Abstract
Alternative splicing is thought to be regulated by nonspliceosomal RNA binding proteins that modulate the association of core components of the spliceosome with the pre-mRNA. Although the majority of metazoan genes encode pre-mRNAs that are alternatively spliced, remarkably few splicing regulators are currently known. Here, we used RNA interference to examine the role of >70% of the Drosophila RNA-binding proteins in regulating alternative splicing. We identified 47 proteins as splicing regulators, 26 of which have not previously been implicated in alternative splicing. Many of the regulators we identified are nonspliceosomal RNA-binding proteins. However, our screen unexpectedly revealed that altering the concentration of certain core components of the spliceosome specifically modulates alternative splicing. These results significantly expand the number of known splicing regulators and reveal an extraordinary richness in the mechanisms that regulate alternative splicing.
Pre-mRNA splicing involves the removal of introns and ligation of the flanking exons. This reaction is catalyzed by the spliceosome, a macromolecular machine composed of five RNAs and hundreds of proteins (1). Alternative splicing generates multiple mRNAs from a single gene, thus increasing proteome diversity (2). Alternative splicing also plays a key role in the regulation of gene expression in many developmental processes ranging from sex determination to apoptosis (3), and defects in alternative splicing have been linked to many human disorders (4). In general, alternative splicing is regulated by proteins that associate with the pre-mRNA and function to either enhance or repress the ability of the spliceosome to recognize the splice site(s) flanking the regulated exon (5). Whether a particular alternative exon will be included or excluded from an mRNA in each cell is thought to be determined by the relative concentration of a number of positive and negative splicing regulators and the interactions of these factors with the pre-mRNA and components of the spliceosome (5).
Although at least 74% of human genes encode alternatively spliced mRNAs (6), relatively few splicing regulators have been identified. Much of our insight into the mechanisms of splicing regulation was initially obtained by genetic analysis of the sex determination pathway in Drosophila (3). These experiments have identified three proteins, Sex-lethal (SXL), Transformer (TRA), and Transformer 2 (TRA2), that tightly regulate the alternative splicing of five genes, Sex-lethal, transformer, male specific lethal-2, fruitless, and doublesex. Subsequent biochemical experiments helped to elucidate the mechanisms by which SXL, TRA, and TRA2 function in this pathway. Aside from these examples, a simple genetic system to analyze specific alternative splicing events has not been available. Here we describe an RNA interference (RNAi) screen in cultured Drosophila cells designed to identify RNA-binding proteins that regulate alternative splicing of pre-mRNAs transcribed from endogenous genes.
Methods
Construction of a Drosophila RNA-Binding Protein Double-Stranded RNA (dsRNA) Library. Gene-specific primers were designed to amplify 300 Drosophila melanogaster genes from Flybase release 3.0 that are annotated as encoding proteins containing known RNA binding motifs. RT-PCR was performed with these primers to amplify cDNA fragments from total RNA isolated from S2 cells. PCR products were cloned into pCRII-TOPO vector (Invitrogen) and sequenced. The sequence of each cDNA fragment was blast searched against the D. melanogaster genome to ensure that no other genes shared regions of at least 19 consecutive nucleotides of identity. In total, 250 genes were cloned and are summarized in the supporting information, which is published on the PNAS web site.
Each vector was used as a PCR template with M13 forward and reverse primers to generate dsRNA transcription templates. The PCR products were separately transcribed with T7 and SP6 RNA polymerase (AmpliScribe kits, Epicentre Technologies, Madison, WI) in 20-μl reactions, followed by DNase I treatment to remove the template DNA. Integrity of both the PCR products and single-stranded RNA was monitored by agarose gel electrophoresis. The two RNA strands were annealed in 100 mM NaCl/20 mM Tris·HCl, pH 8.0/1 mM EDTA by incubation at 78°C for 10 min followed by a slow cooling at room temperature for 30 min.
