Abstract
The switch from an out-crossing to a self-fertilizing mating system is one of the most prevalent evolutionary trends in plant reproduction and is thought to have occurred repeatedly in flowering plants. However, little is known about the evolution of self-fertility and the genetic architecture of selfing. Here, we establish Arabidopsis thaliana as a model for genetic analysis of the switch to self-fertility in the crucifer family, where the ancestral out-crossing mode of mating is determined by self-incompatibility (SI), a genetic system controlled by the S locus. We show that A. thaliana ecotypes exhibit S-locus polymorphisms and differ in their ability to express the SI trait upon transformation with S-locus genes derived from the obligate out-crosser Arabidopsis lyrata. Remarkably, at least one ecotype was reverted to a stable, self-incompatible phenotype identical to that of naturally self-incompatible species. These ecotype differences are heritable and reflect the fixation in different A. thaliana populations of independent mutations that caused or enforced the switch to self-fertility. Their continued analysis promises to identify the loci that were the targets of natural selection for selfing and to contribute to a mechanistic understanding of the SI response.
The mating system adopted by a plant species determines the patterns of genetic variation in and between populations and thus has profound consequences for the rate and mode of evolutionary change (reviewed in ref. 1). Out-crossing has several well known advantages, and, accordingly, flowering plants have evolved several effective mechanisms that promote cross-fertilization. However, the selfing mode of reproduction is very common in flowering plants, and, apparently, it has evolved repeatedly in some taxa (2–5). Several explanations have been proposed to explain the evolution of self-fertilizing (autogamous) from out-crossing (allogamous) mating systems (3); these explanations include reproductive assurance when mating partners or pollinators are scarce (6, 7) and the transmission advantage of selfing (8). However, this issue has rarely been addressed empirically in genetically tractable systems, and the genetic events that underlie evolutionary switches to self-fertility are largely unknown.
The model plant Arabidopsis thaliana is a highly self-fertile species (9), with no reports of obligate out-crossing in any of its hundreds of accessions collected from various geographical locations. Nevertheless, an out-crossing mode of mating is thought to be the ancestral condition in the genus Arabidopsis and, more generally, in the crucifer (Brassicaceae) family. In this family, out-crossing is determined by a genetic self-incompatibility (SI) system controlled by haplotypes of the S locus (reviewed in ref. 10). In self-incompatible crucifers, the recognition and subsequent inhibition of self-related pollen by the stigma epidermis is effected through allele-specific interactions (11, 12) between two highly polymorphic S-locus-encoded proteins: the S-locus receptor kinase (SRK), a transmembrane protein displayed at the stigma epidermal surface, and the S-locus cysteine-rich protein (SCR), a small protein that is localized in the pollen coat. In self-pollination, SRK–SCR interaction leads to activation of the receptor and triggers a poorly understood signal transduction cascade within the stigma epidermal cell that results in inhibition of pollen hydration, germination, and tube growth (10).
Previous studies have supported the hypothesis that A. thaliana, which is thought to have diverged from its closest out-crossing relative, Arabidopsis lyrata, ≈5 million years ago (13, 14), was propelled into an autogamous mode of reproduction as a result of mutations that disrupt the SI system. Indeed, the A. thaliana accession Col-0 contains a nonfunctional S locus with inactive alleles of the SRK and SCR genes (15). Furthermore, transformation of A. thaliana Col-0 plants with just two transgenes, the SRKb and SCRb genes isolated from the Sb haplotype of A. lyrata, imparted a self-incompatible phenotype during a narrow window of floral development (16), namely, in very young flowers and in mature floral buds that are about to enter into anthesis (flower stages 13 and early stage 14, according to ref. 17). The SI phenotype of Col-0 transformants harboring the two transgenes was transient, however, and the ensuing breakdown of SI that accompanied floral maturation allowed self-pollination and resulted in ample seed set (16).
