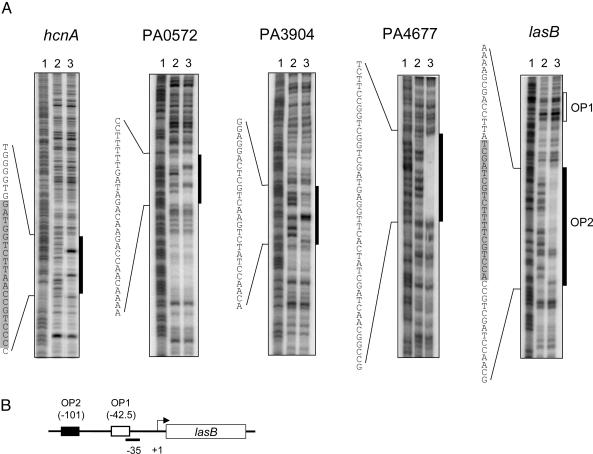
Furthermore, our gel-shift analysis revealed different modes of binding (Fig. 5): As shown above for the rsaL promoter, LasR bound the hcnA, PA0572, and PA3904 promoters with no cooperativity (Hill coefficients ≈ 1; Table 1), and a single shifted band was observed. In contrast, LasR bound the lasB and PA4677 promoters with positive cooperativity (Hill coefficients > 1), and at least two shifted bands were observed. DNase I protection analysis of these promoters showed that the footprinting patterns correlated with the modes of binding observed in our gel-shift experiments (Fig. 6). LasR protected a rather small region on the hcnA, rsaL, PA0572, and PA3904 promoters that showed noncooperative binding, but LasR protected a significantly larger region on the lasB and PA4677 promoters that showed cooperative binding. Some of the sequences within the protected regions bore little resemblance to the presumed las–rhl box consensus. Remarkably, however, in a clustering analysis, all of the sequences of las-specific promoters formed two distinct groups according to their modes of binding (Fig. 7).
Fig. 6.
DNase I protection analysis of various quorum-controlled promoters. (A) End-labeled fragments ≈280 bp in length were incubated without LasR (lane 2) or with LasR (lane 3) in the presence of 5 μM 3OC12-HSL. Lane 1 shows a Maxam–Gilbert A+G sequencing ladder of the respective probe. The hcnA and PA3904 fragments were end-labeled on the coding strand, and the lasB, PA0572, and PA4677 fragments were end-labeled on the noncoding strand. The black boxes indicate the regions protected by LasR, and the open box shows the position of the predicted OP1 sequence. The shaded areas within the protected sequences indicate predicted 20-bp las–rhl boxes. (B) Promoter region of the lasB gene showing the relative locations of OP1 and OP2.
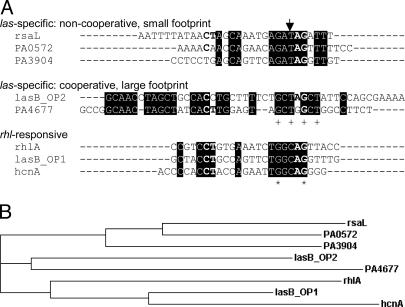
Fig. 7.
Sequence analysis. (A) Alignment of binding sequences in quorum-controlled promoters performed with clustalw (www.ebi.ac.uk/clustalw). Depicted are the sequences protected from DNase I digestion as determined from Figs. 1D and 6. The sequence of the rhlA promoter that did not bind LasR but presumably binds RhlR was extrapolated based on the location of a predicted las–rhl box sequence and the footprinting pattern that LasR exhibits when it binds to promoters with no cooperativity. Elements of the CT-[N]12-AG motif previously thought to be absolutely conserved (12, 31) are shown in bold. The arrow designates a base position potentially conferring las vs. rhl sequence specificity. + indicates base positions identical in all five las-specific sequences. Asterisk indicates base positions identical in all eight sequences shown. (B) Phylogram (hierarchical clustering) of the sequences shown in A.
Our gel-shift and footprinting analysis of the lasB promoter also showed that LasR primarily binds to one of two sites that were thought to be involved in LasR-3OC12-HSL-mediated activation of lasB (13, 14). LasR bound to operator 2 (OP2) cooperatively with high affinity (Kd = 8.9 pM), and to OP1 noncooperatively with much lower affinity (Kd = 160 pM) (Fig. 5). The lasB footprint in Fig. 6 did not show appreciable protection of OP1 because the assay was designed to qualitatively assess the extent of protection resulting from high-affinity binding of LasR. When we modified the assay by using much lower DNA concentrations (reaching the sensitivity limits of the assay), a footprint of ≈26 bp that contained the predicted OP1 sequence could be detected (data not shown). Interestingly, our clustering analysis predicted that the low-affinity OP1 is primarily a RhlR, rather than LasR, binding site (Fig. 7), which is consistent with the observation that lasB also responds to RhlR-C4-HSL (31, 32). The OP1 sequence clusters with the las–rhl box-like sequence of the rhl-specific gene rhlA, which we assume to be a RhlR-binding site and did not show any binding to LasR.
