Abstract
Inorganic polyphosphate (poly P), a chain of hundreds of phosphate residues linked by ATP-like bonds, is found in every cell in nature and is commonly produced from ATP by poly P kinases (e.g., PPK1). Dictyostelium discoideum, the social slime mold, possesses a PPK activity (DdPPK1) with sequence similarity to bacterial PPKs. We find here a previously unrecognized PPK (DdPPK2) in D. discoideum with the sequences and properties of actin-related proteins (Arps) that are similar to muscle actins in size, properties, and globular-filamentous structural transitions. Significantly, the unique actin inhibitors, phalloidin and DNase I, also inhibit synthesis of poly P by DdPPK2. Thus, this particular Arp complex is an enzyme that can polymerize into an actin-like filament concurrent with its synthesis of a poly P chain in a fully reversible reaction.
Keywords: actin-related protein, Dictyostelium discoideum
Inorganic polyphosphate (poly P) is a chain of many tens or hundreds of phosphate residues linked by the same phosphoanhydride, “high-energy” bonds as in ATP. Poly P is produced abiotically in volcanic exudates and deep-sea steam vents but likely was a phosphorylating agent and catalyst of peptide-bond formation in prebiotic times (1). Most significantly, poly P is synthesized in every bacterial, fungal, plant, and animal cell. Long known but lacking any defined functions, poly P had been regarded a “molecular fossil” (2).
By isolating enzymes that make and use poly P and by manipulating genes, we have demonstrated that poly P is essential for bacterial responses to stresses and starvation and for survival (3). A family of enzymes, poly P kinases (PPKs), make poly P reversibly from the terminal phosphate of ATP: nATP ↔ poly Pn+ nADP (4). PPK1 has been found in nearly 100 bacterial species, including some 20 major pathogens (5–7). PPK1-null mutants of the pathogens are deficient in growth, motility, quorum sensing, biofilm formation, and virulence (6). The active kinase site of PPK1 of Pseudomonas aeruginosa, the agent of fatal pneumonia in cystic fibrosis and of septicemia in burn patients, is a target of effective antibiotics; the active site is conserved widely intact, as is that of Bacillus anthracis, the cause of anthrax (W. Xu and S. Lee, personal communication).
Resuming our earlier studies of poly P synthesis in eukaryotic cells (8), in which poly P was found to be abundant in the nuclei and vacuoles among subcellular organelles, we have focused on the slime mold Dictyostelium discoideum because it is the only eukaryote known to have a PPK homologous to bacterial PPK1. In addition, D. discoideum has a vacuole, the acidocalcisome that contains large amounts of poly P and is responsible for Ca2+ flux in cells; this vacuole is also prominent in Trypanosoma cruzi, Leishmania major, Toxoplasma gondii, the alga Chlamydomonas reinhardtii, and the prokaryote Agrobacterium tumefaciens (9–12).
M. Sims and E. R. Katz (unpublished data) first observed that resistance of D. discoideum mutants to polyene antibiotics mapped in a genetic locus homologous to bacterial PPK1 and that extracts had reduced levels of PPK activity. D. discoideum PPK1 (DdPPK1)† has an amino acid similarity of 43% and an identity of 31% to Escherichia coli PPK. We find that mutants of DdPPK1 (AX2M1) still retain poly P at a level of 15% of the wild type because of PPK activity at ≈30% of the DdPPK1 level (H. Zhang, unpublished work). The latter activity, DdPPK2, probably associated with the acidocalcisomes, can be extracted by strong salt at an elevated pH and purified to apparent homogeneity from lysates of the mutant lacking DdPPK1 or from wild-type cells.
Materials and Methods
Organisms and Growth Conditions. D. discoideum AX203a was grown in HL5 medium (13); D. discoideum AX2M1 was grown in HL5 supplemented with blasticidin (5 μg/ml).
Reagents. Sources were [γ-32P]ATP and [32Pi] from Amersham Biosciences; nonradiolabeled oligonucleotides, poly P, BSA, ovalbumin, DNase I, and phalloidin from Sigma; Mono Q and Superose 12 FPLC columns from Pharmacia LKB; protein standards for SDS/PAGE from Bio-Rad; silver staining kit from ICN; ADP, ATP, and the ATP bioluminescence assay kit CLS II from Roche Molecular Biochemicals.
