Abstract
Alphonsea glandulosa sp. nov. is described from Yunnan Province in south-west China. It is easily distinguished from all previously described Alphonsea species by the possession of glandular tissue at the base of the adaxial surface of the inner petals. Nectar was observed throughout the flowering period, including the pistillate phase and subsequent staminate phase. Small curculionid beetles were observed as floral visitors and are inferred to be effective pollinators since they carry pollen grains. A phylogenetic analysis was conducted to confirm the placement of this new species within Alphonsea and the evolution of the inner petal glands and specialized pollinator reward tissues throughout the family.
Introduction
The genus Alphonsea Hook. f. & Thomson (Annonaceae) currently comprises 27 species of shrubs or trees, distributed in wet tropical lowland forests across south and south-east Asia, from India to the Philippines. It is characterized by flowers with saccate petals, ‘miliusoid’ stamens (sensu [1]) and multi-seeded fruits [2]. Keßler [2] recognized 23 species in his taxonomic treatment of the genus, although four additional species were subsequently described from Vietnam [3], Papua New Guinea [4], Borneo [5], and Peninsular Malaysia [6]. Six species have been recorded from China (A. boniana Finet & Gagnep., A. hainanensis Merr. & Chun, A. mollis Dunn, A. monogyna Merr. & Chun, A. squamosa Finet & Gagnep., and A. tsangyuanensis P. T. Li), distributed in Guangdong, Guangxi, Guizhou and Yunnan Provinces [7,8], although A. squamosa has been treated as a synonym of A. boniana [2].
Two indigenous trees growing in Xishuangbanna Tropical Botanical Garden and in a small forest patch near Man-zhang Reservoir in Meng-la County, Yunnan Province, were easily identifiable as conspecific and belonging to Alphonsea, but could not be assigned to any previously described species. The two trees are readily distinguished from all other Alphonsea species as they have multiple flowers (often 5 to 9) in each inflorescence and have glandular tissue at the base of the inner petals. We propose that these two trees represent a hitherto undescribed species, which we describe and name here as Alphonsea glandulosa. Molecular phylogenetic analyses based on six chloroplast regions were carried out to ascertain the systematic position of the species.
Materials and Methods
Ethics statements
The new species reported in this study was collected from Xishuangbanna Tropical Botanical Garden, Yunnan Province, China, which permitted our field work in the Garden. The other collecting location, Man-zhang Reservoir, Meng-la County, Yunnan, is not a protected area. Since this species is currently undescribed, it is inevitably not currently included in the Chinese Red Data Book [9].
Phylogenetic analysis
Taxon and DNA region sampling
Sequences of 60 species of Annonaceae were downloaded from GenBank, and supplemented with newly generated sequences of the new species, as well as the Chinese species Alphonsea mollis and A. monogyna. The final dataset included 43 accessions representing 31 genera from subfam. Malmeoideae, 15 accessions representing 15 genera from subfam. Annonoideae, four accessions of four genera in subfam. Ambavioideae, and one species of Anaxagorea A. St.-Hil. (subfam. Anaxagoreoideae). Six chloroplast DNA (cpDNA) regions (matK, ndhF, psbA-trnH, rbcL, trnL-F, and ycf1) were amplified for the three species. Genera reported to have inner petal glands were included (see Results and Discussion), with at least one representative species for each. The voucher information and GenBank accession numbers are provided in S1 Table.
DNA extraction, amplification and sequencing
Genomic DNA was extracted from herbarium materials using a modified cetyl trimethyl ammonium bromide (CTAB) method [10]. A single amplification protocol was used for amplification of the chloroplast regions, viz.: template denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 sec; primer annealing at 50°C for 1 min; and primer extension at 72°C for 1 min, followed by a final extension step at 72°C for 10 min. The primers used were those from [11,12]. PCR products were visualized using agarose gel electrophoresis. Successful amplifications were purified, and sequenced commercially.
Alignment and phylogenetic analyses
Sequences were assembled and edited using Geneious ver. 5.4.3 [13] and pre-aligned with the MAFFT [14] plugin in Geneious using the automatic algorithm selection and default settings, with subsequent manual checking and optimization. One inversion of 15 positions in psbA-trnH was identified and reverse complemented in the alignment, following a strategy previously applied [15] to retain substitution information in the fragments.
Maximum parsimony (MP) analyses of the six combined regions were conducted using PAUP ver. 4.0b10 [16]. All characters were weighted equally, with gaps treated as missing data. The most parsimonious trees were obtained with heuristic searches of 1,000 replicates of random stepwise sequence addition, tree bisection-reconnection (TBR) branch swapping with no limit to the number of trees saved. Bootstrap support (BS) was calculated following [17], with 10,000 simple stepwise addition replicates with TBR branch swapping and no more than 10 trees saved per replicate.
Bayesian analysis was performed using the NSF Extreme Science & Engineering Discovery Environment (XSEDE) application of MrBayes ver. 3.2.2 [18,19] provided by the CIPRES Science Gateway [20,21]. Six partitions based on DNA region identity were used and the parameters for each locus were allowed to evolve independently using the unlinked setting. MrModeltest ver. 2.3 [22] was used to determine the best-fit likelihood model for each locus and the concatenated matrix using the Akaike Information Criterion: the general time-reversible model with a gamma distribution of substitution rates (GTR+G) was chosen for the matK, psbA-trnH and trnL-F regions; and the GTR+I+G model with a proportion of invariant sites was selected for the ndhF, rbcL and ycf1 regions. Two independent Metropolis-coupled Markov chain Monte Carlo (MCMCMC) analyses were run. Each search used three incrementally heated and one cold Markov chain, and was run for 10 million generations and sampled every 1,000th generation. The mean branch length prior was set from the default mean (0.1) to 0.01 (brlenspr = unconstrained: exponential (100.0)) to reduce the likelihood of stochastic entrapment in local tree length optima [23,24]. Convergence was assessed using the standard deviation of split frequencies, with values < 0.01 interpreted as indicating good convergence. The first 25% of samples (2,500 trees) were discarded as burn-in, and the post-burn-in samples summarized as a 50% majority-rule consensus tree.