RNAi Screen. S2 cells were grown at 27°C in Drosophila-SFM (Invitrogen), supplemented with 1× penicillin/streptomycin and 2 mM l-glutamine. Twenty micrograms of each dsRNA was added to ≈1 × 106 cells in six-well culture dishes. After 2 days, an additional 20 μg of dsRNA was added to each well, and the cells were incubated for 2 additional days. After a total of 4 days, total RNA was isolated by using TRIzol (Invitrogen) and subjected to reverse transcription with SuperScript II (Invitrogen). PCR was used to amplify the alternatively spliced regions. In each case, one of the two primers was 32P-end-labeled, and the PCR products were analyzed by PAGE or single-stranded conformation polymorphism gel electrophoresis (7) and quantitated by using the Cyclone Phosphorimager System (PerkinElmer). Each dsRNA that significantly affected at least one alternative exon was considered as a positive and was repeated in a second round of screening. All data from the dsRNA-treated cells were compared to untreated cells and analyzed by using cluster 3.0 and java treeview run on Macintosh osx. In some cases, the efficiency of RNAi was monitored by Western blotting or RT-PCR (see supporting information).
Analysis of Down Syndrome Cell Adhesion Molecule (Dscam) Splicing in ps and mushroom body expressed (mub) Mutant Flies. The ps2 (ps2/TM6B-Ubx-lacZ) flies were obtained from D. Andrew (The Johns Hopkins University, Baltimore), and mub04093 (P mub04093ry506/TM3,ryRKSb1Ser-1) flies were obtained from the Bloomington Stock Center (Bloomington, IN). mub04093 females were crossed to ps2 males to generate +/+, +/ps2, +/mub04093, and mub04093/ps2 flies. Total RNA was isolated from males of each genotype by using TRIzol, and Dscam exon 4 alternative splicing was analyzed by RT-PCR as described above.
Results and Discussion
An RNAi Screen to Identify Splicing Regulators. The Drosophila genome (8) contains ≈300 genes that encode proteins containing known RNA-binding motifs (9) and several additional genes that encode other components of the spliceosome (10). We cloned 250 cDNA fragments representing ≈70% of known Drosophila RNA-binding proteins and several additional spliceosomal components (see supporting information). Each clone was then used as a template for dsRNA synthesis. After treating Drosophila S2 cells with each of the 250 dsRNAs, we performed RT-PCR to examine the splicing of 19 alternative exons in three different endogenous genes. Each dsRNA was tested once in the first round of screening; all dsRNAs that significantly altered the splicing of at least one exon were retested with newly synthesized dsRNA. Each dsRNA that tested positive in the first screen yielded qualitatively similar results in the second screen. Because very few antibodies were available to the targeted proteins, in most cases we did not monitor the efficiency with which each targeted protein was depleted. However, in those cases that we did examine, by either Western blotting or RT-PCR, the RNAi was found to be efficient (see supporting information).
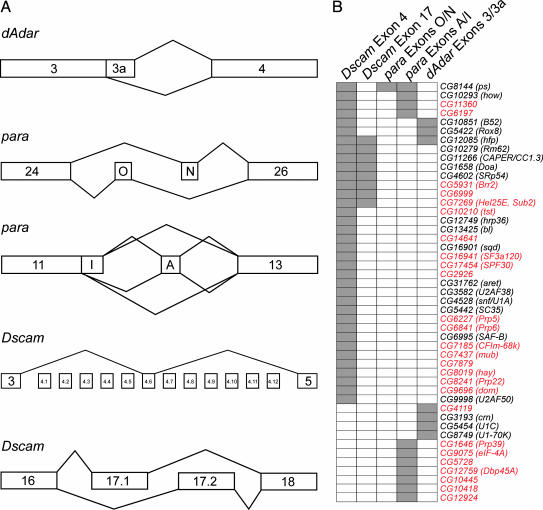
We chose three functionally diverse genes, dAdar, paralytic (para), and Dscam, to monitor alternative splicing (Fig. 1A). Each gene is expressed both in S2 cells and the nervous system of the fly, and each encodes numerous alternatively spliced mRNAs. In addition, these genes represent many of the different types of alternative splicing patterns, including alternative 5′ splice sites, cassette exons, and mutually exclusive exons. Moreover, two of the genes, para and Dscam, contain multiple alternatively spliced regions within the same pre-mRNA, providing the opportunity to examine the possibility that specific proteins may coordinately regulate the splicing of multiple exons in a single pre-mRNA. The first gene we chose, dAdar, encodes an adenosine deaminase that functions in RNA editing (11). dAdar contains two alternative 5′ splice sites in exon 3, of which the downstream 5′ splice site (3a) contains a C at position +2 (underlined) in the intron rather than the nearly invariant U (GA/GCAAGU). The second gene chosen, para, encodes a voltage-gated action potential sodium channel, contains 13 alternative exons, and can potentially generate 1,536 different mRNA isoforms (12). We examined the splicing of two different regions of para. The first encompasses two cassette exons, O and N, which are alternatively spliced in a mutually exclusive manner. The second region encompasses exons A and I, which represent a cassette exon and an alternative 5′ splice site, respectively. The final gene we examined, Dscam, encodes an axon guidance molecule and can generate 38,016 different isoforms (13). In this study, we examined the alternative splicing of the exon 4 cluster, which contains 12 mutually exclusive alternative exons, and the exon 17 region, which contains two mutually exclusive alternative exons. As a control, we have also examined the splicing of a constitutive intron in the housekeeping gene GAPDH (see supporting information).
