Abstract
The linked IL-4 and IL-13 cytokine genes, which are activated and silenced in T helper (Th) 2 and Th1 cells, respectively, are flanked by the equivalently expressed RAD50 and KIF3A genes. A scan of DNase I hypersensitivity and DNA methylation across ≈100 kb of the KIF3A/IL-4/IL-13/RAD50 cluster revealed differences in chromatin structure between Th1 and Th2 cells at the 3′ end of the RAD50 gene, a region previously shown to contain a locus control region (LCR) regulating Th2-specific expression of IL-4 and IL-13. Naïve CD4 T cells did not exhibit any DNase I hypersensitivity in this region, but stimulation under either Th1 or Th2 conditions caused rapid development of three hypersensitive sites. An additional hypersensitive site developed rapidly only under Th2 conditions, through a mechanism dependent on signal transducers and activators of transcription 6 (STAT6) but not GATA3. Our data point to a physical separation in the actions of STAT6 and its downstream effector GATA3 during Th2 differentiation: STAT6 directly remodels the RAD50 LCR, whereas GATA3 acts only in the vicinity of the IL-4 gene. We suggest that the RAD50 LCR has a complex and dual role in Th1 and Th2 differentiation, communicating early T cell antigen receptor and cytokine signals to the IL-4/IL-13 locus in both differentiating cell types.
Differentiation of precursor naïve T cells into mature cytokineproducing cells provides a useful paradigm for studying gene transcription (1–3). Differentiation is initiated by stimulation of naïve T cells through the T cell antigen receptor (TCR) and influenced by a large number of genetic and environmental variables including the cytokine milieu (4, 5). Sustained TCR stimulation in the presence of IL-4 yields differentiated T helper (Th)2 cells, which silence the IFNγ gene while activating the IL-4, IL-5, and IL-13 genes. Conversely, stimulation of naïve T cells in the presence of IL-12 yields differentiated Th1 cells, which show the opposite pattern of cytokine expression, silencing the IL-4, IL-5, and IL-13 genes but transcribing IFNγ at high levels upon secondary stimulation (6, 7). IL-12 and IL-4 act by means of the transcription factors signal transducers and activators of transcription (STAT) 4 and STAT6 (8). Th2 cells battle parasites and extracellular pathogens and participate in the pathogenesis of asthma and allergic disease, whereas Th1 cells are critical mediators of immunity against intracellular pathogens and have major roles in inflammation and autoimmunity.
The IL-4, IL-5, and IL-13 genes are linked closely in an evolutionarily conserved cytokine gene cluster, which occupies syntenic regions of mouse chromosome 11 and human chromosome 5 (9, 10). The cluster is located within an ≈220-kb genomic region that is highly conserved in all vertebrate species; it includes the noncytokine gene RAD50 and is bounded by genes encoding the kinesin KIF3A and the transcription factor IRF1 (9, 10). The chromatin changes occurring in the vicinity of the IL-4 and IL-13 genes during Th1/Th2 differentiation have been analyzed exhaustively (3). In Th2 cells, the changes include development of characteristic DNase I hypersensitivity (HS) patterns, DNA demethylation, histone hyperacetylation, and increased restriction enzyme accessibility (11–17). These changes are initiated rapidly by antigen stimulation but are transient unless maintained and reinforced by concomitant stimulation with the polarizing cytokines. The lineage-specific transcription factors GATA3 and T-bet are critical for Th2 and Th1 differentiation, respectively; when ectopically expressed, these proteins promote expression of the relevant cytokines, suppress transcription of the inappropriate cytokine genes, and mediate many of the chromatin structural changes seen during differentiation (3, 5, 18).