DdPPK2 Assay. DdPPK2 activity measured the production of acid-insoluble [32P]-poly P essentially as described in ref. 4, with minor modifications. The reaction mixture (250 μl) contained 50 mM Hepes-KOH (pH 7.2), 50 mM ammonium sulfate, 4 mM MgCl2, 0.5 M KCl, 1 mM [γ-32P]ATP (2,000 cpm/nmol), 2 mM creatine phosphate, 20 μg/ml creatine kinase, 3 mM poly P (Pi residues) prepared as described in ref. 4, and DdPPK2 as indicated. After 15 min at 37°C, the reaction was stopped by the addition of 250 μl of 7% HClO4 and 50 μl of 10% BSA. Total [32P]-poly P was collected on a Whatman GF/C glass-fiber filter, washed with a solution of 1 M HCl and 0.1 M sodium pyrophosphate and then ethanol, and measured in a liquid scintillation counter. One unit of enzyme is defined as the amount that incorporates 1 pmol of [32P] into acid-insoluble poly P per minute at 37°C. Analysis of product of DdPPK2 used 20% PAGE with 7 M urea as described in refs. 8 and 14.
The DdPPK2 reverse reaction was assayed in a 0.1-ml reaction mixture (50 mM Tris·HCl, pH 7.4/40 mM ammonium sulfate/4 mM MgCl2/5 μM ADP/24,000 units of DdPPK2) incubated at 37°C for 30 min and then at 90°C for 2 min. The reaction mixture was diluted 1:100 in 100 mM Tris·HCl (pH 8.0) and 4 mM EDTA, of which 0.1 ml was added to 0.1 ml of luciferase reaction mixture. Luminescence was measured in a luminometer [Mono-light 2010 (Analytical Luminescence Laboratory, San Diego) or Topcount (Packard)]. A standard curve for ATP (1–5 pmol) in 100 mM Tris·HCl (pH 8.0) and 4 mM EDTA was used for comparison. Concentration of poly P is given in terms of Pi residues. [32P]poly P values are based on specific radioactivity and susceptibility to hydrolysis with exopolyphosphatase (PPX) (15).
Purification of DdPPK2 from D. discoideum AX2M1 Cells. Cells (6 liters) were grown in HL5 supplemented with blasticidin (5 μg/ml), harvested at a density of 2 × 106 cells per ml, centrifuged at 2,000 × g for 15 min, washed twice with 20 mM Tris·HCl (pH 7.4), and stored at –80°C. Cells were resuspended in buffer A {50 mM Tris·HCl, pH 7.4/2 mM DTT/2 mM MgCl2/0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)} and a mixture of protease inhibitors (Sigma IV, Sigma). Cells were then broken by sonication in a Branson Sonifier 250 (three pulses of 1 min each). Then, KCl was added to the crude extract to a final concentration of 0.8 M to solubilize the PPK activity and then centrifuged at 38,000 × g for 30 min. The soluble fraction contained 95% of the PPK activity. Ammonium sulfate was added to a level of 30% saturation, and the sample was centrifuged. The pellet was resuspended in buffer A (20 ml) and dialyzed twice against 2 liters of buffer B (20 mM Tris·HCl, pH 9/2 mM DTT/2 mM MgCl2/0.1% CHAPS); the elevated pH was crucial for preventing precipitation of the activity in the dialysis tube. Chromatography was conducted on an HPLC Mono Q column (5 ml) (Amersham Pharmacia); the flow-through was reloaded on the column three times to bind all of the PPK activity. Elution was performed with a continuous gradient of KCl (0–500 mM) in buffer B; PPK activity appeared at 500 mM KCl. A second Mono Q column was equilibrated with 500 mM KCl; elution was performed with KCl between 500 mM and 1 M; PPK activity appeared as a single peak.
Gel Filtration Chromatography. Gel filtration chromatography was performed on a Tosoh G2000 SW-XL column on a PerkinElmer 888-PECHROM in 50 mM Hepes-NaOH (pH 8), 1 mM DTT, 0.5 mM EDTA, and 150 mM Na2SO4. The column was previously loaded with standards of molecular mass in kDa (BioRad): thyroglobulin (670), bovine γ-globulin (158), chicken ovalbumin (44), equine myoglobin (17), and vitamin B12 (1.35).