Fig. 1.
Overview of the RNAi screen. (A) Organization of the alternatively spliced regions analyzed. In the dAdar gene, the alternative splicing of exon 3a, which involves alternative 5′ splice site selection, was analyzed. In the para gene, alternative splicing of two regions, exons O and N and exons A and I, was analyzed. In Dscam, the exon 4 and 17 clusters, which contain 12 and 2 mutually exclusive exons, respectively, were analyzed. (B) Overview of the regulatory proteins identified and the alternative splicing events they control. Each dsRNA that had an effect on the splicing of at least one of the five regions analyzed is shown on the left. The dsRNAs are organized based on the alternatively spliced regions they affect. Genes that have not previously been shown to be involved in the regulation of alternative splicing are highlighted in red. These data show that (i) none of the dsRNAs affected the splicing of all pre-mRNAs tested, (ii) the alternative splicing of different regions in the same pre-mRNA can be coordinately regulated by the same proteins, and (iii) different alternatively spliced regions within the same pre-mRNA can also be regulated by distinct proteins.
Of the 250 proteins targeted by RNAi, 47 influenced alternative splicing of at least one exon (Fig. 1B). Of the proteins that had an effect, 26 had not previously been implicated in alternative splicing (Fig. 1B). Importantly, none of the dsRNAs affected the alternative splicing of all pre-mRNAs tested, and splicing of a constitutive intron in GAPDH was not affected by any of the dsRNAs (see supporting information). Thus, depletion of these proteins by RNAi did not result in global splicing defects. Rather, each of the 47 proteins identified had a specific effect on the splicing of individual alternative exons.
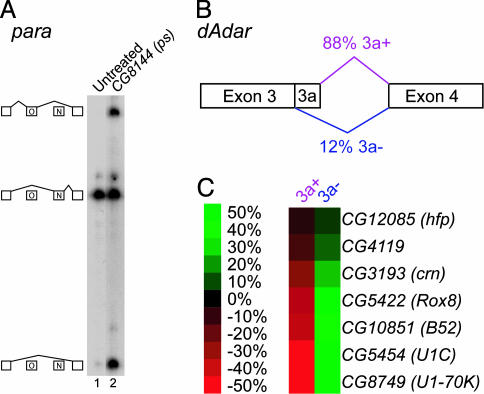
We found striking differences in the number of proteins that affected the splicing of the five different alternatively spliced regions examined. Remarkably, alternative splicing of para exons O and N was affected by only one of the 250 dsRNAs tested, pasilla (ps) (Fig. 2A). In untreated S2 cells, 95% of para transcripts include exon N, <0.5% of transcripts include exon O, and 5% of transcripts skip both alternative exons. Treatment with ps dsRNA dramatically increased the abundance of the exon O and exon-skipping isoforms. Thus, PS appears to function as a regulator of para alternative splicing, but may also control the stability of different mRNA isoforms. PS is most similar to human Nova-1 and Nova-2, which are the targets of the autoimmune neurodegenerative syndrome paraneoplastic opsoclonus-myoclonus ataxia (POMA) (14) and have been shown to regulate alternative splicing by binding to intronic YCAY repeats. PS most likely has the same RNA-binding preference as Nova-1 and Nova-2, because every amino acid within Nova that is directly involved in sequence-specific contacts with the RNA is identical or highly conserved in PS (see supporting information). Interestingly, YCAY repeats that are conserved in several Drosophila species are located in the intron downstream of exon N (data not shown).
Fig. 2.
RNA binding proteins involved in alternative splicing of para and dAdar.(A) Single-stranded conformation polymorphism gel of RT-PCR products from untreated S2 cells (lane 1) and cells treated with ps dsRNA (lane 2). The identity of each band is shown schematically on the left side of the gel. (B) The dAdar gene in the region containing exon 3a. Normally, 88% of transcripts include exon 3a in Drosophila S2 cells. (C) Representation of the effect of each dsRNA on dAdar alternative splicing. The data are represented as the change in the percentage of each isoform in treated cells compared to untreated cells. The magnitude of each effect is indicated by the color scale, where green indicates an increase in the relative abundance of an isoform and red indicates a decrease in the relative abundance of an isoform.
