Abstract
Chromatin is the substrate for many processes in the cell nucleus, including transcription, replication, and various DNA repair systems, all of which require the formation of multiprotein machineries on the chromatin fiber. We have analyzed the kinetics of in vivo assembly of the protein complex that is responsible for nucleotide excision repair (NER) in mammalian cells. Assembly is initiated by UV irradiation of a small area of the cell nucleus, after which the accumulation of GFP-tagged NER proteins in the DNA-damaged area is measured, reflecting the establishment of the dual-incision complex. The dynamic behavior of two NER proteins, ERCC1-XPF and TFIIH, was studied in detail. Results show that the repair complex is assembled with a rate of ≈30 complexes per second and is not diffusion limited. Furthermore, we provide in vivo evidence that not only binding of TFIIH, but also its helicase activity, is required for the recruitment of ERCC1-XPF. These studies give quantitative insight into the de novo assembly of a chromatin-associated protein complex in living cells.
Keywords: DNA repair, ERCC1-XPF, in vivo kinetics, TFIIH
Nucleotide excision repair (NER) is continuously safeguarding genomic integrity by means of its ability to repair several types of helix-distorting DNA damage. For the removal of, for instance, UV-induced photoproducts, placental mammals fully rely on NER (1). This multiprotein system, involving >30 gene products, recognizes damaged DNA and excises the injury as an oligonucleotide from the affected strand, after which DNA polymerase action resynthesizes the gap and the remaining nick is ligated (2–4). We have detailed knowledge of the in vitro NER mechanism, mainly from biochemical studies. However, how it operates in the context of the living cell nucleus has remained largely unsolved.
In general, studies of the dynamics of nuclear processes have been performed by using photobleaching procedures, providing information about protein mobility and exchange rates of components of protein complexes (5, 6). In vivo studies of chromatin-associated processes are hampered by a lack of methods that allow analysis of de novo assembly kinetics in the nuclei of living cells, and, as a result, the dynamic behavior of the involved complexes could be investigated only under steady-state conditions. Taking advantage of the UV-inducible nature of NER, we have analyzed in detail the dynamics of the formation of the double-incision complex by visualizing protein accumulation at sites of DNA damage immediately after UV irradiation. Results obtained for the NER complex may serve as a paradigm for the assembly of other chromatin-associated multiprotein systems, such as transcription and replication complexes.
Methods
Cell Culture. We used 43-3B Chinese hamster ovary (CHO) cells stably expressing ERCC1-GFP as described in ref. 7. We stably transfected 27.1 cells with enhanced GFP (EGFP) (Clontech) cloned in-frame to the C terminus of the XPB subunit of TFIIH, as described in ref. 8. Cells were grown at 37°C under 5% CO2 in a 1:1 mixture of Ham's F10 medium and DMEM supplemented with 10% FCS, 1% glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (all from Invitrogen). Cells were cultured in glass-bottomed Petri dishes (MatTek, Ashland, MA) and were used at 60–80% confluency. Primary human fibroblasts VH25 (normal NER proficient) and XP131MA (XPB helicase mutant) were grown on Alcian blue-coated (Sigma-Aldrich) coverslips in Ham's F10 medium containing 15% FCS and the above-mentioned supplements under a 2.5% CO2 atmosphere.
Local UV Irradiation. For immunofluorescent labeling, the primary fibroblasts were locally UV-irradiated through the 5-μm-diameter pores of a polycarbonate membrane filter (Millipore) as described in ref. 9. For imaging of living cells, local UV irradiation was performed as follows. The filter (5-μm pores) was coated with Alcian blue to allow it to stay in close vicinity of cells when immersed. The Petri dish was filled with serum-free culture medium (DMEM) without phenol red, and a small piece of filter (≈5 mm in diameter) was positioned onto the cells. A glass ring (6 mm in diameter with a 2-mm opening and a height of 2.7 mm) was carefully placed on top of the filter to secure the filter on the cells. Subsequently, the Petri dish was sealed with a lid that contained a quartz window and held the ring and the filter in place. The dish was transferred to the prewarmed microscope stage (see below) and was allowed to thermally stabilize. Irradiation was carried out by using an UV source [a flat array of four Philips (Eindhoven, The Netherlands) TUV 9W PL-S lamps] above the microscope stage. UV irradiation was corrected for absorption by the medium, which was measured spectrophotometrically, resulting in a final UV dose of 120 J/m2 (254 nm) onto the filter (UV irradiation was performed for 12 s at 10 J/m2·s).