The in vivo roles of two clusters of Th2-specific HS sites in the IL-4/IL-13 locus, CNS-1 and CNS-2 (V)/VA, have been investigated by targeted deletion; both regions were shown to function as strong enhancers in vivo (9, 19–21). The functions of selected HS sites also were tested in transgenic mice by coupling them, individually or in combination, to a proximal IL-4 promoter-luciferase cassette that is poorly active on its own (21). All of the tested regions were capable of enhancing reporter activity in Th2 cells, but all of them, including a “minilocus” that contained all of the HS sites in the IL-4 locus, remained subject to position effect variegation (21). This result suggested that additional cis-elements, lying outside the conventional IL-4 locus, were needed to confer Th2 specificity of cytokine expression and protection from repressive chromatin effects. When a similar approach was used in bacterial artificial chromosome transgenic mice, an ≈25-kb region remotely located at the 3′ end of the RAD50 gene was shown to confer position-independent, copy number-dependent, and Th2-selective luciferase reporter activity (22). This characteristic is the defining feature of a locus control region (LCR), a regulatory element with the ability to maintain an open chromatin configuration in its own vicinity and in the vicinity of its regulated genes, even if the transgene that bears it happens to integrate into heterochromatic regions of DNA (23, 24). However, this element had a relatively local effect, conferring copy number-dependent expression on the neighboring IL-13 and IL-4 genes but not on the distant IL-5 gene.
Here, we have taken an independent approach to identifying putative cis-regulatory elements outside the Th2 cytokine gene cluster. By using systematic DNase I HS mapping in conjunction with a long-range method for identifying heavily methylated regions of DNA, we have scanned a large segment (≈100 kb) of the KIF3A/IL-4/IL-13/RAD50 cluster for chromatin structure differences between Th1 and Th2 cells. We find that the 3′ end of the KIF3A gene shows no differences in DNase I HS, DNA methylation, and histone H3 modification when naïve CD4 precursor T cells and differentiated Th1 and Th2 cells are compared, whereas the 3′ end of the RAD50 gene shows striking differences among these three cell types. Both the constitutive and the inducible patterns of DNase I HS in the RAD50 3′ region differ among naïve, Th1, and Th2 cells; moreover, all of the identified HS sites correspond to regions of high-sequence conservation (>75%) between mouse and human. We show that during early T cell differentiation, the RAD50 3′ region is a target for STAT6 but not for GATA3. Our results indicate that STAT6 and GATA3 synergize to influence IL-4/IL-13 transcription by acting at widely separated regions in the cytokine locus and suggest that the RAD50 3′ region dynamically influences cytokine transcription, not only at the earliest stages of Th1/Th2 differentiation but also in fully differentiated cells.
Methods
Mice. Mice were maintained in pathogen-free conditions in barrier facilities at the Center for Animal Resources and Comparative Medicine at Harvard Medical School. All mouse protocols were approved by the Center for Blood Research and Harvard Medical School. Three- to 4-week-old STAT6–/– (25), IL-4–/– (26), BALB/cJ, and C57BL/6J inbred strains were obtained from The Jackson Laboratory. BALB/cJ DO11.10 TCR transgenic mice bred onto the TCRα–/– background were kindly provided by Richard Locksley (University of California, San Francisco).
Cell Culture and in Vitro Th Differentiation. The murine T cell clones D5 and D10 were maintained as described in ref. 11. Naïve CD4 T cells (>90% Mel14hi) were purified by using magnetic beads from young (ages 3–4 weeks) mice and differentiated under Th1 (IL-12, anti-IL4) and Th2 (IL-4, anti-IFN-γ) polarizing conditions with anti-CD3 (2C11 antibody, Pharmingen) and anti-CD28 (37.51 antibody, Pharmingen) as described in ref. 11.
DNase I HS Analysis. Isolation and DNase I digestion of nuclei and purification of genomic DNA were performed as described in ref. 11. Before DNase I HS analysis, Th1 and Th2 cells were left resting, stimulated with phorbol 12-myristate 13-acetate (PMA) (20 nM) and ionomycin (2 μM) for 1.5–2 h, or pretreated with cyclosporin A (2 μM) for 20 min before PMA and ionomycin stimulation. Overlapping BamHI and KpnI (enzymes from New England Biolabs, Beverly, MA) genomic fragments spanning KIF3A to RAD50 were chosen for Southern blot analysis. Probes were designed to hybridize to the ends of the parental bands created by restriction enzyme digest.