Our screen identified seven proteins that are required for utilization of the dAdar exon 3a 5′ splice site (Fig. 2 B and C). Of these, CG10851 (B52), CG8749 (U1–70K), CG5454 (U1C), and CG5422 (Rox8) are particularly intriguing, given their known RNA-binding preferences and protein interaction properties. Rox8, the ortholog of human TIA-1, binds to U-rich intronic sequences and promotes the binding of U1 small nuclear ribonucleoprotein (snRNP) to weak 5′ splice sites (15). Indeed, the nonconsensus 5′ splice site of exon 3a is followed by a U-rich sequence, and both of these elements are highly conserved in a variety of Drosophila species (T. Koyejo and R.A.R., unpublished data). U1C is a component of U1 snRNP, and has been shown to physically interact with Rox8/TIA-1 (15). U1–70K is another component of U1 snRNP that has been shown to interact with serine/arginine-rich (SR) proteins such as B52 (16). SR proteins, in turn, function by binding to exon sequences, where they interact with and recruit general splicing factors to adjacent splice sites (17). Thus, our results suggest that a network of protein–RNA and protein–protein interactions involving Rox8 and B52 function to recruit U1 snRNP to the weak 5′ splice site of exon 3a.
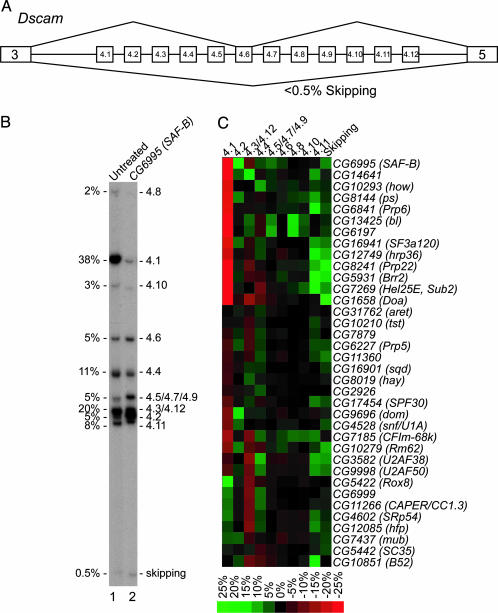
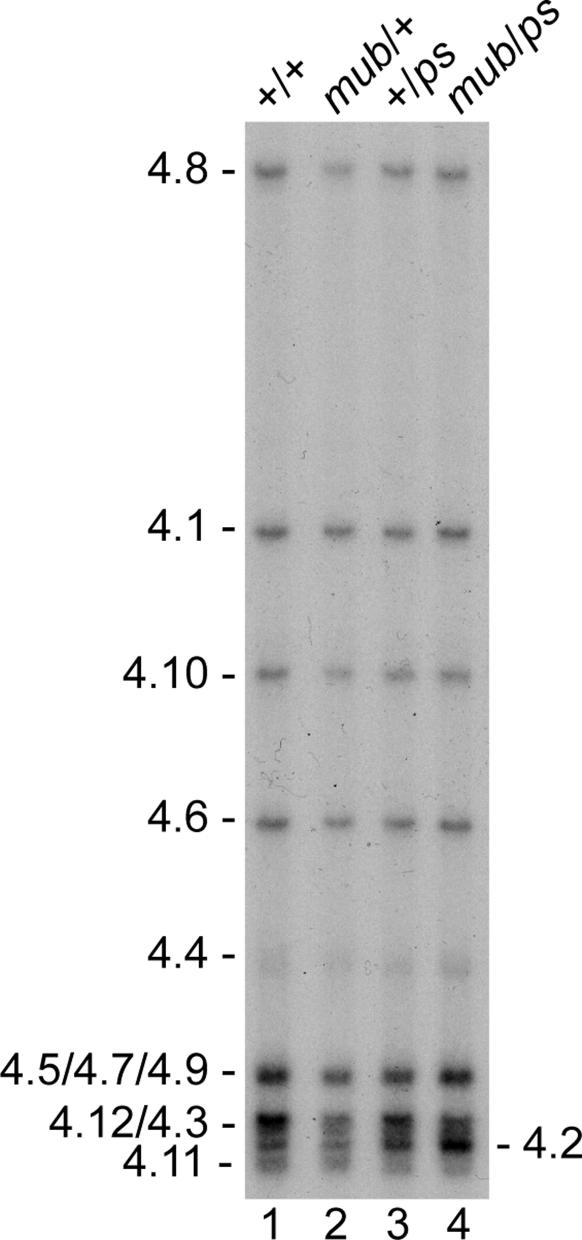
PS and MUB Regulate Dscam Alternative Splicing in Flies. Whereas only one protein affected the alternative splicing of para exons O and N, 36 proteins had an effect on alternative splicing of the exon 4 region of Dscam (Fig. 3). Two proteins that affect Dscam exon 4 alternative splicing are ps, which also affected alternative splicing of para exons O and N, and CG7437 (mub). Depletion of ps and mub both increased the inclusion of exon 4.2 (Fig. 3C). We and others have shown that exon 4.2 is one of the most developmentally regulated exons in Dscam (7, 18), although its physiological significance is unknown. To test whether these proteins regulate splicing in vivo, we analyzed exon 4 splicing in ps2 and mub04093 mutant flies. Because both ps and mub are essential genes (19, 20), and the PS and MUB proteins interact physically (21), we analyzed exon 4 splicing in ps2/mub04093 double heterozygotes. Exon 4.2 inclusion increased ≈1.5-fold in the ps2 and mub04093 single heterozygotes compared to wild type and 2-fold in the ps2/mub04093 double heterozygotes (Fig. 4). These results demonstrate that ps and mub control Dscam exon 4.2 alternative splicing in the fly, and validate that the RNAi screen performed in cultured S2 cells can identify proteins that are normally involved in regulating alternative splicing in the animal.