Morphological observations
The morphological description of the new species was based on the examination of fresh materials and dried herbarium specimens. Morphological comparisons with other species in Alphonsea were based on studies of herbarium specimens (from herbaria HITBC, IBSC, K, KUN, PE and SING; institutional acronyms follow the Index Herbariorum [25]), specimen photographs and a literature survey.
Flowers were fixed in FAA (70% alcohol, formaldehyde and glacial acetic acid in a ratio of 90:5:5) for 24 hrs and subsequently stored in 70% alcohol. Samples examined using scanning electron microscopy were dehydrated using an ethanol series, and critical-point dried with a Leica EM CPD300 Automated Critical Point Dryer (Leica, Wetzlar, Germany). The dried materials were later attached to metal stubs using adhesive carbon tabs, sputter-coated with gold/palladium, and viewed using a JSM-6360LV scanning electron microscope (JEOL, Tokyo, Japan) at 5 kV.
For anatomical observations, the fixed inner petals were dissected and transferred into 70% ethanol, stained with Ehrlich’s hematoxylin, dehydrated in ethanol series, infiltrated with xylene, embedded in paraffin wax and serially sectioned at thickness of 10 μm, using a rotary microtome. Subsequently, mounted slides were examined and photographed under a LEICA DM5500 B microscope equipped with a LEICA DFC550 digital camera.
Nomenclature
The electronic version of this article in Portable Document Format (PDF) in a work with an ISSN or ISBN will represent a published work according to the International Code of Nomenclature for algae, fungi, and plants, and hence the new names contained in the electronic publication of a PLOS article are effectively published under that Code from the electronic edition alone, so there is no longer any need to provide printed copies [26].
In addition, the new name contained in this work has been submitted to IPNI, from where it will be made available to the Global Names Index. The IPNI LSID can be resolved and the associated information viewed through any standard web browser by appending the LSID contained in this publication to the prefix http://ipni.org/. The online version of this work is archived and available from the following digital repositories: PubMed Central and LOCKSS.
Results and Discussion
Phylogenetic analysis
The concatenated alignment of the 63-terminal dataset consisted of 7,399 characters. The MP heuristic search retrieved 24 most parsimonious trees of 3,968 steps (consistency index, CI = 0.66; retention index, RI = 0.70).
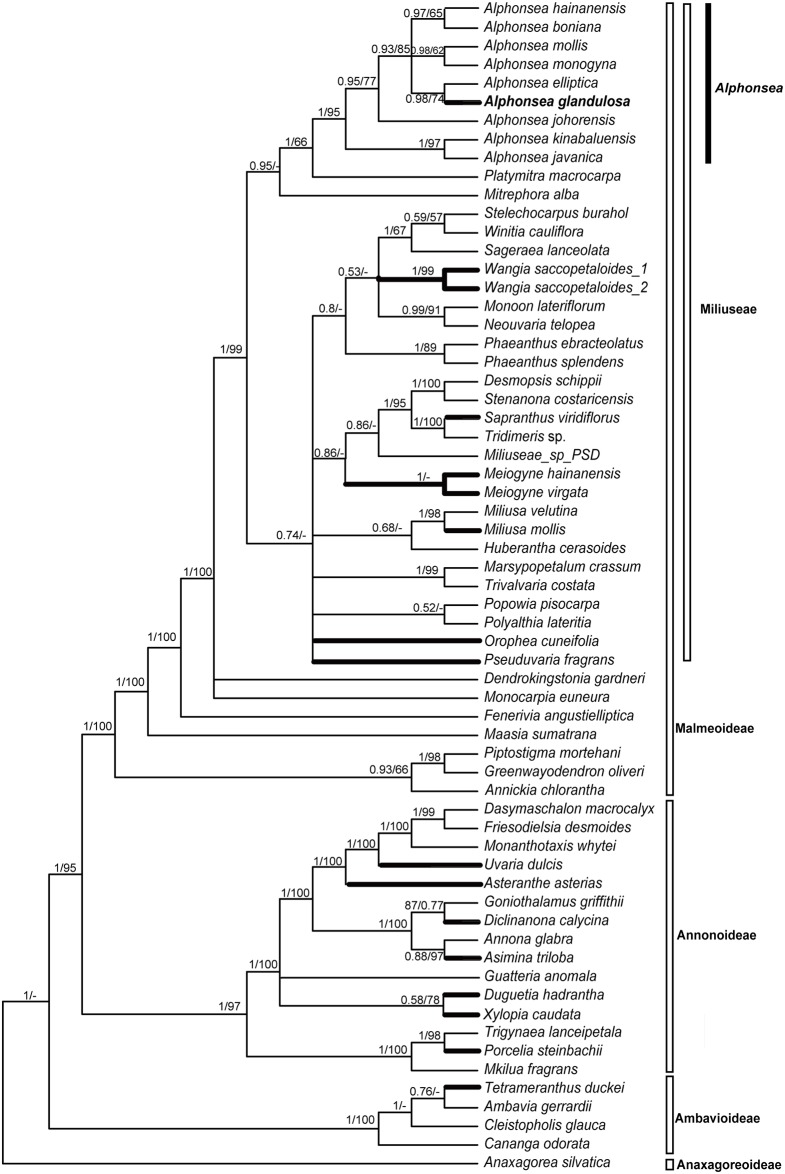
The MP and Bayesian analyses are topologically similar, differing mainly in the relative MP bootstrap (BS) and posterior probability (PP) values for particular groups (Fig 1). The new species, Alphonsea glandulosa, is deeply nested within the Alphonsea clade and retrieved as sister to A. elliptica. Although these results confirm that the new species unequivocally belongs to the genus Alphonsea, limitations in the extent of taxon sampling within the genus (nine out of 28 species; 32%) preclude any definitive conclusion regarding which species is phylogenetically closest to A. glandulosa.
Fig 1. Bayesian 50% majority-rule consensus tree under partitioned models (cpDNA data: matK, ndhF, psbA-trnH, rbcL, trnL-F and ycf1; 63 taxa).
Numbers at the nodes indicate Bayesian posterior probabilities and maximum parsimony bootstrap values (> 50%), in that order. Thick lines indicating the clades in which glands or specialized pollinator reward tissues have evolved.