DNA Methylation. DNA methylation analysis by Southern blotting using a McrBC titration digest was performed as described (27).
Chromatin Immunoprecipitation (ChIP). ChIP was performed as described (28). Antibodies to dimethyl-H3-K4 and dimethyl-H3-K9 were from Upstate Biotechnology (Lake Placid, NY). A STAT6 polyclonal antibody (M-200) was obtained from Santa Cruz Biotechnology. Primer sequences are given in Supporting Methods, which is published as supporting information on the PNAS web site.
ELISA Analysis of Cytokine Production. ELISA was performed as described in ref. 20. Primary and biotinylated secondary pairs of antibodies for IL-4 and IFN-γ both were used at a concentration of 1 μg/ml (Pharmingen).
Retroviral Constructs and Retroviral Transduction. Control retroviral vector (GFP-RV), GATA3 expression vector (GATA3-RV), and the retroviral infection protocol of activated naïve T cells are described in ref. 29. The STAT6-VT mutant was constructed as described in ref. 30, subcloned into the GFP-RV vector, upstream of the IRES-GFP cassette. Retrovirally infected Th1 cells (GFP+) were sorted by using a MoFlo Cell Sorter (Cytomation, Fort Collins, CO) and cultured for 2 days before cytokine analysis by ELISA.
vista Analysis. Human and mouse RAD50 sequence comparison was performed as described in ref. 31.
Results and Discussion
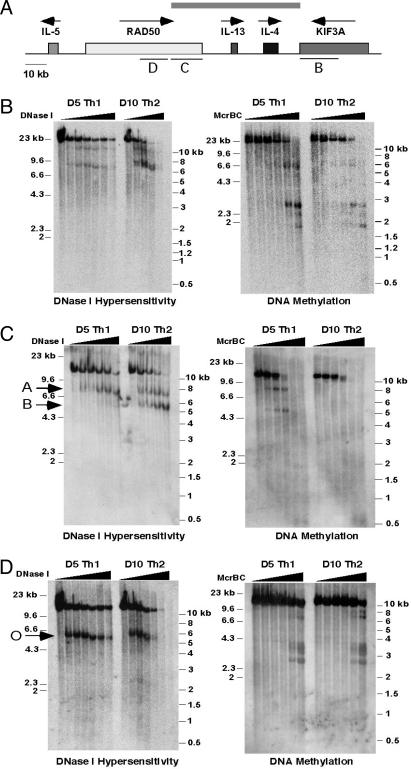
Chromatin Structure Differences Between Differentiated Th1 and Th2 Cells Extend into the Neighboring RAD50 Gene. We used DNase I HS (11, 13) and McrBC titration (27) to investigate how far the differences in chromatin structure between Th1 and Th2 cells extended in ≈100 kb of the KIF3A/IL-4/IL-13/RAD50 locus (Fig. 1). The expected differences in DNase I HS and DNA methylation (11, 13, 14, 17) were observed within the IL4 gene, in the IL-4/IL-13 intergenic region, and across the IL-13 gene (data not shown). An ≈23-kb region containing the 3′ end of the KIF3A gene (region B in Fig. 1A) showed two HS sites in Th1 and Th2 clones as well as in precursor naïve CD4 T cells (Fig. 1B Left and data not shown), indicating no differential chromatin remodeling in this region during Th1 and Th2 differentiation. In the ≈16-kb RAD50/IL-13 intergenic region, DNA methylation differences between Th1 and Th2 cells persisted, but no constitutive or inducible DNase I HS sites were observed (data not shown). Surprisingly, within the RAD50 gene, chromatin structure differences were observed between Th1 and Th2 cells (Fig. 1C). An ≈12-kb KpnI fragment containing the very 3′ end of the RAD50 gene displayed two HS sites, the RAD50-A site common to resting Th1 and Th2 cells and RAD50-B, a “Th2-specific” HS site of much stronger intensity in Th2 cells than in Th1 cells (Fig. 1C Left). These sites correspond, within 100–200 bp, to two regions that are heavily DNA-methylated only in Th1 cells (Fig. 1C Right). The differences in chromatin structure were observed in a total of 20 kb at the 3′ end of the RAD50 gene (region C in Fig. 1A); beyond this region, the differences stopped because region D of Fig. 1A contained a single common HS site (RAD50-O) and displayed identical DNA methylation patterns in Th1 and Th2 cells (Fig. 1D).