Fig. 3.
Proteins that regulate Dscam exon 4 alternative splicing. (A) Organization of the Dscam exon 4 cluster. (B) A representative single-stranded conformation polymorphism gel of RT-PCR products from RNA isolated from untreated S2 cells and cells treated with CG6995 (dSAF-B) dsRNA, which resulted in a dramatic alteration in the pattern of alternative splicing pattern for the exon 4 cluster. The percentage of each isoform in untreated cells is shown on the left side of the gel. (C) Quantitative representation of the change in the percentage of each isoform in dsRNA-treated cells compared to untreated cells for all dsRNAs that had a significant effect.
Fig. 4.
ps and mub regulate Dscam exon 4.2 alternative splicing in the fly. Single-stranded conformation polymorphism gel showing the pattern of the Dscam exon 4 cluster alternative splicing in ps and mub mutant adult male flies. +/+, mub/+, +/ps, and mub/ps represent wild type, mub04093 heterozygous mutant, ps2 heterozygous mutant, and mub04093/ps2 double heterozygous mutant, respectively.
How might ps and mub regulate exon 4.2 splicing? As mentioned earlier, PS is most similar to human Nova-1 and Nova-2 (14), which function as brain-specific splicing regulators by binding directly to intronic YCAY repeats (14). The intron immediately upstream of Dscam exon 4.2 contains several YCAY repeats, many of which are conserved in related Drosophila species (see supporting information). Moreover, this is the only place in the Dscam exon 4 region that contains tandem YCAY repeats. We suggest that PS binds to these intronic YCAY repeats and represses the inclusion of 4.2 exon. The striking similarities between PS and Nova and the observation that PS regulates the splicing of three of the regions we examined suggest that PS is an important splicing regulator in Drosophila.
MUB is homologous to the human poly(C)-binding protein (22). Initially identified based on its high level of expression in mushroom body (22), the center of learning and memory in insects, mub was recently isolated as a suppressor of neurodegeneration induced by expression of ataxin-1 (23). Notably, exon 4.2 is underrepresented in the Dscam mRNAs expressed in mushroom body (24). Because the depletion of mub results in an increase in exon 4.2 inclusion, we suggest that the high level of MUB expressed in mushroom bodies functions to inhibit exon 4.2 inclusion. Because PS and MUB physically interact and ps2/mub04093 double heterozygotes include exon 4.2 at a higher frequency than either single heterozygote, PS and MUB may function together to repress exon 4.2 inclusion.