Morphological comparisons
Alphonsea glandulosa flowers have saccate petals (Fig 2A–2C), ‘miliusoid’ stamens in which the connective does not extend over the thecae (Fig 3C), and hairy ovaries with up to 13 biseriate ovules (Fig 3D). These characters corroborate its placement in the genus Alphonsea.
Fig 2. Flower and fruit morphology of Alphonsea glandulosa.
A, Branch, showing leaf-opposed inflorescence position, and lanceolate leaves. B, Abaxial view of the inflorescence, showing 5–6 carpels per flower. C, Adaxial view of the inflorescence, showing pubescent pedicels with one densely pubescent medial bract (arrowed). D, Fruit with subglobose monocarps. E, Single monocarp, dissected to show seed arrangement.—Photos: A–E, Yun-hong Tan.
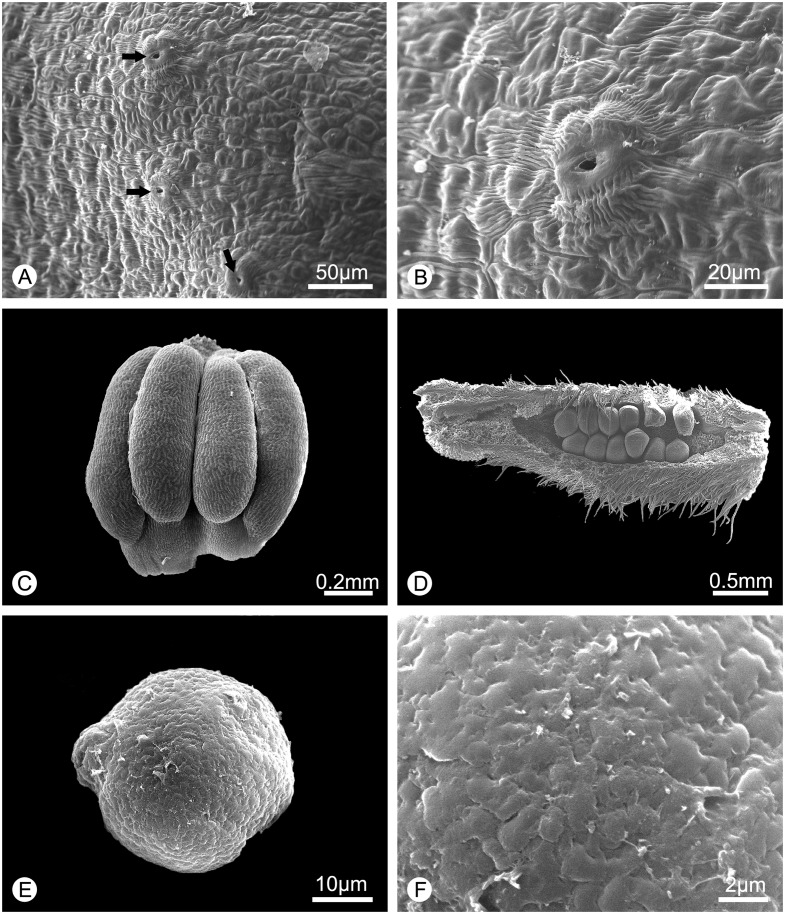
Fig 3. Morphology of the surface of the inner petal glands, stamen, carpel and pollen of Alphonsea glandulosa (scanning electron micrographs).
A, Surface of the glandular tissue, showing the nectary stomata (arrowed). B, Close-up of the nectary stomata. C, Stamen. D, Carpel, dissected to show biseriate ovules. E, Pollen grain. F, Rugulate pollen exine ornamentation.
Alphonsea glandulosa is unusual in the genus, however, in having multiple flowers (often 5 to 9) in each inflorescence (Fig 2A–2C). Although most Alphonsea species have inflorescences with only 1–4 flowers, there are two species, A. philastreana (Pierre) Finet & Gagnep. and A. ventricosa (Roxb.) Hook. f. & Thomson, which have more than four flowers per inflorescence [2].
Alphonsea glandulosa can easily be distinguished from A. ventricosa, even from vegetative characters: A. ventricosa has large leaves (12–27 cm by 4–8 cm) that are distinctly thick and coriaceous, whereas the leaves of the new species are smaller (6–19 cm by 3–5.5 cm) and slightly coriaceous. A. ventricosa furthermore differs in having flowers with more stamens (40–50, in four whorls) and more carpels (10–12) per flower, and in having fruits with larger monocarps (up to 6 cm long, 4 cm in diameter) and longer stipes (ca. 3 cm). Alphonsea glandulosa has flowers with 26–35 stamens in three whorls and 4–7 carpels per flower, fruits that are 2–4 cm long and 1.5–3 cm in diameter, and stipes up to 10 mm long.
In contrast, Alphonsea glandulosa is vegetatively more similar to A. philastreana. The two species differ, however, in the number of secondary veins on each side of leaf, the indumentum of the abaxial surface of the flower buds, pedicel length, the number of carpels per flower, and the shape of the stigma. Alphonsea glandulosa has 8–18 nerves on each side of the leaf, whereas A. philastreana has 8–9 [2,27]. The abaxial surface of flower buds of A. glandulosa is greyish to yellowish pubescent, whereas that of A. philastreana is densely rusty tomentose [27,28]. Pedicel length and the number of carpels per flower are useful characters for species delimitation: Keβler [2], for example, frequently used these characters in his key to species of Alphonsea. Although the name A. philastreana was validated using a protologue description based on flower buds and hence difficulties were encountered when drawing comparisons, the difference in the length of the pedicel in the two species is nevertheless very significant when buds of a similar size are compared: the pedicel of flower buds that are ca. 3–4 mm in diameter, for example, is 10–20 mm long in A. glandulosa (Fig 2B and 2C) but only ca. 3 mm long in A. philastreana [2,28]. Although previous descriptions of A. philastreana state that it has (4–)6 carpels per flower and the stigma is globose [27–29], Keβler [2] counted only three carpels in all studied buds and found that the stigma is actually two-headed (deeply bilobed). The two-headed stigma may have misled previous authors into counting six carpels from above. In contrast, A. glandulosa has between four and seven carpels (Figs 2B and 4B), and the stigma is globose to shallowly bilobed. Ovule numbers of the two species are also different: A. glandulosa has 10–13 ovules per carpel, whereas A. philastreana has 14–18(–20) [27–29].