Fig. 1.
Identification of previously unrecognized DNase I HS sites and regions of differential DNA methylation between Th1 and Th2 cells in the RAD50 gene. (A) Schematic diagram showing the IL-5/RAD50/IL-13/IL-4/KIF3A gene cluster. Shaded boxes represent genes, and arrows depict the direction of transcription. The gene regions indicated by bars B, C, and D are analyzed in B, C, and D, respectively. The gray bar above the locus indicates the extent of differential chromatin remodeling in Th1 and Th2 cells. (B) Southern analysis showing the positions of DNase I HS sites (Left) and regions of heavy DNA methylation (Right) within an ≈23-kb BamHI region encompassing the KIF3A genes in D5 Th1 and D10 Th2 cells. (C) Same as in B except that the fragment analyzed is a 12-kb KpnI fragment containing the very 3′ region of the RAD50 gene. Arrows indicate the common HS site, RAD50-A, and a Th2-specific HS site, RAD50-B. Both of these HS regions are heavily cytosine-methylated in Th1 but not in Th2 cells. (D) Same as in B except that the fragment analyzed is an ≈12-kb BamHI fragment containing a central region of the RAD50 gene. The arrow indicates the common HS site RAD50-O.
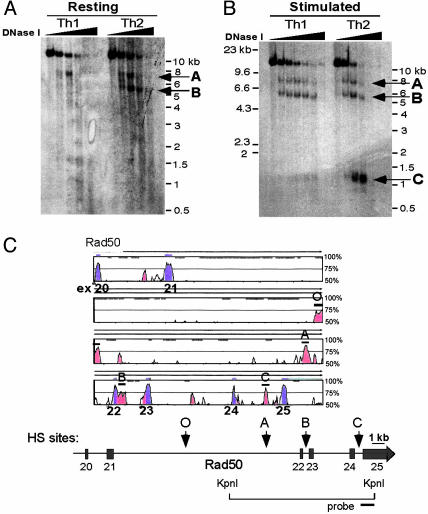
Primary Th1 and Th2 cells, differentiated for 1 week under polarizing conditions, displayed the same differences in DNase I HS in the RAD50 3′ region as did T cell clones (Fig. 2). Surprisingly, stimulation induced new HS sites not only in Th2 cells but also in Th1 cells that have silenced the neighboring IL-13 and IL-4 cytokine genes. Th2 cells displayed a new inducible HS site, RAD50-C, whereas Th1 cells exhibited an inducible HS site at the precise position of RAD50-B (Fig. 2 A and B).
Fig. 2.