Imaging and Analysis. Cells were kept on a LSM510 confocal microscope stage (Zeiss) at the appropriate temperature, by using a custom-made heated stage connected to a temperature controller (Medical Systems, Greenvale, NY). The objective (Zeiss Plan-Neofluar ×100, 1.3 numerical aperture) was kept at the right temperature (either 37 or 27°C; see text) with a Bioptechs (Butler, PA) objective heater. One equatorial image was taken to determine position and relative fluorescence intensity of the cell nuclei (25 mW Ar laser, 0.5% laser power at 488 nm, pinhole open, fast-scanning mode). Going up in the z-direction, a backscattered image of the filter mask was obtained (HeNe laser at 633 nm) to determine the position of the pores in the filter mask. Images of the filter and the cells were overlaid to locate nuclei that were situated under a filter pore. The distance between nucleus and filter was measured at 633 nm excitation and had to be <7 μm to obtain a well defined damaged area. A preirradiation image was acquired, after which the sample was exposed to UV light for 12 s. Immediately after irradiation, images were collected (0.5% laser power at 488 nm, pinhole open, 512 × 512-pixel images, two times averaged) over 15- to 20-s intervals for several minutes, allowing the accumulation of GFP-tagged NER proteins in the UV-damaged area to reach a plateau. The accumulation of fluorescence from GFP was quantified with Zeiss lsm510 software (version 2.5) and photoshop 6 (Adobe Systems, San Jose, CA). For every time point, the average fluorescence intensities in the UV-damaged area and in an equal-size undamaged area in the same nucleus were measured and corrected for background fluorescence outside the cell and for photobleaching. The GFP signal in the undamaged nuclear area represents the pool of the NER proteins that is not involved in DNA repair, e.g., diffusing freely, and that is assumed to be present also in the damaged region (7, 8). This value was subtracted from the fluorescence in the damaged area, resulting in the fluorescence intensity that represents the GFP-tagged protein bound to damaged DNA. At least eight cells were imaged and quantified for every condition. The amount of bound protein was normalized as the fraction of the total amount of that protein in the nucleus. The start of UV irradiation was defined as t = 0 s.
Immunofluorescent Labeling. After UV irradiation, primary fibroblasts were cultured for 3 min, after which immunolabeling was carried out as described in ref. 10. Primary antibodies for the detection of TFIIH were mouse IgG monoclonal anti-XPB and mouse IgG monoclonal anti-p62 (both gifts from J. M. Egly, Institut de Génétique et de Biologie Moléculaire et Cellulaire, Illkirch, France). ERCC1 was detected by using rabbit IgG polyclonal anti-ERCC1. Secondary antibodies were goat anti-mouse coupled to Cy2 and goat anti-rabbit coupled to Cy3 (Jackson ImmunoResearch). Images were obtained by using a Zeiss Axioplan 2 fluorescence microscope, equipped with a cooled Hamamatsu (Bridgewater, NJ) C5935 CCD camera.