Sequencing. The sample was loaded on an SDS/PAGE gel (12% acrylamide/bis) and stained with Coomassie blue. The band was cut from the gel and subjected to sequence analysis at the Harvard Microchemistry Facility (Cambridge, MA) by microcapillary, reverse-phase HPLC nanoelectrospray tandem MS (liquid chromatography–MS/MS) on an LCQ Deca XP Plus quadrupole ion trap mass spectrometer (Finnigan-MAT, San Jose, CA).
Electron Microscopy. Samples were applied to a 400-mesh carbon/formvar-coated Cu grid, allowed to settle for 30 sec, washed with two drops of water, and stained with uranyl acetate (1%) for 1 min. Samples were analyzed in a JEOL JEM-1230 transmission electron microscope at 80 kV; pictures were taken with a digital imaging camera (Gatan, Pleasanton, CA).
Precipitation Assays. The protein sample was dialyzed in 10 mM Tris·HCl (pH 7.4), 0.5 mM ATP, 0.5 mM DTT, and 20 μM MgCl2 before starting the reaction. DdPPK2 (5 μg) was assayed for poly P synthesis in 100 μl of reaction mixture (to which 2 mM EGTA had been added); the reaction was stopped by addition of HClO4 as described above. The conversion from the globular form (G) to the filamentous form (F) was followed by centrifugation at 45,000 rpm in an Optima TLX ultracentrifuge (Beckman Coulter) in a TL 100.2 rotor for 30 min. The supernatants were loaded on a 12% SDS/PAGE gel.
Results
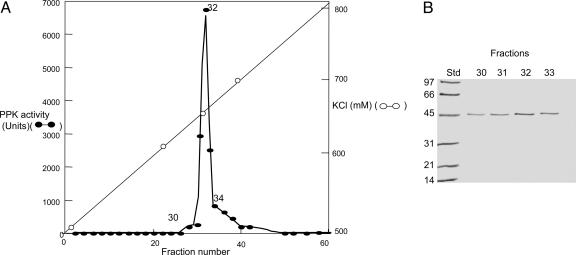
Isolation of DdPPK2 from D. discoideum. Purification procedures require the presence of high salt levels (e.g., 0.5 M KCl) throughout (Fig. 1 and Table 1). The peak fraction of poly P synthesis activity eluted from a Mono Q resin (Fig. 1 A), designated as DdPPK2, was obtained in 27% yield after a 975-fold purification and was judged to be a 43-kDa protein by silver staining of an SDS/PAGE gel (Fig. 1B); the single band coincides exactly with the eluted DdPPK2 activity. The specific activity of 70 × 103 units (pmol ATP converted to poly P/min/mg protein) is in the same range as found for several bacterial PPK1s (16). The purification, upon repetition, resulted in a PPK in the same yield, specific activity, and molecular weight. Gel-exclusion analysis at pH 8 in 50 mM Hepes-NaOH (pH 8), 1 mM DTT, 0.5 mM EDTA, and 150 mM Na2SO4 is consistent with a tetramer of 43-kDa subunits (data not shown). The concentrations of DdPPK2 referred to throughout assume a tetramer of 170 kDa. Under assay conditions, the enzyme has a filamentous structure (see below).
Fig. 1.
Mono Q chromatography of DdPPK2. (A) A single peak of PPK activity (fraction 32) was eluted from the resin with 600 mM KCl. (B) Samples showing PPK activity were analyzed by 12% SDS/PAGE and stained with silver; a single band of 43 kDa was observed. Poly P synthesis is expressed as pmol of Pi residues incorporated into poly P per min. Std, molecular markers (kDa).
Table 1. DdPPK2 purification table.
| Activity, units × 10-3 | Protein, mg | Specific activity, units × 10-3/mg | Purification, fold | |
|---|---|---|---|---|
| Extract | 54 | 650 | 0.08 | 1 |
| Ammonium sulfate | 23 | 165 | 0.14 | 1.7 |
| Mono Q, first column | 16 | 2.7 | 6.1 | 76 |
| Mono Q, second column | 14 | 0.2 | 70 | 975 |
DdPPK2 can also carry out the reverse reaction: poly Pn + ADP ↔ poly Pn–1 + ATP. However, the specific activity is 10-fold lower under the conditions tested. Phalloidin inhibits the reverse reaction at the same ratio as found in the forward reaction (see below).