Constitutive and inducible HS sites in the RAD50 3′ region correspond to CNS elements. (A) DNase I HS assay of 1-week-differentiated Th1 and Th2 cells shows that both cell types exhibit HS sites in the RAD50 3′ region. RAD50-A is present in both Th1 and Th2 cells, whereas RAD50-B is constitutively present in resting Th2 cells. (B) Acute stimulation for 2 h with PMA/ionomycin induces a third HS site in Th2 cells, RAD50-C; RAD50-B, which is absent in resting Th1 cells, becomes an inducible HS site upon Th1 stimulation. (C) vista plot of sequence conservation between human and mouse RAD50 genes (≈30 kb from exons 20–25 are shown). Conserved sequences are shown relative to their positions in the mouse genome (horizontal axes), and their percent identity within a sliding 100-bp window is indicated on the vertical axes (range 50–100%). Regions at least 100 bp long that show >75% sequence identity at the nucleotide level are shown in red (noncoding regions) or blue (exons). A thin line above the vista plot indicates noncoding regions of the RAD50 gene that exhibit HS sites; O, A, B, and C refer to RAD50-O, -A, -B, and -C regions, respectively. Below the plot is a schematic locus diagram of the RAD50 gene corresponding to the vista plot, with arrows indicating the locations of HS sites found in this region. The locations of the ≈12-kb KpnI fragment and the probe used for Southern blotting are shown.
All four HS sites, RAD50-O, -A, -B, and -C, corresponded well to conserved noncoding sequence (CNS) regions (Fig. 2C) (9). The high level of evolutionary conservation of the HS sites, their differential development in Th1 and Th2 cells, and the finding that a fragment containing all of the HS sites displays LCR function in transgenic mice (22) indicate that these sites are cis-regulatory elements that influence cytokine expression in differentiating T cells (9, 31–34). There are several precedents for the presence of critical cis-regulatory elements within the transcribed regions of neighboring, functionally unrelated but constitutively transcribed genes. Correct maternal imprinting of the insulin-2 and insulin-like growth factor-2 genes, which lie ≈100 kb upstream of H19, requires the H19 gene and its flanking sequences (35). More recently, a CD4 thymic enhancer and LCR was identified in the ISOT housekeeping gene, located ≈60 kb downstream of the CD4 promoter (36). Recruitment of DNA-binding and chromatin-remodeling factors is likely to be the rate-limiting step in a cellular differentiation program; thus, it might be advantageous to place key regulatory elements within regions that are already accessible and transcribed at the basal level, such as H19 for Igf2, ISOT for CD4, and RAD50 for IL-4 and IL-13. Because such genes already would have an open and accessible chromatin structure in the precursor cells, they might not require specific hypersensitive “marks” to recruit transcription factors and associated chromatin-modifying enzymes at the earliest stages of lineage commitment.
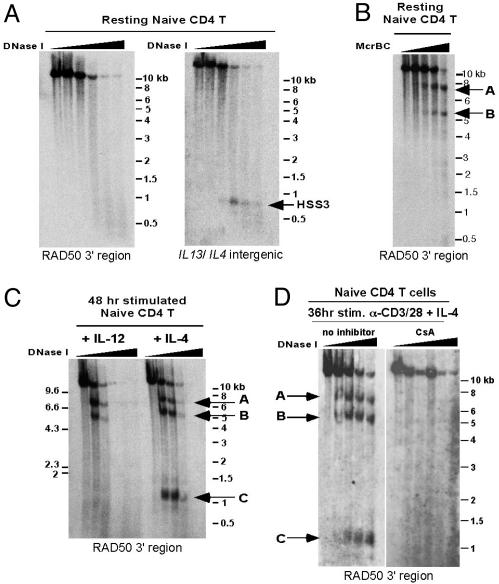
Differential HS Patterns in the RAD50 3′ Region Develop Early During Th Differentiation. DNase I HS analysis using naïve CD4 cells failed to show the presence of any of the HS sites, RAD50-O, -A, -B, and -C (Fig. 3A Left and data not shown). DNA methylation analysis showed that naïve CD4 T cells displayed the same pattern of heavy cytosine-methylation at RAD50-A and -B regions that we had observed previously in Th1 cells (Fig. 3B; compare with Fig. 1C Right). Because these same regions are demethylated in Th2 cells (Fig. 1C Right), our data point to a process of DNA demethylation and HS site development that is tightly associated with Th2 differentiation and that occurs focally at two highly conserved potential cis-regulatory elements in the RAD50 gene.
Fig. 3.