Results and Discussion
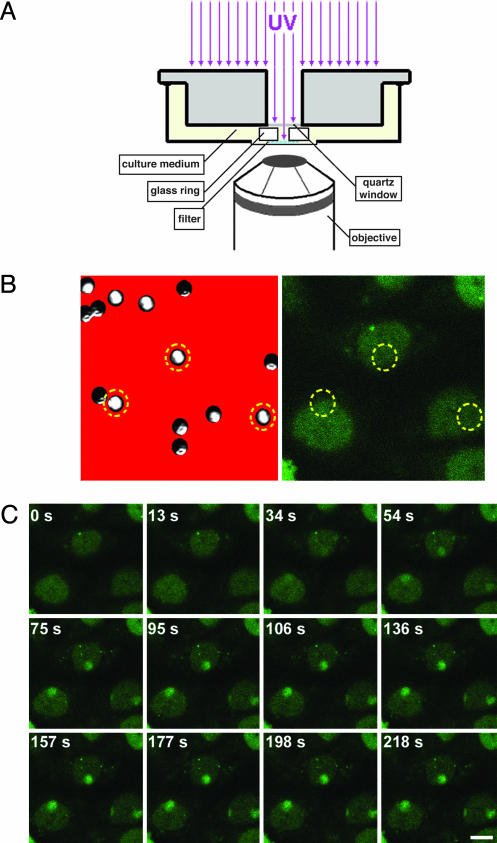
We used repair-deficient CHO cells that had been functionally rescued by stable expression of the wild-type NER protein tagged with GFP (7, 8). To directly measure the uptake of proteins into the NER complex, we locally induced DNA damage in the cell nucleus by UV irradiation through a filter mask with 5-μm pores (9). Cells were cultured in a microscope culture chamber that allowed quantitative measurement of the time course of protein accumulation in the damaged nuclear region (Fig. 1A; see also Methods for a detailed description). The heterodimeric endonuclease ERCC1-XPF generates the 5′ incision in the damaged strand (11). Before UV irradiation, ERCC1-GFP was distributed evenly throughout the nucleoplasm (Fig. 1B). Immediately after local UV irradiation, ERCC1-GFP started to accumulate in the damaged nuclear area (Fig. 1C). The time course of the increase in fluorescence in the damaged area reached a plateau after 2–3 min (Fig. 2 Left), representing a steady-state-like situation in which repair is performed at a constant maximum rate. Nuclei that were UV-irradiated a second time in the damaged nuclear region did not exhibit an increase in their immobilized ERCC1-GFP fraction (data not shown). This observation indicates that the initial DNA damage is sufficient to engage the maximum number of GFP-tagged NER proteins and that, therefore, the plateau corresponds to a saturated repair system [in agreement with previous estimates (9)]. ERCC1-GFP binding is remarkably slow (t1/2 ≈ 65 s) compared with its apparent diffusion rate inside the nucleus (15 ± 5 μm2/s) (7), based on fluorescence recovery after photobleaching experiments, indicating that the rate of recruitment into NER complexes is not diffusion limited.
Fig. 1.
Visualization of ERCC1-GFP recruitment to sites of localized DNA damage. (A) Setup for local UV irradiation of cells growing on a temperature-controlled microscope stage. A monolayer of cells in culture medium is covered with a micropore filter mask that is kept in place by a glass ring. UV irradiation onto the filter is through a quartz window and the opening in the ring. (B) Position of filter pores relative to cell nuclei. (Left) A reflection image of the filter (red) shows the distribution of the micropores. Because not all filter pores are perpendicular to the filter surface, a transmission image identifies pores that properly transmit white light (e.g., pores within the circles). (Right) Image of cells below the filter that express ERCC1-GFP, showing a uniform nuclear distribution before UV irradiation. Circles denote the position of the filter pores. (C) Time series of ERCC1-GFP dynamics at 37°C after local UV irradiation. After a preirradiation image (t = 0 s), cells were UV-irradiated for 12 s. Subsequently, images were taken at the indicated time points after the start of UV irradiation. Cell nuclei exhibited an accumulation of GFP signal in the irradiated nuclear areas (i.e., the areas below the filter pores denoted in B). (Scale bar, 10 μm.)
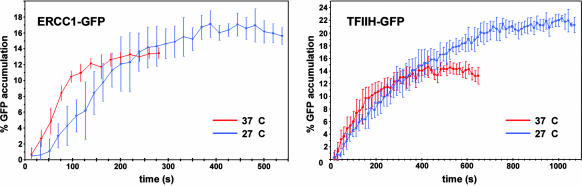
Fig. 2.
Assembly kinetics of ERCC1-GFP and TFIIH-GFP. Time curves show the accumulation of ERCC1-GFP (Left) and TFIIH-GFP (Right) at UV-damaged nuclear areas. Starting at t = 0, cells were irradiated for 12 s, receiving a total dose of 120 J/m2 in the irradiated nuclear area. The subsequent local accumulation of ERCC1-GFP and TFIIH-GFP was measured and is plotted as a percentage of the total GFP fluorescence of the cell nucleus (y axis) vs. time after start of UV irradiation. Experiments were performed at 37°C (red curves) and 27°C (blue curves) and repeated at least eight times. (Error bars represent SD between different experiments.)