Peptide Analysis. Peptides analyzed by MS identified a mixture of sequences for actin-related proteins: Arp1, Arp2, and an actin-like sequence annotated as DDB0188456 in the D. discoideum genome base, each at levels that appear to be 100- to 1,000-fold above those of any other peptides (Table 2).
Table 2. Peptides obtained by MS analysis.
| Arp1 |
| EEELLE |
| DLWVTRQEY |
| EKTCYVAHDPQ |
| VMGLNIENESTYFVGDR |
| SDLDLRKTFYSNIILGGG |
| IQPSEHPVLLTDVPNNPR |
| AGFAGQDPPSHIFQSLVGN |
| PVQPQYTLPDGNVIELGAER |
| Arp2 |
| LRSMLQITY |
| VGQERFQAS |
| CGFAGANFPTA |
| MIECMFEKYGQA |
| HMVFLGGAVLADL |
| ADRDLQQGFYQHITVLSGG |
| Actin-like protein |
| KVENVEI |
| VSDPTECK |
| SYELPDGQVITI |
| EEYDESGPSI |
| RGILTLKYPIERGIITN |
| AGFVGEEAPRSEFPS |
| ELLACSEGLFLPYVFFGTNLDGID |
| TPAIYCENQAVFSIYNSGRMT |
| LSYVSSSDFLWEIDD |
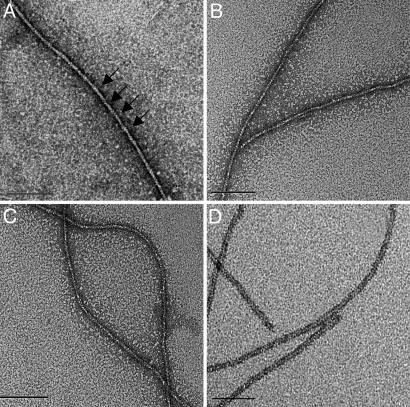
Electron Microscopy. Electron microscopy in 100 mM KCl and 0.5 mM ATP with uranyl acetate staining shows DdPPK2 (Fig. 2 A–C) to be similar to that of chicken muscle F-actin (filamentous actin) (Fig. 2D). As in muscle F-actin, a 36-nm repeat is apparent in the DdPPK2 filaments (shown by arrows in Fig. 2 A).
Fig. 2.
Electron microscopy of filaments of DdPPK2 (A–C) and chicken muscle actin (D). DdPPK2 samples were resuspended in 50 mM Tris·HCl (pH 7.4), 0.1 M KCl, and 0.5 mM ATP and were centrifuged at 20,000 rpm for 30 min in a TL100 ultracentrifuge (Beckman Coulter). All poly P synthesis activity was contained in the pellet. DdPPK2 and chicken muscle actin were resuspended in 50 mM Tris·HCl (pH 7.4), 0.1 M KCl, and 0.5 mM ATP and analyzed. As in muscle actin, a 36-nm repeat is apparent in the DdPPK2 filaments (see arrows in A). (Scale bar, 100 nm.)
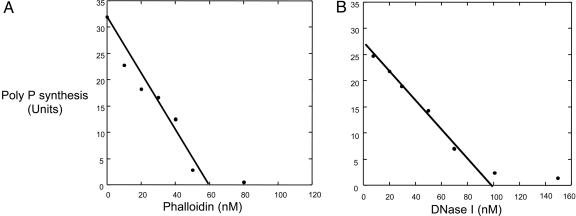
Phalloidin Inhibition. Phalloidin, a toxin from Amanita phalloides, binds G residues in the filament to stabilize it, stops ATP hydrolysis, and prevents the dissociation of the ADP-Pi intermediate. Addition of phalloidin to DdPPK2 promptly blocked poly P synthesis. Assuming that DdPPK2 is a tetramer, complete inhibition is seen at a mole ratio of phalloidin to DdPPK2 of between 1:1 and 1:2 (Fig. 3A). The reverse reaction, the utilization of poly P to make ATP from ADP, was inhibited in the same ratio as in the forward reaction (data not shown). In the inhibition by phalloidin of the common actins, the ratio to G-actin (globular actin) residues has been reported to be 1:1 (17).