RAD50 HS sites are not present in naïve CD4 T cells but are rapidly induced within 48 h during Th differentiation. (A) Naïve CD4 T cells from TCR-transgenic mice were subjected to DNase I HS analysis by using a probe hybridizing to the 12-kb 3′ KpnI fragment of RAD50. No HS sites were observed. As a control, the same membrane was rehybridized with a probe specific for the ≈21-kb KpnI fragment containing the IL-13 gene and part of the IL-4/IL-13 intergenic region to show the presence of HSS3, a hypersensitive site present in naïve CD4, Th1, and Th2 cells (13). (B) DNA methylation/Southern blot analysis shows two regions of heavy cytosine-methylation in the 12-kb KpnI fragment in naïve CD4 T cells (indicated by arrows). The regions coincide with the locations of HS sites RAD50-A and -B, respectively. (C) DNase I HS analysis on naïve CD4 T cells stimulated for 48 h with plate-bound anti-CD3/CD28 in the presence of IL-12 plus neutralizing antibodies to IL-4 (Left) or IL-4 cytokine plus neutralizing antibodies to IFN-γ (Right). Arrows indicate the appearance of HS sites under both stimulation conditions. (D) Same as in C except that naïve CD4 T cells were treated with cyclosporin A (Right) or ethanol control (Left) during the anti-CD3/CD28 and IL-4 cytokine stimulation.
We asked how early the HS sites appeared during Th differentiation. Because of the fragility of the nuclei and the highly decondensed state of the DNA immediately after stimulation, 36–48 h is the earliest time that DNase I HS analyses can be performed on activated naïve T cells. All of the RAD50 HS sites developed within this early time period (Fig. 3C). The RAD50-A and -B sites were observed under both Th1 and Th2 stimulation conditions, whereas RAD50-C appeared only under Th2 polarizing conditions. Pretreatment with cyclosporin A, an inhibitor of the protein phosphatase calcineurin, prevented the formation of all three HS sites, RAD50-A, -B, and -C, in cells stimulated under Th2 conditions (Fig. 3D). Thus, TCR signaling and calcineurin activation are required to induce appearance of all three HS sites in the RAD50 locus; in addition, IL-4 signaling is necessary for induction of RAD50-C.
We compared the pattern of histone modification at RAD50-B and -C in differentiated Th1 and Th2 cells. One-week-differentiated Th2 cells showed significantly higher levels of H3 lysine 4 (K4)-methylation at RAD50-B and -C regions than 1-week-differentiated Th1 cells (see Fig. 7A, which is published as supporting information on the PNAS web site); this modification is associated with actively transcribed genes (37, 38). Conversely, Th1 cells showed high H3-K9-methylation at both RAD50-B and -C (Fig. 7B); this modification is associated with gene silencing and binding of heterochromatin-associated proteins (38). Control experiments confirmed that the IL-4 promoter showed high H3-K4-methylation in Th2 cells, whereas reciprocally, the IFNγ promoter showed high H3-K4-methylation in Th1 cells (Fig. 7A); similarly, the IFNγ 5′ enhancer (28) showed higher H3-K9-methylation in Th2 cells that have silenced the IFNγ gene (Fig. 7B). Thus, although DNase I HS develops rapidly at the RAD50-B region regardless of which cytokines are present, the histone modifications that eventually characterize this site in differentiated T cells depend on the polarizing cytokine.
Northern and RT-PCR analysis show no correlation between RAD50 transcription and chromatin configuration at the 3′ end of the gene: Naïve T cells, which do not exhibit DNase I HS at the RAD50-O, -A, -B, or -C regions, clearly express mRNA encoding for RAD50, a protein essential for DNA repair (data not shown). Similarly, the pattern of HS sites and the histone modification status of the RAD50 3′ region differ in primary Th1 and Th2 cells, but the RAD50 gene is transcribed at similar levels in these two cell types under resting conditions and is equivalently down-regulated upon stimulation (at most a 2-fold difference; data not shown). We conclude that the regulatory activity of the RAD50 3′ region is not directed toward RAD50 transcription itself, but rather influences the process of Th1/Th2 differentiation and transcription of the neighboring cytokine genes.