ERCC1-XPF is thought to be one of the last proteins that is incorporated into the NER double-incision complex. Therefore, its accumulation kinetics is likely to reflect the formation of the complete functional dual-incision complex. How does this hypothesis compare with the behavior of NER components that bind before ERCC1-XPF? The formation of a stretch of unwound DNA is required for ERCC1-XPF to produce the 5′ incision in the injured DNA strand (12, 13). Unwinding is performed through the ATP-dependent helicase activity of TFIIH (14–16). It is, therefore, thought that TFIIH binds to lesions before ERCC1-XPF in vivo. Thus, it is expected that the kinetics of TFIIH binding are the same or faster than those of ERCC1-XPF. To address this hypothesis, we measured the binding kinetics of TFIIH-GFP in CHO cells expressing its XPB subunit coupled to GFP. XPB is the 3′ → 5′ helicase subunit of TFIIH (14). Like ERCC1-GFP, XPB-GFP accumulation started immediately after UV irradiation (Fig. 2 Right). TFIIH recruitment reached a plateau after ≈6 min (t1/2 ≈ 113 s). Like ERCC1-GFP, incorporation of TFIIH, which has an effective diffusion coefficient of 5.9 ± 1 μm2/s, is not a diffusion-limited process (8).
There are ≈30,000 molecules of ERCC1-XPF per cell nucleus (7). Assuming that the TFIIH concentration is the same as that of ERCC1, which is probably an underestimation (17), their assembly rates at UV-damaged DNA can be estimated from the initial slope of the time courses. The tangents show an immobilization of ≈6% and ≈7% of the total fluorescence per minute for ERCC1-GFP and TFIIH-GFP, respectively. Hence, assembly at UV damage occurs at a rate of ≈30 and ≈35 molecules per s for ERCC1-XPF and TFIIH, respectively.
In the absence of DNA damage, NER proteins have been proposed to occur in the nucleoplasm as individual proteins (7, 8, 18), protein complexes (19–21), or a fully preassembled repairosome (22). Measurements on intact cell nuclei provide two types of evidence for a sequential assembly mechanism. First, even in the presence of DNA damage, the mobile populations of several NER proteins appear to diffuse rapidly, indicating that a major fraction is not part of a large complex (7, 8, 18). Second, partially assembled repair complexes have been detected at DNA-damaged sites in the nuclei of NER-deficient mutant cells (10). However, these studies have not ruled out the possibility that different NER factors form possibly transient complexes before their recruitment to damaged DNA. Despite the fact that in this study we measured similar initial assembly rates for ERCC1-GFP and TFIIH-GFP, their overall binding-time courses were dissimilar, as is reflected by the 2-fold difference in t1/2 values for reaching a plateau. To test whether TFIIH and ERCC1 load onto damaged DNA individually, rather than as a complex, we analyzed the effect of temperature on the assembly kinetics. If the two proteins bind as a preformed complex, any temperature effect would be the same for both. ERCC1-GFP and TFIIH-GFP accumulation were measured at 27°C, in addition to the measurements at 37°C. This decrease in temperature only marginally affects diffusion rates, yet it may have a significant effect on the rate of enzymatic processes or other temperature-dependent steps in the NER process. Both ERCC1-GFP and TFIIH-GFP exhibited a higher plateau level at 27°C: an increase from 13.4 ± 1.7 to 17.4 ± 1.9% for ERCC1-GFP and from 14.8 ± 1.1 to 21.6 ± 4.6% for TFIIH-GFP, i.e., an increase of ≈20% and ≈50% for ERCC1-GFP and TFIIH-GFP, respectively. In addition, the establishment of the plateaus was delayed significantly (Fig. 2). Hence, the dissociation rate of NER proteins, e.g., after a repair event, appears to decrease at lower temperature. This finding might indicate the necessity of enzymatic activity for the repair complex to be released (e.g., endonucleic activity by ERCC1-XPF and XPG). The time course of TFIIH binding still exhibited its maximum accumulation rate immediately after UV irradiation (Fig. 2 Right). In contrast, ERCC1-GFP assembly exhibited a lag phase at 27°C (Fig. 2 Left). These data indicate that there is a temperature-sensitive step before ERCC1-XPF binding, but after TFIIH binding. The differences in accumulation between ERCC1-XPF and TFIIH strongly argue against binding of a preassembled complex that contains TFIIH and ERCC1. These results provide in vivo evidence supporting a sequential assembly mechanism of the NER double-incision complex (10, 12).