Coordinate Regulation of Alternative Splicing. Several interesting observations can be made from the large group of splicing regulators identified in our screen. One such observation is that the alternative splicing of different regions within a single pre-mRNA can be regulated by overlapping but distinct sets of proteins (Fig. 1B). For example, all eight proteins involved in Dscam exon 17 splicing also affect Dscam exon 4 splicing. In contrast, 29 of the 36 proteins that regulate Dscam exon 4 splicing had no effect on Dscam exon 17 splicing. Similar results were obtained for para alternative splicing. Thus, the alternative splicing of different regions within a single pre-mRNA may be coordinately regulated by a set of common proteins, but may also be controlled by specific proteins.
Interacting Proteins Have Similar Functions in Splicing Regulation. We also found that interacting proteins tend to have similar effects on splicing. For example, both subunits of the heterodimeric splicing factor U2AF, CG3582 (U2AF38) and CG9998 (U2AF50), have a similar effect on Dscam exon 4 splicing when depleted by RNAi (Fig. 3C). Likewise, CG12085 (hfp), CG4602 (SRp54), and CG11266 (CAPER/CC1.3) encode interacting proteins (21, 25) that have similar affects on Dscam exon 4 (Fig. 3C) and exon 17 alternative splicing (see supporting information). The 47 splicing regulators we identified have recently been shown to interact with at least 81 other proteins (21). Given that interacting proteins tend to have similar effect on alternative splicing, we propose that many of these interacting proteins will also function in the regulation of alternative splicing.
The Coupling of Splicing to Other Nuclear Processes. Several of the proteins identified in our screen as affecting Dscam alternative splicing have been shown to couple splicing with other nuclear processes such as transcription, 3′ end formation, and RNA export (26). For example, CG9696 (domino) is a component of a chromatin remodeling complex (27), and CG8019 (haywire) is the homolog of the human ERCC3 gene, which is associated with Cockayne's syndrome and xeroderma pigmentosum B and is a component of TFIIH (28). Additionally, the human homologs of CG6995 (SAF-B) and CG11266 (CAPER/CC1.3) have been shown to function as estrogen-responsive nuclear receptor transcriptional coactivators (29, 30). Finally, the homolog of human CFIm-68k, CG7185, which had an effect on Dscam splicing, is a component of the 3′ end processing machinery (31). Although we cannot rule out the possibility that these proteins are acting indirectly, depletion of all of these proteins by RNAi specifically affected Dscam alternative splicing, but did not alter the splicing of either para or dAdar (Fig. 1B). Moreover, although each of these proteins altered splicing of the exon 4 region, only one, CAPER/CC1.3, also had an effect on splicing of the exon 17 region (Fig. 1B). Thus, these results are consistent with the idea that splicing is tightly coupled with other aspects of gene expression.
Core Components of the Spliceosome Function as Splicing Regulators. Perhaps the most interesting proteins we identified that affected alternative splicing are core components of the spliceosome (i.e., proteins that are conserved from humans to yeast). These include several components of U1 snRNP [CG4528 (snf/U1A), U1–70K, U1C, and CG1646 (Prp39)], both subunits of U2AF (U2AF38 and U2AF50), a component of U2 snRNP [CG16941 (SF3a120)], a component of the U4/U6 snRNP [CG6841 (Prp6)], CG17454 (SPF30), which was identified as a component of purified spliceosomes (1), and several DExH/D-box proteins [CG8241 (Prp22), CG5931 (Brr2), CG7269 (Hel25E, Sub2), CG6227 (Prp5), and CG10279 (Rm62)]. Although at least one general splicing factor was found to affect the splicing of each gene we tested, none of the general splicing factors affected the splicing of more than one gene (Fig. 1B) or had a detectable effect on the splicing of the constitutive GAPDH intron (see supporting information). Thus, reducing the levels of these general splicing factors by RNAi specifically modulated the inclusion of certain alternative exons.
These results suggest that, in certain cases, changes in the expression levels of core components of the spliceosome might regulate alternative splicing. Consistent with this idea, genome-wide transcription profiling analyses (32, 33) have shown that the mRNA levels of each spliceosomal component identified in our screen fluctuate significantly throughout development and in different tissues (see supporting information). Moreover, the expression patterns of these spliceosomal components differ from one another. For example, Brr2 and SF3a120 are both expressed at low levels in embryos and at high levels during metamorphosis, whereas Hel25E/Sub2 and U1–70K are highly expressed in embryos and are present at low levels during metamorphosis (32) (see supporting information). Differences in the relative concentrations of these spliceosomal components could change the kinetics of spliceosome assembly and/or splice site recognition. Importantly, whereas alternative exons are typically flanked by weak, nonconsensus splice sites, constitutive exons are usually flanked by strong, consensus splice sites (3). Consequently, alternative exons would be more sensitive to alterations in the concentrations of the spliceosomal components than constitutive exons. Consistent with this idea, Query and Konarska (34) recently provided strong evidence in Saccharomyces cerevisiae that substrate selectivity can be modulated by altering the kinetics of spliceosome rearrangement. Thus, fluctuations in the concentrations or activities of core components of the spliceosome may be a common mechanism by which splicing is regulated.