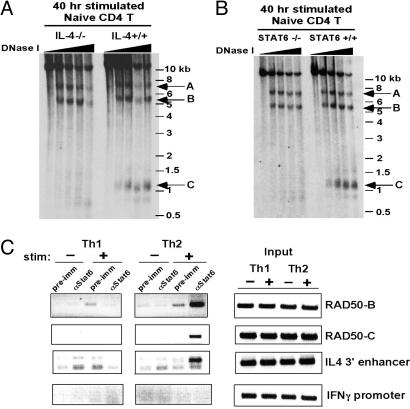
IL-4/STAT6 Signaling Is Required for Development of the RAD50-C HS Site, Whereas GATA3 Is Not. We showed that combined TCR and IL-4/STAT6 signals were essential for early induction of RAD50-C (Fig. 4). Naïve CD4 T cells from wild-type and IL-4–/– mice were stimulated for 40 h in the presence of neutralizing antibodies to IFN-γ; both IL-4+/+ and IL-4–/– cells developed RAD50-A and -B, but only IL-4+/+ cells developed RAD50-C (Fig. 4A). Similarly STAT6–/– T cells stimulated under Th2 polarizing conditions did not show appearance of the RAD50-C site observed in wild-type T cells (Fig. 4B). Sequence inspection and bioinformatics analysis revealed the presence of conserved STAT binding sites in the RAD50-B and -C regions but not in RAD50-A (data not shown). ChIP demonstrated the selective binding of STAT6 to both RAD50-B and -C regions in stimulated Th2 cells (Fig. 4C).
Fig. 4.
Appearance of the RAD50-C HS site in stimulated naïve CD4 T cells depends on IL-4/STAT6 signaling. (A) DNase I HS analysis was performed on naïve CD4 T cells isolated from IL-4–/– or wild-type mice and stimulated for 40 h with plate-bound anti-CD3/CD28 in the presence of neutralizing antibodies to IFN-γ to minimize Th1 differentiation in the culture. No exogenous IL-4 cytokine was added. The RAD50-A and -B HS sites are induced upon stimulation in both IL-4–/– and control T cells, but induction of the RAD50-C site requires IL-4. (B) Same as in A, except that naïve CD4 T cells isolated from STAT6–/– or wild-type mice were stimulated with anti-CD3/CD28 in the presence of exogenous IL-4 and neutralizing antibodies to IFN-γ. (C) ChIP was performed by using chromatin from primary Th1 and Th2 cells, either resting or stimulated with PMA/ionomycin for 1.5 h (the stimulation results in IL-4 production by Th2 cells). (Left) Chromatin was immunoprecipitated with an antibody to STAT6, PCR primers were used to amplify short (≈225 bp) fragments within HS sites RAD50-B and -C, and the IL-4 3′ enhancer and the IFNγ promoter were used as controls. (Right) PCR of input DNA shows equivalent starting material for the assay.
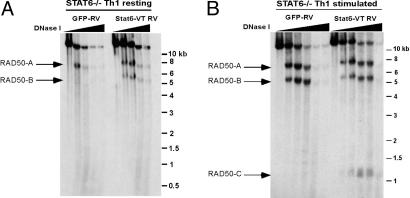
To determine whether STAT6 could directly induce the formation of RAD50-B and -C sites during Th differentiation, we used a constitutively active mutant of STAT6, STAT6-VT (30, 39), with two alanine substitutions in its SH2 domain, which promote STAT6 dimerization and nuclear translocation in the absence of IL-4 receptor signaling. Naïve CD4 T cells from STAT6–/– mice were isolated, activated for 40 h under Th1 polarizing conditions, and then infected with retroviruses encoding STAT6-VT IRES-GFP (STAT6-VT RV) or an IRES-GFP control (GFP-RV); infection efficiency varied from 30% to 60%. The cells then were rested in culture for 4–5 days in the continued presence of IL-12. Because Th1 cells do not basally display HS site RAD50-B, and because in vitro Th2 differentiation is impaired severely in the absence of STAT6, we were able to assess DNase I HS in the bulk population without sorting for GFP+ cells.