Discussion
Purified, native LasR, like A. tumefaciens TraR (21), appears to be a dimer in solution that contains two tightly bound molecules of acyl-HSL. Even prolonged dialysis in the presence of denaturants or detergents failed to remove any 3OC12-HSL from LasR. Similar conditions resulted in the slow release of 3OC8-HSL from TraR (21), whereas mere dilution was sufficient to readily release 3OC6-HSL from V. fischeri LuxR (25). These results suggest a correlation between side-chain length of the acyl-HSL and binding affinity to its cognate LuxR-type regulator: the longer the acyl side chain, the tighter the binding. This simplistic idea is consistent with information from the crystal structure of TraR bound to 3OC8-HSL and DNA (33, 34). The acyl-HSL is fully embedded within the protein, and the fatty acyl side chain makes several van der Waals contacts with mostly nonconserved hydrophobic residues lining the acyl-HSL binding pocket. Consequently, acyl-HSLs with longer side chains might interact more extensively with the hydrophobic core of the protein than those with shorter side chains, resulting in a tighter association.
Because LasR was nonfunctional when expressed in the absence of its cognate signal, it is conceivable that 3OC12-HSL, in analogy to the role of 3OC8-HSL in TraR function (21, 22), is essential for the folding of nascent LasR into its active conformation. LasR has been a major focus of attention for the development of therapeutic quorum-sensing inhibitors (35–41), and the nature of the interaction of 3OC12-HSL with LasR has implications for their potential mechanism of action. Effective quorum-sensing inhibition would require active cellular protein synthesis for the competitive incorporation of a compound that renders LasR inactive, rather than displacement of the native acyl-HSL from the mature protein.
We have shown that purified LasR binds specifically and with high affinity to las-responsive quorum-controlled promoters. LasR bound some promoters cooperatively, and bound to others noncooperatively. The presence of multiple shifted bands and extended regions protected from DNase I digestion suggested that cooperative binding may involve the formation of higher-order multimers of LasR. The observed binding affinities to individual promoter sequences correlated with neither the magnitude nor the timing of a response (Table 1). In a previous study, we hypothesized that the differential binding of LasR to quorum-controlled promoters may govern the timing of quorum-controlled gene expression ranging from early logarithmic to stationary phase (8). Our data suggest that the activation of quorum-controlled promoters is more complex. Many other factors, such as RNA polymerase binding, the interaction of LasR with RNA polymerase, and the binding of other transcription factors, likely contribute to promoter strength and timing of induction.
There was also no good correlation between observed binding, specificity, and the presence of a proposed las–rhl box. Binding occurred at some but not all of the predicted las–rhl box-like sequences and at unexpected binding sites with little resemblance to the established consensus sequence. These discrepancies are in part due to misinterpretations of genetic experiments and limited knowledge for building a las–rhl box consensus sequence. For example, the las-specific gene PA1897 (qsc102) appeared to be directly activated in E. coli when LasR was overexpressed in the presence of 3OC12-HSL (12). Here, we have shown that LasR does not bind to the PA1897 promoter in vitro (Fig. 5). Reexamination of the E. coli experiment also showed that there is no appreciable activation of this promoter when LasR is expressed at lower levels. Instead, the third P. aeruginosa LuxR homolog QscR (17), which is itself regulated by LasR, directly activates PA1897 in this system (J.-H. Lee and E.P.G., unpublished results). Thus, the activation observed initially was likely cross-talk caused by high levels of LasR, which consequently led to a misinterpretation of the specificity determinants of quorum-controlled promoters (12). Furthermore, it was assumed that LasR- and RhlR-binding sites in quorum-controlled promoters possess a high degree of dyad symmetry because other LuxR family members, LuxR, TraR, and CarR, all bind to DNA sequences with nearly perfect dyad symmetry (20, 21, 25). Only one consensus sequence had been proposed for both LasR and RhlR because there appeared to be no discernable difference in the recognition sequences of las- and rhl-specific promoters and because promoters with relaxed specificity appeared to possess one sequence that served as the LasR- as well as RhlR-binding site (7, 8, 12, 14, 15, 31).