Fig 4. Glandular tissue, nectar and flower visitors of Alphonsea glandulosa.
A, Nectar at pistillate phase. B, Nectar at the end of staminate phase. C, Morphology of the glandular tissue after FAA fixation. D, Morphology of the glandular tissue after rehydration from dried specimens. E, Small curculionid beetles observed visiting the flowers. F, Bee observed visiting the flower.—Photos: A, C, F, Yun-Yun Shao; B, D, E, Bine Xue.
Amongst the Alphonsea species sampled in our phylogenetic study, A. elliptica is retrieved as sister to A. glandulosa (Fig 1). The differences between these two species are nevertheless clear: the inflorescences of A. glandulosa are composed of (3–)5–9(–13) flowers, whereas those of A. elliptica are generally single-flowered (rarely with up to three flowers) [2,30]. Alphonsea glandulosa furthermore has stamens in three whorls, whereas A. elliptica has stamens in four whorls [2].
The most distinctive diagnostic character of Alphonsea glandulosa, however, is the possession of nectar glands at the base of the inner petals (Fig 4A–4D). The gland is clearly visible irrespective of preservation technique: in fresh (Fig 4A and 4B) and FAA-fixed (Fig 4C) material the gland is clearly ridge-shaped, and in dried specimens it is apparent as a distinct groove (Fig 4D); it is even obvious in small flower buds.
Anatomically, the nectar glands consist of four distinct tissues (Fig 5A), similar to those described for other species [31–33]: (i) epidermis; (ii) subepidermal secretory parenchyma: several layers of small cells with densely staining cytoplasm; (iii) ground parenchyma: several layers of larger cells, more loosely packed than those of the secretory parenchyma; and (iv) vascular bundles. The anatomical structure is distinct from the non-glandular part of the inner petals (Fig 5B), which only consists of epidermis, several layers of homogeneous parenchyma and a few vascular bundles. The surface of the nectar glands also differs from the surrounding epidermis: nectar stomata, which are raised slightly above the epidermis with an aperture for nectar secretion [32], are found across the surface of the glandular tissue (Fig 3A and 3B), but are otherwise absent from the non-glandular part of the inner petals. The ultrastructure of the glands is similar to those of Pseuduvaria froggattii (F.Muell.) Jessup [34, 35].
Fig 5. Anatomical structure of the inner petal glands and adjacent non-glandular part of the inner petals (paraffin sections).
A, Section of the inner petal nectary gland, showing the nectary epidermis (E), secretory parenchyma (SP), ground parenchyma (GP), and vascular tissue (V). B, Section of the non-glandular part of the inner petals, showing the epidermis (E), parenchyma (P) and vascular tissue (V).
To conclude, both the molecular and morphological data support the placement of the new species in Alphonsea. It differs from all previously described species, and therefore represents a new species.
Evolution and function of nectar glands
Nectar glands have never previously been recorded in Alphonsea [2,36], and it therefore appears that within the genus nectar glands are autapomorphic for A. glandulosa.
Glandular or specialized pollinator food reward tissues are found in several other genera in the family on different parts of the inner petals, often on the lower part of the adaxial surface, viz. Asimina Adans. p.p. [36,37], Asteranthe Engl. & Diels [36], Diclinanona Diels [36,38], Duguetia A.St.-Hil. p.p. [36,39,40], Meiogyne Miq. [11,36,41], Miliusa Lesch. ex A.DC. p.p. [42–44], Pseuduvaria Miq. p.p. [35,36], Sapranthus Seem. [36,45], Tetrameranthus R.E.Fr. [36,46,47] and Wangia X.Guo & R.M.K.Saunders [12]. Paired glands or specialized food tissues are sometimes located along the margins of the inner petals, as found in some Asimina p.p. [36], Diclinanona p.p. [36,38], Porcelia Ruiz & Pav. p.p. [36,48], Uvaria L. p.p. (those species previously classified in Anomianthus Zoll. and Ellipeiopsis R.E.Fr.) [36,49], and Xylopia L. p.p. [36]. In Orophea Blume p.p. [36,50,51] and Pseuduvaria p.p. [35,36] a pair of glands or specialized food tissues are sometimes present underneath the mitre, i.e. on the upper part of the adaxial surface of the inner petals. In Miliusa p.p., thickened specialized tissue is found running inside along the adaxial bilateral midline of the inner petals [42–44]. Van Heusden [36] also stated that Enicosanthum Becc. species (now included in Monoon [52]) possess inner petal glands, although we have not been able to confirm this observation. Nevertheless, most observations of glandular or specialized tissues were based on the examination of herbarium specimens. Field observations of Asimina and Pseuduvaria spp. confirm the secretion of nectar [34,37,53]. For Sapranthus and Meiogyne spp., however, no liquid secretions have been observed (George Schatz, pers. comm); it is therefore debatable whether the specialized tissues in these genera represent true glands that secrete an exudate, and further field observation and anatomical comparisons are required.
Chatrou et al. [54] recognized four subfamilies in their recent subfamilial and tribal classification of the Annonaceae, viz. subfam. Anaxagoreoideae, Ambavioideae, Annonoideae and Malmeoideae (Fig 1). The genera mentioned above with glands or specialized tissues on the inner petals are distributed in all the subfamilies except subfam. Anaxagoreoideae. These genera are classified in up to seven different tribes, indicating that this character is likely to have evolved independently on multiple occasions (Fig 1). A summary of the occurrences of the glands and specialized tissues across the family is provided in Table 1. In the tribe Miliuseae Hook.f. & Thomson, which includes Alphonsea, inner petal glands and specialized tissues have evolved independently in seven genera (Table 1; Fig 1).