Fig. 3.
Phalloidin and DNase I each inhibit DdPPK2 activity. DdPPK2 (0.1 μM, assuming a tetramer) was assayed for poly P synthesis with stoichiometric concentrations of phalloidin (A) and DNase I (B). A phalloidin–DdPPK2 molar ratio ≈1:1–1:2 achieved full inhibition; for DNase I the ratio was 1:1. Poly P synthesis is expressed as pmol of Pi residues incorporated into poly P per min.
DNase I Inhibition. DNase I binds free muscle G-actin monomers (Kd in the nM range) in an equimolar ratio and blocks their assembly into filaments; the concomitant ATP hydrolysis, intrinsically low on G-actin, is also blocked (18). Addition of DNase I to DdPPK2 also inhibited poly P synthesis at an equimolar ratio, assuming DdPPK2 to be a tetramer (Fig. 3B).
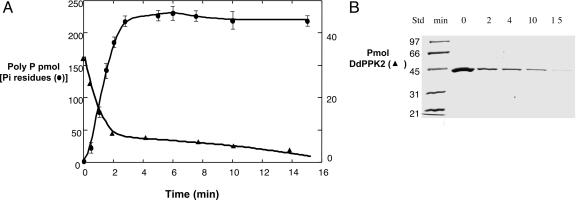
Kinetics of the DdPPK2 Reaction. The course of the reaction was followed by measuring the amount of poly P made and the amount of globular DdPPK2 that remained in the supernatant after high-speed centrifugation. Most of the poly P is produced in the first 2 min of the reaction. The ratio of poly P to DdPPK2 is ≈4:1 assuming a tetrameric state and 1:1 assuming that each subunit is active. This finding is consistent with poly P synthesis being a single turnover event coupled to assembly (Fig. 4).
Fig. 4.
Poly P synthesis linked to conversion of DdPPK2 oligomers to filaments. DdPPK2 (G form) was prepared by dialysis in 10 mM Tris·HCl (pH 7.4), 0.5 mM ATP, 0.5 mM DTT, and 20 μM MgCl2 before starting the reaction. DdPPK2 (5 μg) was assayed for poly P synthesis in 100 μl of reaction mixture; the reaction was stopped as described in Materials and Methods. The sample was centrifuged in an Optima TLX ultracentrifuge at 45,000 rpm in a TL 100.2 rotor for 30 min. The supernatant obtained at each time was loaded on 12% SDS/PAGE gel. (A) Poly P (residues of Pi) and G oligomers at indicated times. (B) SDS/PAGE (12%) of the supernatant fractions collected at indicated times. Std, molecular markers (kDa).
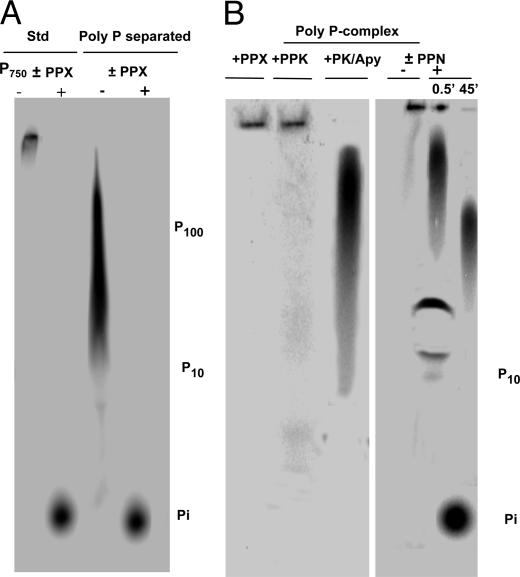
Complex of Nascent Poly P with the Actin-Like Filament. The poly P product of DdPPK2, when separated from the filament by acid or proteinase K, is made up of chains that range in size from 20 to 500 residues, much like the product of the DdPPK1 reaction; the chains are >95% hydrolyzed by ScPPX1, the PPX isolated from Sacharomyces cerevisiae, (Fig. 5A) but not by proteinase K, apyrase, or DNase I. The nascent chains of poly P and filament are associated as a complex as judged by the retardation of poly P on gel electrophoresis (Fig. 5B). In this complexed state, the poly P is unsusceptible to PPX or extension by PPK. However, the poly P can be hydrolyzed by endopolyphosphatase (PPN) (Fig. 5B). Thus, the ends of the nascent poly P chain core are masked, whereas the main body of the chain is not.