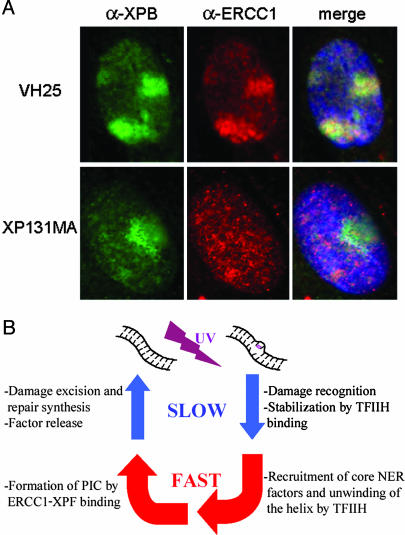
The presence of a temperature-dependent step before ERCC1-GFP binding suggests that an enzymatic reaction occurs before ERCC1-XPF can bind. The ATP-dependent helicase activity of TFIIH is the only known enzymatic activity that is required to form a functional double-incision complex. Hence, not only binding of TFIIH, but also its helicase activity, may be necessary for recruitment of ERCC1-XPF. To investigate this possibility, we carried out immunofluorescent double labeling on wild-type fibroblasts and NER-deficient XP131MA fibroblasts. XP131MA cells contain a mutation in their XPB subunit that has been shown to result in impaired helix opening in NER (12). In VH25 wild-type cells, TFIIH and ERCC1 both amassed at sites of local UV damage 3 min after UV irradiation, as expected (Fig. 3A Upper). In the helicase mutant, TFIIH was still able to bind to UV-damaged DNA, as could be shown by using antibodies against its p62 core subunit (data not shown) and against the XPB subunit (Fig. 3A Lower). In contrast, the XP131MA mutant cells did not show any accumulation of ERCC1 in the damaged area after 3 min. This result suggests that TFIIH helicase activity, rather than just TFIIH binding, must precede binding of ERCC1-XPF.
Fig. 3.
Recruitment of ERCC1-XPF to DNA damage requires TFIIH helicase activity. (A) TFIIH helicase mutant cells exhibit impaired damaged-DNA binding of ERCC1-XPF. Primary human fibroblasts were locally UV-irradiated through a micropore filter mask and subsequently cultured for 3 min before fixation. After immunofluorescent double labeling, nuclei of wild-type cells (VH25, Upper) exhibited nuclear accumulations of TFIIH (green) and ERCC1-XPF (red) in the DNA-damaged areas. In XPB helicase mutant fibroblasts (XP131MA, Lower), UV-damaged areas displayed accumulation of TFIIH (green), but no recruitment of ERCC1-XPF was detected. Merged images also show DNA counterstaining by 4′,6-diamidino-2-phenylindole in blue. (B) Dynamics of an in vivo NER cycle on its chromatin template. Clockwise from top left, events are as follows (see text for details). Upon induction of DNA damage by UV light, early events including lesion recognition and ensuing recruitment of TFIIH are relatively slow compared with the subsequent steps of preincision complex (PIC) formation. Before repairosome assembly can be accomplished by binding of ERCC1-XPF, the double helix around the injury is unwound rapidly by TFIIH helicase activity. The following cascade of events that lead to release of the NER factors is again comparatively time-consuming.
Our in vivo data support a model in which NER factors assemble into the incision complex at sites of DNA damage in a sequential and interdependent manner. In addition to the TFIIH requirement, ERCC1-XPF binding has been found to depend also on the presence of the XPA protein. In contrast, the 3′ endonuclease XPG can load onto damaged DNA in cells that lack XPA (10). Therefore, ERCC1-XPF recruitment is assumed to be the last step in NER complex formation. The genome-wide removal of UV-induced cyclobutane pyrimidine dimers is essentially absent in CHO cells (23). Although cyclobutane pyrimidine dimers are repaired by transcription-coupled NER, this repair subpathway contributes only a minor fraction to the total NER activity at any given time [<1% (Leon Mullenders, personal communication)]. Thus, the binding rate of ERCC1-GFP of ≈30 molecules per s is likely to reflect the in vivo rate of formation of the complete and functional dual-incision complex at (6–4) photoproducts.