How self-fertility evolved in A. thaliana is a question of great interest. Recent studies have uncovered significant natural variation among A. thaliana geographical accessions (ecotypes) for many traits, including flowering time, circadian rhythm, response to light, seed size, and response to pathogens (18–20). However, little is known about the differentiation of ecotypes with respect to reproductive processes in general and pollination in particular. Because extant natural variation for a particular trait often reflects the evolutionary history of that trait, we initiated a survey of different A. thaliana ecotypes to assess the degree of polymorphism in genes required for SI and to determine whether some natural isolates of the species have retained the potential to sustain a stable SI response when transformed with functional SI recognition genes.
Materials and Methods
Plant Growth and Transformation. Plants of the A. thaliana ecotypes listed in Table 1 were grown in a growth chamber under standard conditions, with cold treatments to induce seed germination and flowering as required for each ecotype.
Table 1. The SI response in plants of seven A. thaliana ecotypes transformed with A. Iyrata SRKb and SCRb genes.
| Transformed ecotype (Arabidopsis Biological Resource Center stock no.) | No. of transgenic plants | Intensity of SI in stage-13 buds* | Self-pollination in mature flowers | Open-pollinated seed set |
|---|---|---|---|---|
| Col-0 (CS1092) | 15 | High | Compatible | + |
| Mt-0 (CS1380) | 10 | Low | Compatible | + |
| Nd-0 (Cs1390) | 10 | Low | Compatible | + |
| No (CS1394) | 12 | Low | Compatible | + |
| RLD (CS913) | 12 | High | Compatible | + |
| Ws-0 (CS915) | 10 | High | Compatible | + |
| C24 (CS906) | 20 | High | Incompatible | - |
High, 0-10 pollen tubes per self-pollinated stigma; Low, variable number of pollen tubes per self-pollinated stigma, ranging from <50 on some stigmas to >100 on others.
The previously described plant transformation vector, a pBIN-PLUS derivative containing the SRKb gene (16), was modified by insertion of a 1.1-kb fragment containing the SCRb gene. This fragment was amplified from a bacterial artificial chromosome derived from the Sb haplotype of A. lyrata (15) by using the primers 5′-GGGGTACCGACAGATTTGATTGGTTAGTT-3′ and 5′-GGGGTACCCCAGGCCTACATAGAACATGT-3′, which included terminal KpnI sites to facilitate cloning. The resulting SRKb–SCRb plasmid was used for Agrobacterium-mediated transformation of A. thaliana ecotypes by the floral dip method (21) or by the root explant method (22). Transformants were identified by selection on kanamycin.
Molecular Genetic Analysis of Transgenic Plants. The presence of the transgenes in primary transformants and their kanamycin-resistant progenies, the independent derivation of transgenic families, and the identification of transgenic plants in which the SRKb–SCRb transgenes were integrated at a single chromosomal locus were accomplished by DNA gel blot analysis as described in ref. 16. For plants expressing a developmentally stable SI response, homozygotes were generated and maintained over several generations by manual pollination of the self-compatible stigmas of immature floral buds. Plants homozygous for a transgene locus were identified as those plants that, upon selfing, produced only kanamycin-resistant progenies in ≈100 seedlings tested.
Expression of SRKb and SCRb was analyzed by gel blot analysis of pistil and anther poly(A)+ RNA, respectively, as described in ref. 15. Blotted RNA was hybridized first with an SRKb or SCRb probe and then with an actin probe. After normalization by using the actin hybridization signal, the relative amounts of SRKb and SCRb transcripts in SRKb–SCRb transformants and A. lyrata SaSb plants were quantitated with im-agequant software (Molecular Dynamics).
Pollination Assays. Developing floral buds were staged as described by Smyth et al. (17). For developmental studies of SI in SRKb–SCRb plants, pollination assays were performed by using stigmas from floral buds at various stages of maturation, from stage 12 through floral maturity. Stigmas were examined for absence of contaminating pollen under a stereoscope, and appropriate pollen was manually applied to their surface. Two hours after pollination, flowers were fixed for 30 min in a 3:1 mixture of ethanol and acetic acid, softened for 30 min in 1 M NaOH at 65°C, washed for 30 min in water, stained in decolorized aniline blue, and mounted on slides for examination by epifluorescence microscopy (23). Under these conditions, an incompatible response is typically manifested by the total or near-total absence of pollen grains on the stigma surface, because pollen grains that are not anchored by a pollen tube are dislodged during fixation, softening, and washing treatments. All pollination assays were performed by using three stigmas per replicate and were repeated on three different dates.