Table 1. The occurrence of inner petal glands and specialized pollinator reward tissues in genera, tribes and subfamilies across the family Annonaceae.
| Subfamilies | Tribes | Genera |
|---|---|---|
| Ambavioideae | – | Tetrameranthus |
| Annonoideae | tribe Annoneae Endl. | Asimina p.p., Diclinanona |
| tribe Bocageeae Endl. | Porcelia p.p. | |
| tribe Duguetieae Chatrou & R.M.K.Saunders | Duguetia p.p. | |
| tribe Monodoreae Baill. | Asteranthe | |
| tribe Uvarieae Hook.f. & Thomson | Uvaria p.p. | |
| tribe Xylopieae Endl | Xylopia p.p. | |
| Malmeoideae | tribe Miliuseae Hook.f. & Thomson | Alphonsea p.p., Meiogyne, Orophea, Pseuduvaria p.p., Sapranthus, Wangia, Miliusa p.p. |
Pollination ecology studies have been undertaken for several species that possess glandular or specialized inner petal tissues, viz.: Asimina obovata (Willd.) Nash and A. pygmaea (W. Bartram) Dunal, which are pollinated by large scarabaeid beetles [37]; Pseuduvaria froggattii, pollinated by Drosophilidae and other flies [34]; Pseuduvaria mulgraveana Jessup, pollinated by small diurnal nitidulid beetles [53]; and Sapranthus palanga R.E.Fr., pollinated by tenebrionid, nitidulid, and scarabaeid beetles and apid bees [45]. In Asimina obovata and A. pygmaea, beetles were observed to consume the corrugated inner petal tissues that were sometimes also observed to secrete a small volume of exudate [36]. In Pseuduvaria froggattii and P. mulgraveana, the secretion of nectar from the inner petal glands was recorded in detail and shown to be concomitant with the anthetic changes: nectar was present throughout the pistillate and staminate phases in hermaphroditic flowers of both species, and in staminate flowers of P. mulgraveana [34,53]. Pollinators were observed consuming the nectar [34,53], indicating that the nectar glands function as a food reward for pollinators throughout anthesis.
As with all hermaphroditic-flowered Annonaceae species [55], Alphonsea glandulosa is protogynous. In A. glandulosa, abundant sweet nectar was observed to be secreted from the beginning of the pistillate phase until the end of the staminate phase (Fig 4A and 4B). Around three to eight small curculionid beetles were found inside the flowers (Fig 4E), and pollen was observed on their bodies, indicating that they are likely to be effective pollinators. The nectar is likely to provide a food reward to those beetles, as the beetles were observed to consume it. One opportunist bee was also observed visiting the flower (Fig 4F), although it is unlikely that the bee could effectively pollinate the flower because of the presence of the floral chamber formed by the inner petals, which would restrict access by the bees [56].
Taxonomic treatment
Alphonsea glandulosa Y.H. Tan & B. Xue, sp. nov. [urn:lsid:ipni.org:names:77159533–1] (Figs 2, 3, 4, 5 and 6)
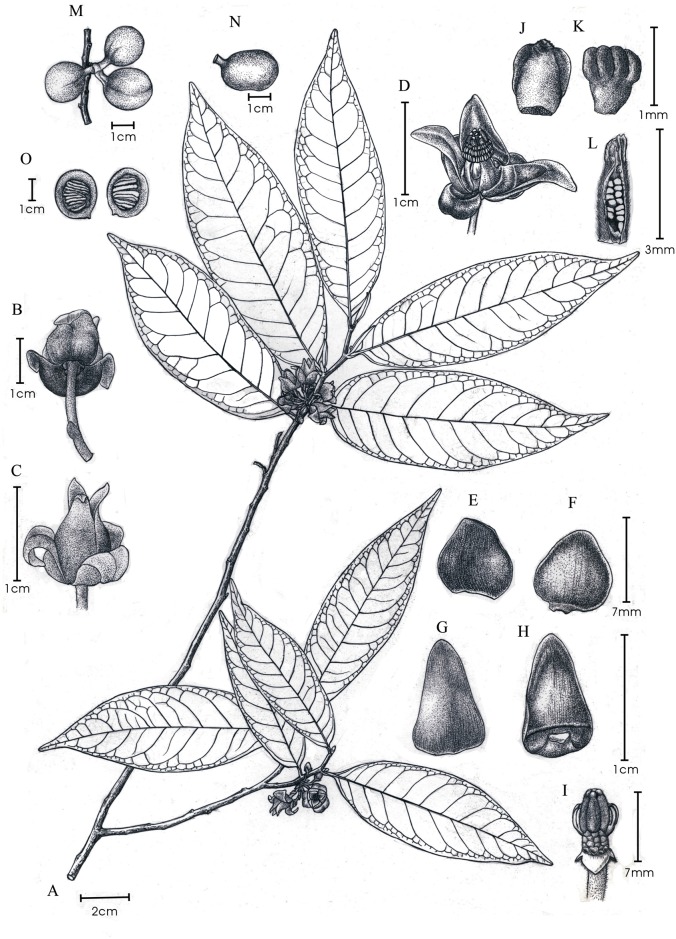
Fig 6. Alphonsea glandulosa sp. nov.
A, Flowering branch. B–D, Flower (B, Lower view. C, Lateral view. D, Top view, showing six carpels and three rows of stamens). E–F, Outer petal (E, Abaxial view. F, Adaxial view). G–H, Inner petal (G, Abaxial view. H, Adaxial view, with nectar glands at the base). I, Flower with petals removed, showing the attachment of stamens and carpels. J–K, Stamen (J, Adaxial view. K, Abaxial view). L. Carpel, longitudinal section, showing ovule arrangement. M, Fruit, composed of separate monocarps. N, Single monocarp. O, Longitudinal section of monocarp, showing seed arrangement. Drawn by Yun-xiao Liu.
Type: CHINA. Yunnan Province, Meng-la County, Meng-lun, Man-zhang Reservior, 21°55′53″N, 101°10′58″E, alt. 625 m. Y. H. Tan 10145, 2016.04.07 (fl.) (holotype HITBC; isotypes IBSC, KUN)
Diagnosis
Alphonsea glandulosa is unique amongst Alphonsea species in having a nectar gland at the base of the adaxial surface of each inner petal. It is most similar to A. philastreana (Pierre) Finet & Gagnep., but differs in having a greater number of secondary veins on each side of the leaf, greyish to yellowish pubescent flower buds, longer pedicels, a greater number of carpels per flower, a smaller number of ovules per carpel, and globose to shallowly bilobed stigmas.