For global genomic NER, DNA damage probably is recognized by the XPC-HR23B heterodimer, which seems to be the major requirement for TFIIH to associate with DNA damage (10, 24). Our results indicate that these early events, possibly accompanied by changes in chromatin structure (25), are relatively slow, i.e., on the minutes scale. Subsequently, the double-incision complex is assembled in multiple steps, including the binding of RPA, XPG, XPA, helix opening by TFIIH, and the binding of ERCC1-XPF. Because we observed similar initial assembly rates for TFIIH and ERCC1-XPF incorporation, it can be concluded that once TFIIH is bound, the complete incision complex is established rapidly. In turn, the ensuing steps of damage removal and resynthesis that lead to dissociation of the NER complex is again in the minutes range, as is reflected by the long residence times of NER proteins at sites of DNA damage observed with photobleaching studies (7, 8, 18). Fig. 3B summarizes these dynamics of events during a full NER cycle.
In contrast to NER, a variety of proteins that are part of other chromatin-associated complexes, such as those responsible for transcription initiation or heterochromatinization, have been found to exchange rapidly at their target sites, i.e., at the seconds scale (26, 27). Here, we show that the de novo assembly of the NER complex on the chromatin fiber in vivo is slow, with a t1/2 value of ≈1 min. It would be interesting to compare this finding with the in vivo assembly rates of other nuclear machineries. The NER mechanism has many similarities to other chromatin-related processes and even shares a variety of its factors with several of these pathways. Sequential assembly of protein complexes at specific genomic sites has been suggested for many major nuclear processes, including base excision repair (28), DNA replication (29), transcription initiation for RNA polymerase I (26) and RNA polymerase II (30), and recently also for DNA double-strand-break repair (31). Hence, the NER system can serve as a paradigm for other chromatin-associated protein processes.
Acknowledgments
This research was supported by The Netherlands Organization for Scientific Research in Earth and Life Sciences Grant 805-33-443-P.
Author contributions: M.J.M., E.M.M.M., A.B.H., J.H.J.H., W.V., and R.v.D. designed research; M.J.M., T.B., C.D., F.A.G., and M.V. performed research; M.J.M., T.B., and E.M.M.M. contributed new reagents/analytical tools; M.J.M., A.B.H., J.H.J.H., W.V., and R.v.D. analyzed data; and M.J.M. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CHO, Chinese hamster ovary; NER, nucleotide excision repair.
References
- 1.Friedberg, E. C., Walker, G. C. & Siede, W. (1995) DNA Repair and Mutagenesis (ASM Press, Washington, DC).
- 2.de Laat, W. L., Jaspers, N. G. J. & Hoeijmakers, J. H. J. (1999) Genes Dev. 13, 768–785. [DOI] [PubMed] [Google Scholar]
- 3.Araújo, S. J. & Wood, R. D. (1999) Mutat. Res. 435, 23–33. [DOI] [PubMed] [Google Scholar]
- 4.Costa, R. M., Chigancas, V., Galhardo, R. d. S., Carvalho, H. & Menck, C. F. (2003) Biochimie 85, 1083–1099. [DOI] [PubMed] [Google Scholar]
- 5.Houtsmuller, A. B. & Vermeulen, W. (2001) Histochem. Cell. Biol. 115, 13–21. [DOI] [PubMed] [Google Scholar]
- 6.Dundr, M. & Misteli, T. (2001) Biochem. J. 356, 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houtsmuller, A. B., Rademakers, S., Nigg, A. L., Hoogstraten, D., Hoeijmakers, J. H. J. & Vermeulen, W. (1999) Science 284, 958–961. [DOI] [PubMed] [Google Scholar]