Fig. 5.
The poly P product of DdPPK2. (A) The separated poly P product of the ATP–DdPPK2 reaction was obtained as follows. The reaction mixture was digested with DNase I/RNase (20 μg/ml each) and apyrase (Apy) (0.5 units) for 30 min at 37°C and then heat-inactivated at 85°C. The sample was then digested with proteinase K (PK) (350 μg/ml) for 30 min at 37°C, extracted with phenol/chloroform, and precipitated with ethanol. PAGE analysis was performed as described in ref. 8 with [γ-32P]ATP, [32P]poly P750 (synthesized with E. coli PPK1), and [32Pi] as size markers. Electrophoresis was at 300 V until the dye was 7–8 cm from the origin. The migration distance by [γ-32P]ATP corresponds to a poly P chain length of ≈P10 (31). PPX was isolated from S. cerevisiae. (B) The poly P-complex product of the ATP–DdPPK2 reaction was obtained as follows. The reaction mixture was treated directly with PPX or PPK, PK (350 μg/ml), and Apy or was incubated with 1,000 units of yeast endopolyphosphatase (PPN) for various amounts of time.
Discussion
A previously unrecognized PPK has been found in D. discoideum, probably located in the acidocalcisome, a vacuole rich in poly P and Ca2+ and responsible for Ca2+ flux in cells. A similar vacuole is also present in pathogenic Trypanosomes, Leishmania, and Toxoplasma sp. and in the alga C. reinhardtii. The enzyme solubilized by salt has been purified 1,000-fold in 27% yield (Table 1). Analysis of the single 43-kDa band by MS yielded the remarkable result that the enzyme, DdPPK2, is likely a complex of three actin-related proteins: Arp1, Arp2, and an unreported Arpx (Table 2). Actins, a family of proteins conserved in all eukaryotes, interact with members of the myosin family and contribute to cellular motility (19). The 43-kDa globular form of actin (G) is converted reversibly to filaments (F) of hundreds of G monomers (20). Arps have been found that perform diverse and significant roles in processes of vesicle motility, mitosis, actin-filament dynamics, and modulation of chromatin structure (21, 22). These Arps show an amino acid identity of only ≈60% compared with the common muscle actin.
Arp1 is the major subunit in the dynactin complex, a multi-subunit complex initially identified as a factor that promotes dynein-mediated movement of membrane vesicles along microtubules. A remarkable feature of Arp1 is that it is the only actin-related protein known to polymerize into filaments (21, 23, 24); it does this slowly at a low ionic strength that does not support the filament formation of conventional actins. Under conditions that favor actin assembly, Arp1 polymerizes rapidly with no appreciable lag. Arp2 has been well characterized as part of the Arp2/3 complex, comprising two Arps and five other subunits and playing a role in cellular motility by initiating polymerization of new actin filaments (25).
It is important to note that the concentration of DdPPK2 used in the assays is 100 nM, a level at or below the critical concentration (Cc) of the conventional actin obtained from most organisms. By contrast, the Cc of Arp1 has been determined to be more than two orders of magnitude lower than actin Cc values (21), consistent with the low DdPPK2 concentrations that are active in the assays (Fig. 3).
Thus, DdPPK2 resembles the actin family in molecular weight, peptide sequence similarity, physical properties, filamentous structure, and the inhibition by phalloidin and DNase I. Furthermore, DdPPK2, as a small oligomer (possibly a tetramer under assay conditions), bypasses the rate-limiting initial steps by which monomeric G residues become trimers or tetramers before their rapid conversion to a filament.