Description
Trees to 15–20 m tall, ca. 25–30 cm dbh. Bark brownish, fissured. Young twigs green, puberulent, soon become greyish and glabrous. Petioles 3–8 mm long, 1–2 mm in diameter, transversely densely striate; leaf laminae narrlowly elliptic, elliptic or ovate, 6–19 × 3–6.7 cm, base cuneate, apex acuminate, slightly coriaceous, abaxially sparsely pubescent to glabrescent, adaxially glabrous; midrib impressed and glabrous above, raised and hairy to glabrous below; secondary veins 8–18 on each side of the leaf, parallel, diverging at 45–60° from midrib, anastomosing within margin, distinctly raised below; tertiary veins reticulate, prominent abaxially. Inflorescences leaf-opposed or supra-axillary; (3–)5–9(–13) flowers per inflorescence (Fig 2A–2C). Peduncles absent or up to 3 mm long (Fig 2A–2C). Pedicels 10–20 mm long, 1–1.5 mm in diameter, pubescent, with one densely pubescent median bract (Fig 2A–2C). Sepals ovate, 1.5–2 × 1.5–2.5 mm, hairy abaxially, glabrous adaxially; outer petals ovate, 10–14 × 6–8.5 mm, base acute, apex acute, tomentose abaxially, sparsely hairy to glabrous adaxially; inner petals narrower, 10–14 × 5–8 mm wide, hairy abaxially, glabrous adaxially, with glandular tissue near the base (Fig 4A–4D), apparent as ridge in fresh (Fig 4A and 4B) and FAA-fixed (Fig 4C) material, and as distinct groove in dried specimens (Fig 4D). Stamens ‘miliusoid’ with very short connective prolongation not extending over pollen sacs, 26–35 per flower, ca. 1 mm long, in 3 whorls (Figs 3C, 4A and 4F). Carpels 4–7 per flower, ca. 3 mm long, with short brown hairs (Figs 2B and 4B); ovules 10–13 per carpel, biseriate (Fig 3D). Fruiting pedicels 7–20 mm long, 3 mm in diameter; monocarps 1–7 per fruit, subglobose to cylindrical, ca. 2–4 cm long, 1.5–3 cm in diameter, yellow when mature, greyish pubescent, smooth, sometimes with a longitudinal ridge or groove, apex rounded; stipe to 10 mm long, ca. 4 mm thick (Fig 2D and 2E). Seeds flattened-ellipsoid, up to 13 per monocarp (Fig 2E). Pollen grains solitary, subspherical, 30–40 μm in diameter, rugulate (Fig 3E and 3F).
Etymology
The specific epithet reflects the presence of nectar glands at the base of the adaxial surface of each inner petal.
Additional specimens examined (paratypes)
China. Meng-la, Yunnan, 2009-03-25, Chun-fen Xiao C100647 (HITBC); 2015-04-29, B. Xue 188 (IBSC); 2016-04-26, B. Xue 265 (IBSC, KUN); 2016-04-27, B. Xue 268 (IBSC, SING).
Distribution
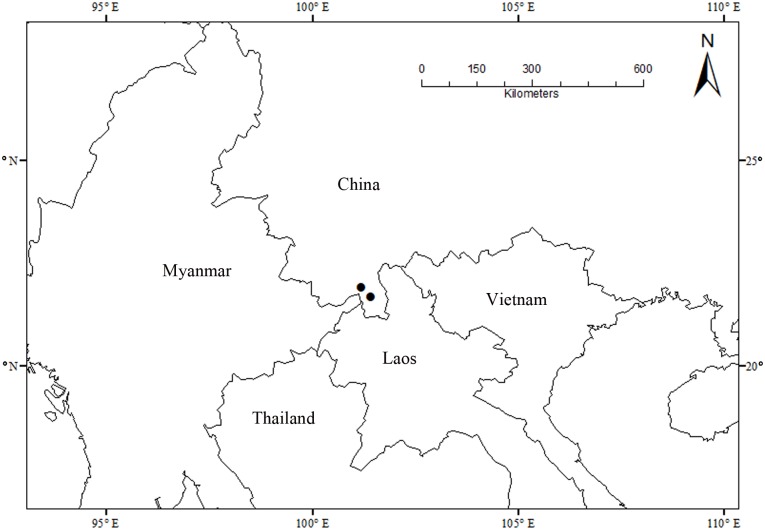
Only known from two localities in Yunnan, China (Fig 7).
Fig 7. Distribution of Alphonsea glandulosa.
Ecology and phenology
In evergreen forests. Flowering specimens collected in March to May, and fruiting specimens in June to July.
IUCN Conservation Status
Only two individuals were found in Meng-la County, Yunnan Province. The primary forests in Xishuangbanna have been under severe pressure from agricultural expansion over the last 30 years, and below 900 m elevation most unprotected forest has been replaced by rubber plantations [57]. The tree growing in the forests close to Man-zhang Reservoir in Meng-la County is located at the edge of a rubber plantation. One of the authors, Yun-Hong Tan, has undertaken an extensive field survey in Xishuangbanna, but was unable to locate other individuals. Perhaps because of the dearth of individuals, the level of fruitset in the two trees is poor. On the basis of current IUCN red list categories and criteria [58], we therefore recommend critically endangered status, CR D.
Supporting Information
(XLSX)
Acknowledgments
We are grateful to the curators of IBSC, K, KUN, HITBC, PE and SING herbaria for permission to access their collections. The first author thanks the H.M. Burkill Research Fellowship and the Singapore Botanical Gardens for offering the opportunity to study their Annonaceae specimens in SING in 2015. We also thank George Schatz and two anonymous reviewers for critically reading our manuscript and suggesting improvements.
Data Availability
All relevant data are within the paper and its Supporting Information files, with DNA sequences available for download from GenBank (http://www.ncbi.nlm.nih.gov/genbank/).
Funding Statement
Financial support was provided by a grant from National Natural Science Foundation of China (www.nsfc.gov.cn; grant no. 31400199) awarded to Bine Xue, and a special pilot program of the Fourth National Survey on Chinese Materia Medica Resources from Traditional Chinese Medicine Industry Research (http://www.zyzypc.com.cn/; grant no. 201207002). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of manuscript.