Our in vitro data show that the recognition sequences of las-specific promoters exhibit little overall sequence conservation and a marked lack of dyad symmetry. This finding suggests that LasR might contact its recognition sequences also at positions of degenerate homology, analogous to the transcriptional regulator OxyR (42). The sequence heterogeneity and differences in binding may be a reflection of the large number and diversity of genes regulated by LasR (8). Sequence identity is higher within each binding class, resulting in the clustering of sequences according to their mode of binding (Fig. 7). The sequences predicted to be RhlR binding sites retain a higher degree of dyad symmetry, and they form a cluster separate from all of the sequences of las-specific genes (including lasB OP2). This cluster includes two binding sites within promoters that respond to both LasR and RhlR in vivo (14, 15, 31). These sites, lasB OP1 and hcnABC, bound LasR with comparatively low affinity in vitro, suggesting that there is indeed some overlap in sequence specificity for relaxed promoters. Sequence alignment also identified one conserved candidate position as a LasR versus RhlR specificity determinant. At this position, all las-specific sequences have a T, whereas all rhl-responsive sequences have a C (Fig. 7).
Taken together, our sequence analysis revealed discernable differences in sequence composition for at least three classes of quorum-controlled promoters: las-specific promoters that exhibit noncooperative binding, las-specific promoters that exhibit cooperative binding, and rhl-responsive promoters. The in vitro approach taken in this study has led to a reassessment of the conclusions thus far exclusively drawn from in vivo data. A genome-scale identification of additional direct binding sites of LasR and RhlR, along with the in vitro DNA-binding analysis used here, will allow us to completely define the sequence requirements and specificity determinants of quorum-controlled promoters.
Acknowledgments
We thank S. Ramaswamy for his assistance with dynamic light scattering and Myron Crawford for amino acid analysis. This study was supported by National Institute of General Medicine Grant GM59026 and the W. M. Keck Foundation.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 20, 2004.
Abbreviations: 3OC12-HSL, 3-oxo-dodecanoyl-homoserine lactone; LRPB, LasR purification buffer; OP1, operator 1; OP2, operator 2.
See accompanying Biography on page 15830.
References
- 1.Taga, M. E. & Bassler, B. L. (2003) Proc. Natl. Acad. Sci. USA 100, Suppl. 2, 14549–14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler, B. L. (2002) Cell 109, 421–424. [DOI] [PubMed] [Google Scholar]
- 3.Fuqua, C., Parsek, M. R. & Greenberg, E. P. (2001) Annu. Rev. Genet. 35, 439–468. [DOI] [PubMed] [Google Scholar]
- 4.Whitehead, N. A., Barnard, A. M., Slater, H., Simpson, N. J. & Salmond, G. P. (2001) FEMS Microbiol. Rev. 25, 365–404. [DOI] [PubMed] [Google Scholar]
- 5.Rumbaugh, K. P., Griswold, J. A. & Hamood, A. N. (2000) Microbes Infect. 2, 1721–1731. [DOI] [PubMed] [Google Scholar]
- 6.Smith, R. S. & Iglewski, B. H. (2003) Curr. Opin. Microbiol. 6, 56–60. [DOI] [PubMed] [Google Scholar]
- 7.Wagner, V. E., Bushnell, D., Passador, L., Brooks, A. I. & Iglewski, B. H. (2003) J. Bacteriol. 185, 2080–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuster, M., Lohstroh, C. P., Ogi, T. & Greenberg, E. P. (2003) J. Bacteriol. 185, 2066–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pesci, E. C., Pearson, J. P., Seed, P. C. & Iglewski, B. H. (1997) J. Bacteriol. 179, 3127–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latifi, A., Foglino, M., Tanaka, K., Williams, P. & Lazdunski, A. (1996) Mol. Microbiol. 21, 1137–1146. [DOI] [PubMed] [Google Scholar]
- 11.Medina, G., Juarez, K., Diaz, R. & Soberon-Chavez, G. (2003) Microbiology 149, 3073–3081. [DOI] [PubMed] [Google Scholar]