S-Locus Polymorphisms. Genomic DNA was isolated as described by Murray and Thompson (24) from leaf tissue of plants belonging to different ecotypes. Samples were screened by PCR using the following four pairs of primers derived from the Col-0 ψSRK and ψSCR sequences: (i) 5′-GGTTTCTTCAGAATC-CTTGGG-3′ and 5′-CAATGTGTCTTGCACTTCTC-3′, (ii) 5′-ATAGGTGCTGCGGTTGCTCTTGCT-3′ and 5′-ATA-ATCTTAAACCTTGAATCTT-3′, (iii) 5′-TGGGTCGGGAT-CACAAAAGAG-3′ and 5′-AAAACCCCGAAGCTGAAAA-CATC-3′, and (iv) 5′-TATTCCCCTTTTCGCTACC-3′ and 5′-ATAGGTGCTGCGGTTGCTCTTGCT-3′.
The products produced in successful amplifications were subjected to automated sequencing at the Cornell University BioResource Center. DNA gel blot analysis was performed by using a probe corresponding to the first exon of Col-0 ψSRK (At4g21370) and a probe corresponding to the Col-0 ψSCR sequence (15). To increase the chance of detecting polymorphisms, the DNA was digested with HincII, resulting in several restriction fragments that hybridize with the ψSRK probe in Col-0 DNA. After electrophoresis and transfer to nylon membranes, the DNA was hybridized at 65°C in 10% (wt/vol) dextran sulfate/330 mM sodium phosphate, pH 7.0/10 mM EDTA/5% (wt/vol) SDS. Washes were performed in a solution containing 0.2× SSC (1× SSC = 0.15 M NaCl/0.015 M sodium citrate, pH 7) and 0.1% (wt/vol) SDS at 65°C.
Results
Ecotype Differences in Expression of the SI Trait. The transgenic plants we had previously generated were produced by transforming plants of the Col-0 ecotype separately with the SRKb and SCRb genes and subsequently crossing individual SRKb and SCRb transformants to generate double transformants (16). To facilitate the production and analysis of transformants expressing SRKb and SCRb, we constructed a transformation vector that carried both genes in tandem along with a kanamycin-resistance selectable marker. This SRKb–SCRb construct was introduced into Col-0 and six other accessions, C24, Mt-0, Nd-0, No, RLD, and Ws-0, by Agrobacterium-mediated transformation (see Materials and Methods). Independent transgenic plants generated in these ecotypes, which varied from 10 to 20 transformants per ecotype (Table 1), were analyzed for the presence of the transgenes by DNA gel blot analysis or DNA amplification using PCR and for the SI phenotype by pollination assays and seed-set monitoring (see Materials and Methods).
Within an ecotype, independent transformants that expressed the SI trait were uniform with respect to the strength and developmental regulation of SI. Among ecotypes, no differences in expression of SI in pollen were observed, and the pollen of all transformants was inhibited on stigmas of SRKb plants but not on those of untransformed control plants (data not shown). However, significant differences were observed among ecotypes in the stigma, relative to both the strength of SI and the persistence of this response through floral maturation (Table 1). Transformants of three ecotypes (Col-0, RLD, and WS-0) exhibited a robust SI phenotype in stage-13 stigmas, with subsequent breakdown of SI in mature flower stigmas as described in ref. 16 for Col-0. Transformants of three other ecotypes (Mt-0, Nd-0, and No) exhibited weak SI in stage-13 stigmas and reversion to self-fertility in mature flower stigmas (Table 1).