A Tool to Study Alternative Splicing. The lack of a robust genetic system to identify proteins that regulate the splicing of specific exons has been a major impediment to elucidating the mechanisms involved in regulating alternative splicing. Instead, one typically first generates a minigene that is accurately spliced in vitro or in vivo, identifies RNA sequences required for the regulation, and subsequently identifies the protein(s) that binds to these elements. The recent discovery of the profound influence of promoter identity and other gene expression processes such as transcription, 3′ processing, and RNA export on alternative splicing (26) highlights the importance of examining the role of splicing regulators on the alternative splicing of endogenous genes. Our RNAi screen achieved this goal and led to the identification of 26 proteins that had not been previously implicated in alternative splicing. Importantly, our approach did not depend on prior knowledge about the locations or identities of any important splicing regulatory elements or the proteins that could be involved in regulating splicing. It is possible that some of the proteins identified in our screen may be acting indirectly. Discriminating between direct and indirect effects will require additional biochemical and genetics experiments for each protein identified. However, the demonstration that two of the genes identified in our RNAi screen, ps and mub, encode proteins that function as splicing regulators in vivo establishes that this approach can be successfully used to identify authentic regulators of a gene of interest. Coupling this RNAi screen with additional genetic and biochemical experiments should provide tremendous insight into the detailed mechanisms involved in regulating alternative splicing. Finally, the fact that we identified 47 potential splicing regulators by analyzing the alternative splicing of only three genes suggests an extraordinary richness of the mechanisms that exists to regulate alternative splicing on a genome-wide level.
Supplementary Material
Acknowledgments
We thank A. Das, K. Hertel, K. Lynch, T. Maniatis, and members of the Graveley laboratory for discussions and comments on the manuscript. We thank D. Andrew for providing the ps2 flies, D. Rio (University of California, Berkeley) for antibodies, and S.L. Zipurksy for communicating results before publication. This work was supported by National Institutes of Health Grants GM062291-04 (to R.A.R.), GM62516-04 (to B.R.G.), and GM67842-02 (to B.R.G.).
Author contributions: J.W.P., K.P., A.M.C., R.A.R., and B.R.G. designed research; J.W.P., K.P., and A.M.C. performed research; J.W.P., K.P., A.M.C., R.A.R., and B.R.G. analyzed data; and J.W.P. and B.R.G. wrote the paper.
Abbreviations: RNAi, RNA interference; dsRNA, double-stranded RNA; snRNP, small nuclear ribonucleoprotein.
References
- 1.Jurica, M. S. & Moore, M. J. (2003) Mol. Cell 12, 5–14. [DOI] [PubMed] [Google Scholar]
- 2.Graveley, B. R. (2001) Trends Genet. 17, 100–107. [DOI] [PubMed] [Google Scholar]
- 3.Black, D. L. (2003) Annu. Rev. Biochem. 72, 291–336. [DOI] [PubMed] [Google Scholar]
- 4.Caceres, J. F. & Kornblihtt, A. R. (2002) Trends Genet. 18, 186–193. [DOI] [PubMed] [Google Scholar]
- 5.Smith, C. W. & Valcarcel, J. (2000) Trends Biochem. Sci. 25, 381–388. [DOI] [PubMed] [Google Scholar]
- 6.Johnson, J. M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P. M., Armour, C. D., Santos, R., Schadt, E. E., Stoughton, R. & Shoemaker, D. D. (2003) Science 302, 2141–2144. [DOI] [PubMed] [Google Scholar]