References
- 1.Mols JB, Gravendeel B, Chatrou LW, Pirie MD, Bygrave PC, Chase MW, et al. Identifying clades in Asian Annonaceae: monophyletic genera in the polyphyletic Miliuseae. Am J Bot. 2004; 91: 590–600. 10.3732/ajb.91.4.590 [DOI] [PubMed] [Google Scholar]
- 2.Keßler PJA. Studies on the tribe Saccopetaleae (Annonaceae)—IV. Revision of the genus Alphonsea Hook. f. & Thomson. Bot Jahrb Syst. 1995; 118: 81–112. [Google Scholar]
- 3.Bân NT. Annonaceae In: Ban NT, Ly TD, Khanh TC, Loc PK, Thin NN, Tien NV & Khoi NK, editor. Flora of Vietnam. Hanoi: Science & Technics Publishing House; [In Vietnamese]; 2000. pp. 5–341. [Google Scholar]
- 4.Okada H. New genus and new species of the Annonaceae from the Malesian wet tropics. Acta Phytotax Geobot. 1996; 47: 1–9. [Google Scholar]
- 5.Turner IM. A new species of Alphonsea (Annonaceae) from Borneo. Gardens’ Bulletin, Singapore. 2009; 61: 185–188. [Google Scholar]
- 6.Turner IM, Utteridge TMA. A new species of Alphonsea (Annonaceae) from Peninsular Malaysia. Blumea. 2015; 59: 206–208 [Google Scholar]
- 7.Tsiang Y, Li PT. Annonaceae In: Tsiang Y, Li PT, editors. Flora Reipublicae Popularis Sinicae. Beijing: Science Press; 1979. pp. 10–175. [Google Scholar]
- 8.Li PT, Gilbert MG. Annonaceae In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Beijing: Science Press; and St. Louis: Missouri Botanical Garden Press; 2011. pp. 672–713. [Google Scholar]
- 9.Fu LK, Chin CM. China Plant Red Data Book, Volume 1: Rare and Endangered Plants. Beijing: Science Press; 1992. 741pp. [Google Scholar]
- 10.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987; 19: 11–15. [Google Scholar]
- 11.Thomas DC, Surveswaran S, Xue B, Sankowsky G, Mols JB, Keßler PJA, et al. Molecular phylogenetics and historical biogeography of the Meiogyne-Fitzalania clade (Annonaceae): generic paraphyly and late Miocene-Pliocene diversification in Australasia and the Pacific. Taxon. 2012; 61: 559–575. [Google Scholar]
- 12.Guo X, Wang J, Xue B, Thomas DC, Su YC, Tan YH, et al. Reassessing the taxonomic status of two enigmatic Desmos species (Annonaceae): Morphological and molecular phylogenetic support for a new genus, Wangia. J Syst Evol. 2014; 52: 1–15. [Google Scholar]
- 13.Drummond A, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, et al. Geneious version 5.1. 2010. http://www.geneious.com/.
- 14.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002; 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pirie MD, Chatrou LW, Mols JB, Erkens RHJ, Oosterhof J. ‘Andean-centred’ genera in the short-branch clade of Annonaceae: testing biogeographical hypotheses using phylogeny reconstruction and molecular dating. J Biogeogr. 2006; 33: 31–46. [Google Scholar]
- 16.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods). Version 4.0b10. Sunderland, Massachusetts: Sinauer Associates; 2003. [Google Scholar]
- 17.Müller K. The efficiency of different search strategies in estimating parsimony jackknife, bootstrap, and Bremer support. BMC Evol Biol. 2005; 5: art. 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001; 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 19.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003; 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 20.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE): New Orleans: IEEE; 2010. pp. 1–8.
- 21.Miller MA, Pfeiffer W, Schwartz T. The CIPRES science gateway: a community resource for phylogenetic analyses. Proceedings of the 2011 TeraGrid Conference: Extreme Digital. New York: ACM; 2011. pp. 41.
- 22.Nylander JAA. MrModeltest, version 2. Evolutionary Biology Centre, Uppsala University; 2004. http://wwwabcse/~nylander/mrmodeltest2/mrmodeltest2html.
- 23.Brown JM, Hedtke SM, Lemmon AR, Lemmon EM. When trees grow too long: investigating the causes of highly inaccurate Bayesian branch-length estimates. Syst Biol. 2010; 59: 145–161. 10.1093/sysbio/syp081 [DOI] [PubMed] [Google Scholar]
- 24.Marshall DC. Cryptic failure of partitioned Bayesian phylogenetic analyses: lost in the land of long trees. Syst Biol. 2010; 59: 108–117. 10.1093/sysbio/syp080 [DOI] [PubMed] [Google Scholar]
- 25.Thiers B. Index Herbariorum: A global directory of public herbaria and associated staff. New York Botanical Garden's Virtual Herbarium; 2015. Database. http://sweetgum.nybg.org/ih/.