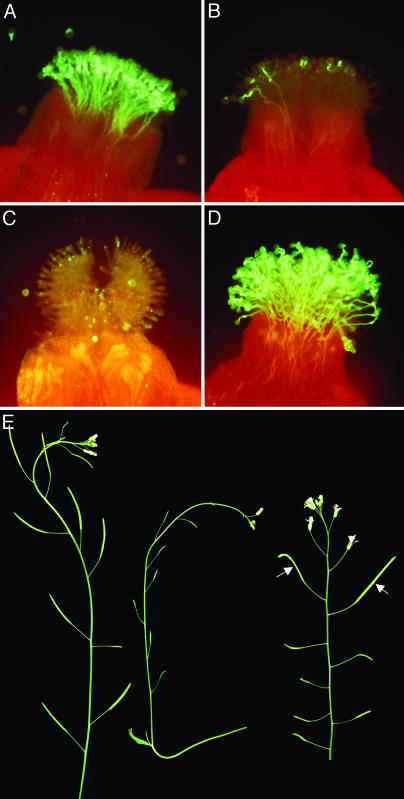
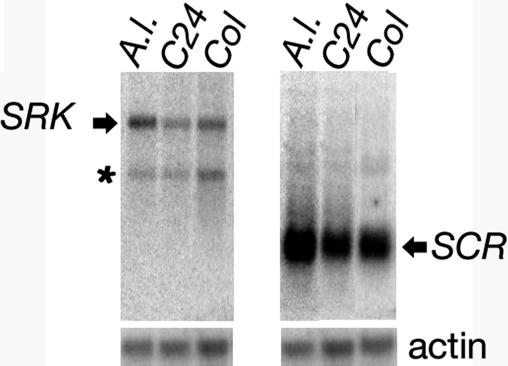
Only transformants of the C24 ecotype exhibited a robust and stable SI response that was identical to that of the A. lyrata donor in its developmental regulation (Table 1 and Fig. 1). In these plants, as in transformants of all other ecotypes, immature stage-12 stigmas were self-compatible (Fig. 1A), and the SI response was first evident in stage-13 stigmas (Fig. 1B). However, this SI response persisted in mature transgenic C24 flower stigmas, and these stigmas inhibited self-pollen tube development (Fig. 1C) but allowed the development of non-self-pollen tubes (Fig. 1D). This stable SI phenotype resulted in self-sterile plants (Fig. 1E) that produced few seeds and, consequently, exhibited delayed senescence relative to untransformed controls. Importantly, differences in the stability of the SI phenotype among A. thaliana accessions were not due to differences in the stability or accumulation of SRKb transcripts. As shown in Fig. 2, the steady-state levels of SRKb transcripts in C24 SRKb–SCRb transformants were no different from those of the Col-0 SRKb–SCRb transformants, and in both sets of transformants, SRKb transcript levels were only ≈40% of the levels observed in the stigmas of A. lyrata SaSb heterozygotes. Similarly, the levels of SCRb transcripts in anthers of Col-0 and C24 transformants were equivalent but lower than those in A. lyrata anthers (Fig. 2). The robust SI phenotype of C24 transformants indicates that these reduced transcript levels are above the threshold required for conferring SI in stigma and pollen.
Fig. 1.
SI in C24 SRKb–SCRb transformants. (A–D) Developmental regulation of the SI response. Self-pollination results in ample pollen tube growth in immature stage-12 stigmas (A) but not in stage-13 stigmas (B). Mature flower stigmas are self-incompatible (C) but are compatible with pollen from wild-type untransformed plants (D). (E) Inflorescences of transformants and wild-type untransformed plants. On the left is an inflorescence of a nontransformed plant with elongated, seed-filled siliques. Shown in the center is an inflorescence of a transformed plant with undeveloped, empty siliques. On the right is an inflorescence of a transformed plant in which manual self-pollination at stage 12 of floral development resulted in seed set (arrows).
Fig. 2.
Expression of the SRKb and SCRb transgenes in SRKb–SCRb transformants. The RNA gel blots show SRKb transcripts in the pistils (Left) and SCRb transcripts in the anthers (Right) of A. lyrata SaSb (A.l.) and A. thaliana C24 and Col-0 SRKb-SCRb transformants. In the SRK panel, the arrow indicates the full-length 3.0-kb SRKb transcript, and the * indicates the alternative 1.6-kb transcript that corresponds to the first exon of SRKb (16). Hybridization with an actin probe served as a loading control.