- 12.Whiteley, M. & Greenberg, E. P. (2001) J. Bacteriol. 183, 5529–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rust, L., Pesci, E. C. & Iglewski, B. H. (1996) J. Bacteriol. 178, 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson, R. M., Zimprich, C. A. & Rust, L. (1999) J. Bacteriol. 181, 6264–6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pessi, G. & Haas, D. (2000) J. Bacteriol. 182, 6940–6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devine, J. H., Shadel, G. S. & Baldwin, T. O. (1989) Proc. Natl. Acad. Sci. USA 86, 5688–5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chugani, S. A., Whiteley, M., Lee, K. M., Argenio, D. D., Manoil, C. & Greenberg, E. P. (2001) Proc. Natl. Acad. Sci. USA 98, 2752–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.You, Z., Fukushima, J., Ishiwata, T., Chang, B., Kurata, M., Kawamoto, S., Williams, P. & Okuda, K. (1996) FEMS Microbiol. Lett. 142, 301–307. [DOI] [PubMed] [Google Scholar]
- 19.Stevens, A. M., Dolan, K. M. & Greenberg, E. P. (1994) Proc. Natl. Acad. Sci. USA 91, 12619–12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welch, M., Todd, D. E., Whitehead, N. A., McGowan, S. J., Bycroft, B. W. & Salmond, G. P. (2000) EMBO J. 19, 631–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu, J. & Winans, S. C. (1999) Proc. Natl. Acad. Sci. USA 96, 4832–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu, J. & Winans, S. C. (2001) Proc. Natl. Acad. Sci. USA 98, 1507–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin, Y., Luo, Z. Q., Smyth, A. J., Gao, P., Beck von Bodman, S. & Farrand, S. K. (2000) EMBO J. 19, 5212–5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearson, J. P., Pesci, E. C. & Iglewski, B. H. (1997) J. Bacteriol. 179, 5756–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urbanowski, M. L., Lostroh, C. P. & Greenberg, E. P. (2004) J. Bacteriol. 186, 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitz, A. & Galas, D. J. (1979) Nucleic Acids Res. 6, 111–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galas, D. J. & Schmitz, A. (1978) Nucleic Acids Res. 5, 3157–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson, J. P., Gray, K. M., Passador, L., Tucker, K. D., Eberhard, A., Iglewski, B. H. & Greenberg, E. P. (1994) Proc. Natl. Acad. Sci. USA 91, 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seed, P. C., Passador, L. & Iglewski, B. H. (1995) J. Bacteriol. 177, 654–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiratisin, P., Tucker, K. D. & Passador, L. (2002) J. Bacteriol. 184, 4912–4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whiteley, M., Lee, K. M. & Greenberg, E. P. (1999) Proc. Natl. Acad. Sci. USA 96, 13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brint, J. M. & Ohman, D. E. (1995) J. Bacteriol. 177, 7155–7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vannini, A., Volpari, C., Gargioli, C., Muraglia, E., Cortese, R., De Francesco, R., Neddermann, P. & Marco, S. D. (2002) EMBO J. 21, 4393–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, R. G., Pappas, T., Brace, J. L., Miller, P. C., Oulmassov, T., Molyneaux, J. M., Anderson, J. C., Bashkin, J. K., Winans, S. C. & Joachimiak, A. (2002) Nature 417, 971–974. [DOI] [PubMed] [Google Scholar]
- 35.Smith, K. M., Bu, Y. & Suga, H. (2003) Chem. Biol. 10, 81–89. [DOI] [PubMed] [Google Scholar]
- 36.Smith, K. M., Bu, Y. & Suga, H. (2003) Chem. Biol. 10, 563–571. [DOI] [PubMed] [Google Scholar]
- 37.Wu, H., Song, Z., Hentzer, M., Andersen, J. B., Molin, S., Givskov, M. & Hoiby, N. (2004) J. Antimicrob. Chemother. 53, 1054–1061. [DOI] [PubMed] [Google Scholar]
- 38.Passador, L., Tucker, K. D., Guertin, K. R., Journet, M. P., Kende, A. S. & Iglewski, B. H. (1996) J. Bacteriol. 178, 5995–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hentzer, M., Riedel, K., Rasmussen, T. B., Heydorn, A., Andersen, J. B., Parsek, M. R., Rice, S. A., Eberl, L., Molin, S., Hoiby, N., et al. (2002) Microbiology 148, 87–102. [DOI] [PubMed] [Google Scholar]
- 40.Hentzer, M., Wu, H., Andersen, J. B., Riedel, K., Rasmussen, T. B., Bagge, N., Kumar, N., Schembri, M. A., Song, Z., Kristoffersen, P., et al. (2003) EMBO J. 22, 3803–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kline, T., Bowman, J., Iglewski, B. H., de Kievit, T., Kakai, Y. & Passador, L. (1999) Bioorg. Med. Chem. Lett. 9, 3447–3452. [DOI] [PubMed] [Google Scholar]
- 42.Tartaglia, L. A., Gimeno, C. J., Storz, G. & Ames, B. N. (1992) J. Biol. Chem. 267, 2038–2045. [PubMed] [Google Scholar]