- 26.McNeill J, Barrie FR, Buck WR, Demoulin V, Greuter W, Hawksworth DL, et al. International Code of Nomenclature for algae, fungi and plants (Melbourne Code). Germany: Koeltz Botanical Books; 2012. [Google Scholar]
- 27.Finet A, Gagnepain F. Annonacees In: Lecomte H, editor. Flore générale de l'Indo-Chine. Paris: Masson; 1907–08. p.123. [Google Scholar]
- 28.Pierre. Flore forestière de la Cochinchine. Paris: O. Doin; 1881. t.16. [Google Scholar]
- 29.Finet A. & Gagnepin F. Contributions à la flore de l'Asie orientale. Bulletin de la Société botanique de France. 1906; 53 Mém. 4 (2): 164. [Google Scholar]
- 30.Hooker JD, Thomson T. Anonaceae In: Hooker JD, editor. The Flora of British India vol 1. Ashford, Kent: L. Reeve; 1872. p.90. [Google Scholar]
- 31.Durkee LT. The ultrastructure of floral and extrafloral nectaries In: Bentley B & Elias T, editors. The biology of nectaries. New York: Columbia University Press; 1983. 1–29. [Google Scholar]
- 32.Nepi M. Nectary structure and ultrastructure In: Nicolson SW, Nepi M, Pacini E, editors. Nectaries and nectar. Dordrecht: Springer; 2007. pp. 129–166. [Google Scholar]
- 33.Abedini M, Movafeghi A, Aliasgharpour M, Dadpour MR. Anatomy and ultrastructure of the floral nectary in Peganum harmala L. (Nitrariaceae). Plant Species Biol. 2013; 28: 185–192. [Google Scholar]
- 34.Silberbauer-Gottsberger I, Gottsberger G, Webber A. Morphological and functional flower characteristics of New and Old World Annonaceae with respect to their mode of pollination. Taxon. 2003; 52: 701–718. [Google Scholar]
- 35.Su YCF, Saunders RMK. Monograph of Pseuduvaria (Annonaceae). Syst Bot Monogr. 2006; 79: 1–204. [Google Scholar]
- 36.van Heusden ECH. Flowers of Annonaceae: morphology, classification, and evolution. Blumea Supplement. 1992; 7: 1–218. [Google Scholar]
- 37.Norman EM, Clayton D. Reproductive biology of two Florida pawpaws: Asimina obovata and A. pygmaea (Annonaceae). Bulletin of the Torrey Botanical Club. 1986; 113: 16–22. [Google Scholar]
- 38.Erkens RHJ, Chatrou LW, Chaowasku T, Westra LYT, Maas JW, Maas PJW. A decade of uncertainty: resolving the phylogenetic position of Diclinanona (Annonaceae), including taxonomic notes and a key to the species. Taxon. 2014; 63: 1244–1252. [Google Scholar]
- 39.Maas PJM, Westra LYT, Chatrou LW. Duguetia. Flora Neotropica Monograph. 2003; 88: 1–274. [Google Scholar]
- 40.van Zuilen CM. Pattens and affinities in the Duguetia alliance (Annonaceae). Molecular and morphological studies. [PhD thesis]. Utrecht: Utrecht University; 1996.
- 41.Xue B, Thomas DC, Chaowasku T, Johnson DM, Saunders RMK. Molecular phylogenetic support for the taxonomic merger of Fitzalania and Meiogyne (Annonaceae): new nomenclatural combinations under the conserved name Meiogyne. Syst Bot. 2014; 39: 396–404. [Google Scholar]
- 42.Chaowasku T, Keßler PJA. Seven new species of Miliusa (Annonaceae) from Thailand. Nord J Bot. 2013; 31: 680–699. [Google Scholar]
- 43.Chaowasku T, Keßler PJA, Chatrou LW. Phylogeny of Miliusa (Magnoliales: Annonaceae: Malmeoideae: Miliuseae), with descriptions of two new species from Malesia. Eur J Taxon. 2013; 64: 1–21. [Google Scholar]
- 44.Mols JB, Kessler PJA. The genus Miliusa (Annonaceae) in the Austro-Malesian area. Blumea. 2003; 48: 421–462. [Google Scholar]
- 45.Olesen JM. Flower mining by moth larvae vs. pollination by beetles and bees in the cauliflorous Sapranthus palanga (Annonaceae) in Costa Rica. Flora. 1992; 187: 9–15. [Google Scholar]
- 46.Westra LYT. Studies in Annonaceae. IV. A taxonomic revision of Tetrameranthus R. E. Fries. Proc K Ned Akad Wet C. 1985; 88: 449–482. [Google Scholar]
- 47.Westra LYT, Maas JM. Tetrameranthus (Annonaceae) revisited including a new species. Phytokeys. 2012; 12: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murray NA. Revision of Cymbopetalum and Porcelia (Annonaceae). Syst Bot Monogr. 1993; 40: 1–121. [Google Scholar]
- 49.Zhou LL, Su YCF, Saunders RMK. Molecular phylogenetic support for a broader delimitation of Uvaria (Annonaceae), inclusive of Anomianthus, Cyathostemma, Ellipeia, Ellipeiopsis and Rauwenhoffia. Syst and Biodivers. 2009; 7: 249–258. [Google Scholar]
- 50.Keßler PJA. Revision der Gattung Orophea Blume (Annonaceae). Blumea. 1988; 33: 1–80. [Google Scholar]
- 51.Keßler PJA. Studies on the tribe Saccopetaleae (Annonaceae)—II. Additions to the genus Orophea Blume. Blumea. 1990; 34: 505–516. [Google Scholar]
- 52.Xue B, Su YCF, Thomas DC, Saunders RMK. Pruning the polyphyletic genus Polyalthia (Annonaceae) and resurrecting the genus Monoon. Taxon. 2012; 61: 1021–1039. [Google Scholar]
- 53.Pang CC, Scharaschkin T, Su YCF, Saunders RMK. Functional monoecy due to delayed anther dehiscence: a novel mechanism in Pseuduvaria mulgraveana (Annonaceae). PLoS ONE. 2013; 8: e59951 10.1371/journal.pone.0059951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chatrou LW, Pirie MD, Erkens RHJ, Couvreur TLP, Neubig KM, Abbott JR, et al. A new subfamilial and tribal classification of the pantropical flowering plant family Annonaceae informed by molecular phylogenetics. Bot J Linn Soc. 2012; 169: 5–40. [Google Scholar]
- 55.Pang CC, Saunders RMK. The evolution of alternative mechanisms that promote outcrossing in Annonaceae, a self-compatible family of early-divergent angiosperms. Bot J Linn Soc. 2014; 174: 93–109. [Google Scholar]
- 56.Saunders RMK. The diversity and evolution of pollination systems in Annonaceae. Bot J Linn Soc. 2012; 169: 222–244. [Google Scholar]
- 57.Xu J, Grumbine RE. Landscape transformation through the use of ecological and socioeconomic indicators in Xishuangbanna, Southwest China, Mekong Region. Ecol Indic. 2012; 36: 749–756. [Google Scholar]
- 58.IUCN. IUCN red list categories and criteria: version 3.1. Second edition Gland, Switzerland and Cambridge, UK: IUCN; 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files, with DNA sequences available for download from GenBank (http://www.ncbi.nlm.nih.gov/genbank/).