Genetic Analysis of Ecotype Differences. To initiate a study of the genetic basis of ecotype differences in expression of SI, we identified two independent transformants carrying single transgene integrations for each of Col-0, RLD, and C24. From these primary transformants, transgenic homozygous SRKb–SCRb lines were generated and propagated, by forced self-pollination in the immature bud stage in the case of the C24 transformants (see Materials and Methods). After determining that the SI phenotypes expressed by the various ecotypes were maintained over the four generations analyzed, a C24 SRKb–SCRb homozygote was crossed to untransformed plants belonging to the Col-0, Mt-0, Nd-0, No, RLD, and WS-0 accessions. In all crosses, F1 plants exhibited stable SI, demonstrating the recessive nature of the unstable SI phenotype. In F2 populations, kanamycin resistance segregated in a 3:1 ratio as expected, and the kanamycin-resistant progenies all expressed SI in stage-13 stigmas, albeit at different levels, but they segregated for stability of SI in mature flower stigmas. In all crosses except the cross to RLD, the ratios of plants exhibiting stable and unstable SI were consistent with segregation at two or more loci required for stable SI. Only in the C24 SRKb–SCRb × RLD cross did kanamycin-resistant F2 progenies exhibit a 3:1 (231:76) ratio of stable-SI and unstable-SI plants, indicative of the segregation of a recessive RLD-derived allele at a single locus. We conclude that the C24 ecotype carries dominant functional alleles at several “SI-modifier” loci required for the stable SI phenotype, whereas the other ecotypes analyzed carry different combinations of loss-of-function alleles of these genes.
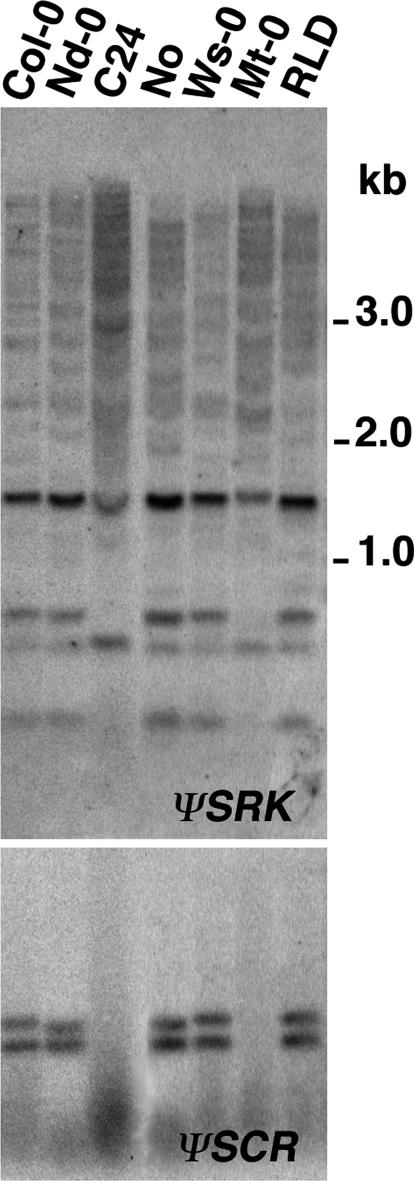
S-Locus Polymorphisms in A. thaliana. The ecotypes used for transformation with the SRKb–SCRb genes were also found to be polymorphic at the S locus, but these polymorphisms bore no correlation to the SI phenotypes exhibited by SRKb–SCRb transformants or the geographical origin of the ecotypes. Amplification with Col-0-derived primers and sequence analysis of amplified products (see Materials and Methods; data not shown), as well as hybridization with Col-0 ψSRK and ψSCR probes (Fig. 3), indicated that Col-0, Nd-0, No, Ws-0, and RLD share the same S haplotype. In contrast, C24 and Mt-0 are inferred to share a different S haplotype on the basis of similar hybridization patterns (Fig. 3) and failure of amplification with Col-0-derived primers. Interestingly, the species as a whole appears to harbor several distinct S haplotypes, and hybridization of 20 other ecotypes with Col-0 ψSRK identified at least eight additional hybridization patterns (Table 2 and Fig. 4, which are published as supporting information on the PNAS web site).