The course of the G (small oligomer)-to-F transition, measured as the assembly of DdPPK2 into sedimentable filaments, is paralleled by the synthesis of poly P (Fig. 4). The poly P synthesis is a single-turnover reaction coupled to the assembly Arp (G) + nATP ↔ Arp (F) + poly Pn + nADP. Given that the ATP binding sites are well separated in the conventional actin filaments, the conformation of the DdPPK2 complex seems to be different enough to support the synthesis of poly P. The specific activity of DdPPK2 in the early course of the synthesis is 5 × 104 units/mg protein, a value observed in assays of the purified sample. These data, along with the actions of phalloidin and DNase I, suggest that filament formation and the synthesis of poly P are tightly linked. Studies are under way to knock out and overexpress the arp genes to determine the molecular mechanisms and structures of DdPPK2 and explain how poly P and filaments are concurrently assembled or disassembled.
With regard to the role of DdPPK2 in cellular processes, we speculate that in certain cellular locations and in response to metabolic needs, the polar growth of actin filaments may conserve ATP energy in the form of poly P as observed in DdPPK2.
A DdPPK2-like activity also appears to be present in C. reinhardtii, a microalga far up the phylogenetic tree from D. discoideum. Partial purification of a poly P-synthesizing activity from membranes of C. reinhardtii shows similar features to those found in DdPPK2 with regard to the salt dependence in the assay and phalloidin inhibition (unpublished data).
Recent studies have established that prokaryotes also possess and depend on skeletons of microfilaments that resemble actin and tubulin (26–28). These features determine cell shape and also are likely to be involved in a variety of cellular functions, some of which might depend on poly P. Of special interest is that in the synthesis of poly P by PPK1 of P. aeruginosa, filamentous assemblies are formed, and this poly P synthesis also is inhibited by phalloidin, although at a stoichiometry different from that for DdPPK2 (M.R.G.-G., H. Hayashi, and A.K., unpublished data).
It is important to note that DdPPK2 was obtained from D. discoideum AX2M1 membranes that contain the acidocalcisomes. Poly P synthesis measured in isolated vacuoles is inhibited 75% by 10 μM carbonylcyanide m-chlorophenylhydrazone (CCCP), a proton-motive-force (PMF) poison (unpublished data). Thus, the actions of the vesicular PMF-driven synthesis of the phosphoanhydride bonds of poly P resemble the synthesis of inorganic pyrophosphate (29) and ATP (30). The G-to-F transitions of DdPPK2 and poly P may play a vital role in the contractility and other motor functions of the D. discoideum vacuole.
Acknowledgments
We are indebted to Professor James Spudich for invaluable advice, Dr. William Lane (Harvard University, Cambridge, MA) for the DdPPK2 sequencing data, Dr. Alex Dunn (Stanford University) for the sample of chicken muscle actin, Dr. Xaobing Shi (Stanford University) for the sample of endopolyphosphatase, Dr. Haiyu Zhang (Stanford University) for the DdPPK1 mutant, and Michiko Hayashi for technical assistance. We are most grateful for the grants from the National Institutes of Health that made this work possible.
Abbreviations: Arp, actin-related protein; poly P, inorganic polyphosphate; PPK, poly P kinase; DdPPK, Dictyostelium discoideum PPK; PPX, exopolyphosphatase.
See Commentary on page 15825.
Footnotes
DdPPK1, of strain AX203a (GenBank accession no. AAD53165), has an amino acid similarity of 43% and an identity of 31% to E. coli PPK1. The null mutant (AX2M1) was used in these studies.