- 7.Celotto, A. M. & Graveley, B. R. (2001) Genetics 159, 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., et al. (2000) Science 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 9.Lasko, P. (2000) J. Cell Biol. 150, F51–F56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mount, S. M. & Salz, H. K. (2000) J. Cell Biol. 150, F37–F44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palladino, M. J., Keegan, L. P., O'Connell, M. A. & Reenan, R. A. (2000) Cell 102, 437–449. [DOI] [PubMed] [Google Scholar]
- 12.Hanrahan, C. J., Palladino, M. J., Ganetzky, B. & Reenan, R. A. (2000) Genetics 155, 1149–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmucker, D., Clemens, J. C., Shu, H., Worby, C. A., Xiao, J., Muda, M., Dixon, J. E. & Zipursky, S. L. (2000) Cell 101, 671–684. [DOI] [PubMed] [Google Scholar]
- 14.Dredge, B. K., Polydorides, A. D. & Darnell, R. B. (2001) Nat. Rev. Neurosci. 2, 43–50. [DOI] [PubMed] [Google Scholar]
- 15.Forch, P., Puig, O., Martinez, C., Seraphin, B. & Valcarcel, J. (2002) EMBO J. 21, 6882–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu, J. Y. & Maniatis, T. (1993) Cell 75, 1061–1070. [DOI] [PubMed] [Google Scholar]
- 17.Graveley, B. R. (2000) RNA 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neves, G., Zucker, J., Daly, M. & Chess, A. (2004) Nat. Genet. 36, 240–246. [DOI] [PubMed] [Google Scholar]
- 19.Seshaiah, P., Miller, B., Myat, M. M. & Andrew, D. J. (2001) Dev. Biol. 239, 309–322. [DOI] [PubMed] [Google Scholar]
- 20.Spradling, A. C., Stern, D., Beaton, A., Rhem, E. J., Laverty, T., Mozden, N., Misra, S. & Rubin, G. M. (1999) Genetics 153, 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giot, L., Bader, J. S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y. L., Ooi, C. E., Godwin, B., Vitols, E., et al. (2003) Science 302, 1727–1736. [DOI] [PubMed] [Google Scholar]
- 22.Grams, R. & Korge, G. (1998) Gene 215, 191–201. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez-Funez, P., Nino-Rosales, M. L., de Gouyon, B., She, W. C., Luchak, J. M., Martinez, P., Turiegano, E., Benito, J., Capovilla, M., Skinner, P. J., et al. (2000) Nature 408, 101–106. [DOI] [PubMed] [Google Scholar]
- 24.Zhan, X.-L., Clemens, J. C., Neves, G., Hattori, D., Flanagan, J. J., Hummel, T., Vasconcelos, M. L., Chess, A. & Zipursky, S. L. (2004) Neuron 43, 673–686. [DOI] [PubMed] [Google Scholar]
- 25.Page-McCaw, P. S., Amonlirdviman, K. & Sharp, P. A. (1999) RNA 5, 1548–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reed, R. (2003) Curr. Opin. Cell Biol. 15, 326–331. [DOI] [PubMed] [Google Scholar]
- 27.Ruhf, M. L., Braun, A., Papoulas, O., Tamkun, J. W., Randsholt, N. & Meister, M. (2001) Development (Cambridge, U.K.) 128, 1429–1441. [DOI] [PubMed] [Google Scholar]
- 28.Mounkes, L. C., Jones, R. S., Liang, B. C., Gelbart, W. & Fuller, M. T. (1992) Cell 71, 925–937. [DOI] [PubMed] [Google Scholar]
- 29.Townson, S. M., Dobrzycka, K. M., Lee, A. V., Air, M., Deng, W., Kang, K., Jiang, S., Kioka, N., Michaelis, K. & Oesterreich, S. (2003) J. Biol. Chem. 278, 20059–20068. [DOI] [PubMed] [Google Scholar]
- 30.Auboeuf, D., Dowhan, D. H., Kang, Y. K., Larkin, K., Lee, J. W., Berget, S. M. & O'Malley, B. W. (2004) Proc. Natl. Acad. Sci. USA 101, 2270–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruegsegger, U., Blank, D. & Keller, W. (1998) Mol. Cell 1, 243–253. [DOI] [PubMed] [Google Scholar]
- 32.Arbeitman, M. N., Furlong, E. E., Imam, F., Johnson, E., Null, B. H., Baker, B. S., Krasnow, M. A., Scott, M. P., Davis, R. W. & White, K. P. (2002) Science 297, 2270–2275. [DOI] [PubMed] [Google Scholar]
- 33.Li, T. R. & White, K. P. (2003) Dev. Cell 5, 59–72. [DOI] [PubMed] [Google Scholar]
- 34.Query, C. C & Konarska, M. M. (2004) Mol. Cell 14, 343–354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.