Fig. 3.
S-haplotype polymorphisms in A. thaliana. Gel blots of genomic DNA from various ecotypes digested with HincII and hybridized with ψSRK (Upper) and ψSCR (Lower) probes derived from the Col-0 S locus. Note that C24 and Mt-0 exhibit a ψSRK hybridization pattern that differs from that of the other ecotypes and that they do not hybridize with the Col-0 ψSCR probe.
Discussion
Our results demonstrate that although all known A. thaliana accessions are self-fertile with no distinguishing feature of their pollination biology, these natural populations exhibit polymorphisms in genes required for the out-crossing mode of mating. In addition to S-locus polymorphisms, they harbor hidden variation for expression of the SI trait, which can be uncovered only after transfer of S-locus recognition genes from self-incompatible A. lyrata. All ecotypes that were transformed with SRKb–SCRb must carry nonfunctional S haplotypes because they show no evidence of SI at any stage of stigma development in the absence of the SRKb and SCRb transgenes. The full reversion of the C24 ecotype to the ancestral out-crossing mode of mating demonstrates that inactivation of the S locus was the single mutational event that caused self-fertility in this ecotype. In contrast, other ecotypes have experienced additional loss-of-function mutations at one or more SI-modifier genes that cause SI to break down in mature flowers and, in some cases, to become attenuated in stage-13 floral buds. The SI response, like other receptor-mediated signaling systems, involves the formation of complexes between receptor, ligand, and possibly other proteins and also requires the activity of largely unknown elements of a downstream signaling cascade (10). Thus, selection for mutations that promote self-fertility in natural populations is expected to act not only on the S-locus recognition genes but also on various components of the signaling complex and response pathway, some of which might be encoded by the SI-modifier loci identified in this study.
The fixation of independent mutations in genes required for SI in different natural populations and the existence of at least one ecotype that still retains functional alleles for all genes required for the expression of a robust and stable SI response suggest that the conversion of A. thaliana to self-fertility is of relatively recent origin. Although the spread of self-fertility alleles in a largely self-incompatible population requires that the benefits of selfing outweigh the costs associated with increased homozygosity and inbreeding depression, alleles that promote selfing can have a selective advantage under appropriate ecological and genetic conditions (25, 26). Thus, stochastic mutations that cause or enforce self-fertility might have arisen in distinct small populations of A. thaliana. Once a mutation causes loss of SI in a particular population, there would be no selective advantage for maintenance of functional alleles at genes with SI-specific functions, allowing the occurrence of secondary mutations in these genes. The primary mutation might inactivate either the S locus or an SI-modifier gene, and the nature, number, and timing of secondary mutations would differ in different populations. Thus, it is possible that some ecotypes carry mutations at one or more SI-modifier genes while retaining a functional S locus, and such an S haplotype might be uncovered in a specieswide survey of SRK and SCR alleles. In any case, continued analysis of A. thaliana ecotypes for S-locus polymorphisms and natural variation in expression of the SI trait conferred by the SRKb–SCRb transgenes presents us with a unique opportunity to investigate the genetics of inbreeding in a genetically tractable species. This analysis, coupled with future isolation of the SI-modifier genes, should provide insight into the molecular events that underlie the evolutionary switch to autogamy in the A. thaliana lineage and also help elucidate the mechanism of self-recognition in the SI response of crucifers.
Supplementary Material
Acknowledgments
We thank Andrew Clark for providing valuable comments on the manuscript. This work was supported by a grant from the National Science Foundation. Seed for various ecotypes was obtained from the Arabidopsis Biological Resource Center at Ohio State University, which is funded by the National Science Foundation.
Author contributions: M.E.N. and J.B.N. designed research; M.E.N., P.L., S.S.-B., N.A.B., and J.B.N. performed research; M.E.N., P.L., S.S.-B., and J.B.N. analyzed data; and M.E.N. and J.B.N. wrote the paper.