References
- 1.Kornberg, A. (1999) Prog. Mol. Subcell. Biol. 23, 1–18. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg, A., Rao, N. N. & Ault-Riché, D. (1999) Annu. Rev. Biochem. 68, 89–125. [DOI] [PubMed] [Google Scholar]
- 3.Rao, N. N. & Kornberg, A. (1999) Prog. Mol. Subcell. Biol. 23, 183–195. [DOI] [PubMed] [Google Scholar]
- 4.Ahn, K. & Kornberg, A. (1990) J. Biol. Chem. 265, 11734–11739. [PubMed] [Google Scholar]
- 5.Kim, K. S., Rao, N. N., Fraley, C. D. & Kornberg, A. (2002) Proc. Natl. Acad. Sci. USA 99, 7675–7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rashid, M. H. & Kornberg, A. (2000) Proc. Natl. Acad. Sci. USA 97, 4885–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rashid, M. H., Rumbaugh, K., Passador, L., Davies, D. G., Hamood, A. N., Iglewski, B. H. & Kornberg, A. (2000) Proc. Natl. Acad. Sci. USA 97, 9636–9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumble, K. D. & Kornberg, A. (1995) J. Biol. Chem. 270, 5818–5822. [DOI] [PubMed] [Google Scholar]
- 9.Marchesini, N., Ruiz, F. A., Vieira, M. & Docampo, R. (2002) J. Biol. Chem. 277, 8146–8153. [DOI] [PubMed] [Google Scholar]
- 10.Ruiz, F. A., Rodrigues, C. O. & Docampo, R. (2001) J. Biol. Chem. 276, 26114–26121. [DOI] [PubMed] [Google Scholar]
- 11.Seufferheld, M., Vieira, M. C., Ruiz, F. A., Rodrigues, C. O., Moreno, S. N. & Docampo, R. (2003) J. Biol. Chem. 278, 29971–29978. [DOI] [PubMed] [Google Scholar]
- 12.Ruiz, F. A., Marchesini, N., Seufferheld, M., Govindjee & Docampo, R. (2001) J. Biol. Chem. 276, 46196–46203. [DOI] [PubMed] [Google Scholar]
- 13.Kranke, J. & Kessin, R. (1977) Proc. Natl. Acad. Sci. USA 74, 2157–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sethuraman, A., Rao, N. N. & Kornberg, A. (2001) Proc. Natl. Acad. Sci. USA 17, 8542–8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wurst, H. & Kornberg, A. (1994) J. Biol. Chem. 269, 10996–11001. [PubMed] [Google Scholar]
- 16.Akiyama, M., Crooke, E. & Kornberg, A. (1992) J. Biol. Chem. 267, 22556–22561. [PubMed] [Google Scholar]
- 17.de la Cruz, E. M. & Pollard, T. D. (1994) Biochemistry 33, 14387–14392. [DOI] [PubMed] [Google Scholar]
- 18.Kabsch, W., Mannherz, H. G., Pai, E. F. & Holmes, K. C. (1990) Nature 347, 37–44. [DOI] [PubMed] [Google Scholar]
- 19.Sanger, J. W. & Sanger, J. M. (1992) Nature 375, 442. [DOI] [PubMed] [Google Scholar]
- 20.Korn, E. D., Carlier, M. & Pantaloni, F. D. (1987) Science 238, 638–644. [DOI] [PubMed] [Google Scholar]
- 21.Shen, X., Ranallo, R., Choi, E. & Wu, C. (2003) Mol. Cell 12, 147–155. [DOI] [PubMed] [Google Scholar]
- 22.Shafer, D. A. & Schroer, T. A. (1999) Annu. Rev. Cell Dev. Biol. 15, 341–363. [DOI] [PubMed] [Google Scholar]
- 23.Binham, J. B. & Schroer, T. A. (1999) Curr. Biol. 9, 223–226. [DOI] [PubMed] [Google Scholar]
- 24.Quintyne, N. J. & Schroer, T. A. (2002) J. Cell Biol. 159, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pollard, T. D. & Beltzner, C. C. (2002) Curr. Opin. Struct. Biol. 12, 768–774. [DOI] [PubMed] [Google Scholar]
- 26.Carballido-López, R. & Errington, J. (2003) Trends Cell Biol. 11, 577–583. [DOI] [PubMed] [Google Scholar]
- 27.Carballido-López, R. & Errington, J. (2003) J. Dev. Cell 4, 19–28. [DOI] [PubMed] [Google Scholar]
- 28.Jones, L. J., Carballido-López, R. & Errington, J. (2001) Cell 104, 913–922. [DOI] [PubMed] [Google Scholar]
- 29.Belogurov, G. A., Turkina, M. V., Penttinen, A., Huopalathi, S., Baykov, A. A. & Lathi, R. (2002) J. Biol. Chem. 277, 22209–22214. [DOI] [PubMed] [Google Scholar]
- 30.Boyer, P. (1997) Annu. Rev. Biochem. 66, 717–749. [DOI] [PubMed] [Google Scholar]
- 31.Ogawa, N., DeRisi, J. & Brown, P. O. (2000) Mol. Biol. Cell 11, 4309–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]