- 8.Hoogstraten, D., Nigg, A. L., Heath, H., Mullenders, L. H. F., van Driel, R., Hoeijmakers, J. H. J., Vermeulen, W. & Houtsmuller, A. B. (2002) Mol. Cell 10, 1163–1174. [DOI] [PubMed] [Google Scholar]
- 9.Moné, M. J., Volker, M., Nikaido, O., Mullenders, L. H. F., van Zeeland, A. A., Verschure, P. J., Manders, E. M. M. & van Driel, R. (2001) EMBO Rep. 2, 1013–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volker, M., Moné, M. J., Karmakar, P., van Hoffen, A., Schul, W., Vermeulen, W., Hoeijmakers, J. H. J., van Driel, R., van Zeeland, A. A. & Mullenders, L. H. F. (2001) Mol. Cell 8, 213–224. [DOI] [PubMed] [Google Scholar]
- 11.Sijbers, A. M., de Laat, W. L., Ariza, R. R., Biggerstaff, M., Wei, Y. F., Moggs, J. G., Carter, K. C., Shell, B. K., Evans, E., de Jong, M. C., et al. (1996) Cell 86, 811–822. [DOI] [PubMed] [Google Scholar]
- 12.Evans, E., Moggs, J. G., Hwang, J. R., Egly, J. M. & Wood, R. D. (1997) EMBO J. 16, 6559–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mu, D., Hsu, D. S. & Sancar, A. (1996) J. Biol. Chem. 271, 8285–8294. [DOI] [PubMed] [Google Scholar]
- 14.Schaeffer, L., Roy, R., Humbert, S., Moncollin, V., Vermeulen, W., Hoeijmakers, J. H. J., Chambon, P. & Egly, J. M. (1993) Science 260, 58–63. [DOI] [PubMed] [Google Scholar]
- 15.Schaeffer, L., Moncollin, V., Roy, R., Staub, A., Mezzina, M., Sarasin, A., Weeda, G., Hoeijmakers, J. H. J. & Egly, J. M. (1994) EMBO J. 13, 2388–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egly, J. M. (2001) FEBS Lett. 498, 124–128. [DOI] [PubMed] [Google Scholar]
- 17.Kimura, H., Tao, Y., Roeder, R. G. & Cook, P. R. (1999) Mol. Cell. Biol. 19, 5383–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rademakers, S., Volker, M., Hoogstraten, D., Nigg, A. L., Moné, M. J., Van Zeeland, A. A., Hoeijmakers, J. H. J., Houtsmuller, A. B. & Vermeulen, W. (2003) Mol. Cell. Biol. 23, 5755–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drapkin, R., Reardon, J. T., Ansari, A., Huang, J. C., Zawel, L., Ahn, K., Sancar, A. & Reinberg, D. (1994) Nature 368, 769–772. [DOI] [PubMed] [Google Scholar]
- 20.Araújo, S. J., Nigg, E. A. & Wood, R. D. (2001) Mol. Cell. Biol. 21, 2281–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wakasugi, M. & Sancar, A. (1999) J. Biol. Chem. 274, 18759–18768. [DOI] [PubMed] [Google Scholar]
- 22.Svejstrup, J. Q., Wang, Z., Feaver, W. J., Wu, X., Bushnell, D. A., Donahue, T. F., Friedberg, E. C. & Kornberg, R. D. (1995) Cell 80, 21–28. [DOI] [PubMed] [Google Scholar]
- 23.Vreeswijk, M. P., van Hoffen, A., Westland, B. E., Vrieling, H., van Zeeland, A. A. & Mullenders, L. H. F. (1994) J. Biol. Chem. 269, 31858–31863. [PubMed] [Google Scholar]
- 24.Yokoi, M., Masutani, C., Maekawa, T., Sugasawa, K., Ohkuma, Y. & Hanaoka, F. (2000) J. Biol. Chem. 275, 9870–9875. [DOI] [PubMed] [Google Scholar]
- 25.Moggs, J. G. & Almouzni, G. (1999) Biochimie 81, 45–52. [DOI] [PubMed] [Google Scholar]
- 26.Dundr, M., Hoffmann-Rohrer, U., Hu, Q., Grummt, I., Rothblum, L. I., Phair, R. D. & Misteli, T. (2002) Science 298, 1623–1626. [DOI] [PubMed] [Google Scholar]
- 27.Becker, M., Baumann, C., John, S., Walker, D. A., Vigneron, M., McNally, J. G. & Hager, G. L. (2002) EMBO Rep. 3, 1188–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nilsen, H. & Krokan, H. E. (2001) Carcinogenesis 22, 987–998. [DOI] [PubMed] [Google Scholar]
- 29.Cardoso, M. C. & Leonhardt, H. (1998) J. Cell. Biochem. 70, 222–230. [PubMed] [Google Scholar]
- 30.Lee, T. I. & Young, R. A. (2000) Annu. Rev. Genet. 34, 77–137. [DOI] [PubMed] [Google Scholar]
- 31.Wolner, B., van Komen, S., Sung, P. & Peterson, C. L. (2003) Mol. Cell 12, 221–232. [DOI] [PubMed] [Google Scholar]