Abbreviations: SCR, S-locus cysteine-rich protein; SI, self-incompatibility; SRK, S-locus receptor kinase.
References
- 1.Charlesworth, D. & Wright, S. I. (2001) Curr. Opin. Genet. Dev. 11, 685–690. [DOI] [PubMed] [Google Scholar]
- 2.Stebbins, G. L. (1957) Am. Nat. 91, 337–354. [Google Scholar]
- 3.Jain, S. K. (1976) Annu. Rev. Ecol. Syst. 7, 469–495. [Google Scholar]
- 4.Lloyd, D. G. (1992) Int. J. Plant Sci. 153, 370–380. [Google Scholar]
- 5.Barrett, S. C. H. (2002) Nat. Rev. Genet. 3, 274–284. [DOI] [PubMed] [Google Scholar]
- 6.Darwin, C. R. (1876) The Effects of Cross and Self-Fertilization in the Vegetable Kingdom (John Murray, London).
- 7.Kalisz, S., Vogler, D. W. & Hanley, K. M. (2004) Nature 430, 884–887. [DOI] [PubMed] [Google Scholar]
- 8.Fisher, R. A. (1941) Ann. Eugen. 11, 53–63. [Google Scholar]
- 9.Abbott, R. J. & Gomes, M. F. (1989) Heredity 62, 411–418. [Google Scholar]
- 10.Kachroo, A., Nasrallah, M. E. & Nasrallah, J. B. (2002) Plant Cell 14, Suppl., S227–S238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kachroo, A., Schopfer, C. R., Nasrallah, M. E. & Nasrallah, J. B. (2001) Science 293, 1824–1826. [DOI] [PubMed] [Google Scholar]
- 12.Takayama, S., Shimosato, H., Shiba, H., Funato, M., Che, F. S., Watanabe, M., Iwano, M. & Isogai, A. (2001) Nature 413, 534–538. [DOI] [PubMed] [Google Scholar]
- 13.Koch, M., Haubold, B. & Mitchell-Olds, T. (2000) Mol. Biol. Evol. 17, 1483–1498. [DOI] [PubMed] [Google Scholar]
- 14.Koch, M., Haubold, B. & Mitchell-Olds, T. (2001) Am. J. Bot. 88, 534–544. [PubMed] [Google Scholar]
- 15.Kusaba, M., Dwyer, K. D., Hendershot, J., Vrebalov, J., Nasrallah, J. B. & Nasrallah, M. E. (2001) Plant Cell 13, 627–643. [PMC free article] [PubMed] [Google Scholar]
- 16.Nasrallah, M. E., Liu, P. & Nasrallah, J. B. (2002) Science 297, 247–249. [DOI] [PubMed] [Google Scholar]
- 17.Smyth, D. R., Bowman, J. L. & Meyerowitz, E. M. (1990) Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alonso-Blanco, C. & Koornneef, M. (2000) Trends Plant Sci. 5, 22–29. [DOI] [PubMed] [Google Scholar]
- 19.Maloof, J. N. (2003) Curr. Opin. Genet. Dev. 13, 576–582. [DOI] [PubMed] [Google Scholar]
- 20.Bergelson, J., Stahl, E., Dudek, S. & Kreitman, M. (1998) Genetics 148, 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clough, S. J. & Bent, A. (1998) Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- 22.Valvekens, D., Van Montagu, M. & Van Lijsebettens, M. (1988) Proc. Natl. Acad. Sci. USA 85, 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kho, Y. O. & Baer, J. (1968) Euphytica 17, 298–302. [Google Scholar]
- 24.Murray, M. G. & Thompson, W. F. (1980) Nucleic Acids Res. 8, 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jarne, P. & Charlesworth, D. (1993) Annu. Rev. Ecol. Syst. 24, 441–466. [Google Scholar]
- 26.Uyenoyama, M. K., Holsinger, K. E. & Waller, D. M. (1993) Oxford Surv. Evol. Biol. 9, 327–381. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.