Under resting conditions, GFP-RV-infected STAT6–/– Th1 cells displayed only HS site RAD50-A, whereas STAT6-VT RV-infected Th1 cells resembled resting Th2 cells in displaying both RAD50-A and -B (Fig. 5A). The RAD50-C HS site did not appear under these resting conditions (Fig. 5A). When the cells were stimulated with PMA and ionomycin, RAD50-B (but not RAD50-C) was induced in the control GFP-RV-infected Th1 cells as expected, whereas RAD50-C was induced only in the cells expressing STAT6-VT (Fig. 5B). Control experiments confirmed that STAT6-VT RV induced IL-4 expression by Th1 cells, which normally silence this cytokine gene, and also induced appearance of the characteristic DNase I HS sites in the IL-4 gene (data not shown).
Fig. 5.
STAT6 is sufficient to induce HS in the RAD50 3′ locus. (A) DNase I HS analysis was performed on 1-week STAT6–/– Th1 cells retrovirally transduced with GFP (GFP-RV) or a constitutively active STAT6 (STAT6-VT RV). The RAD50-A site is constitutive in Th1 cells, whereas expression of STAT6-VT in resting Th1 cells induces formation of the RAD50-B site. (B) Same as in A, except cells were stimulated with PMA/ionomycin for 2 h before analysis. The appearance of the RAD50-B site is seen in stimulated GFP-RV Th1 cells (Left), whereas the RAD50-C site becomes induced with stimulation in STAT6-VT-infected Th1 cells (Right).
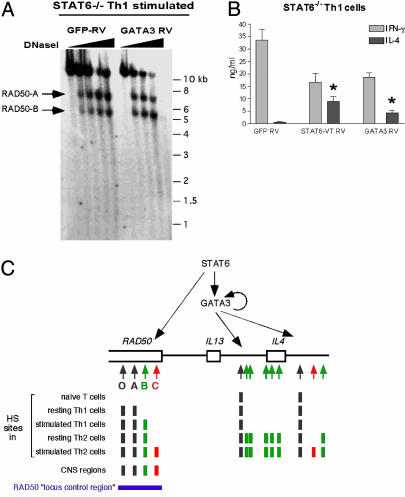
The transcription factor GATA3 is up-regulated by STAT6 activation and plays a critical role in Th2 differentiation (40–42). Surprisingly, overexpression of GATA3 did not induce formation of the Th2-specific HS site RAD50-C (Fig. 6). When differentiating Th1 cells from STAT6–/– mice were transduced retrovirally with a GFP-RV or GATA3-RV (infection efficiency 50–70%), both infected cell populations had comparable levels of T-bet expression, indicative of differentiated Th1 cells, but only GATA3-RV-infected Th1 cells showed GATA3 expression by Western analysis (data not shown). Unlike STAT6-VT-expressing cells, which display both RAD50-A and -B under resting conditions and induce RAD50-C upon stimulation, the GATA3-RV-infected cells displayed only RAD50-A under resting conditions (data not shown) and induced only RAD50-B when stimulated (Fig. 6A). Together, these results show that STAT6, but not GATA3, is involved in induction of the RAD50-C HS site during Th2 differentiation.
Fig. 6.
GATA3 fails to induce Th2-specific chromatin remodeling at the RAD50 3′ region in differentiating Th1 cells. (A) DNase I HS analysis was performed on GFP-RV-(Left) or GATA3-RV-(Right) transduced STAT6–/– Th1 cells stimulated for 2 h with PMA/ionomycin. Arrows indicate the appearance of HS sites. (B) Analysis of secreted IL-4 and IFN-γ produced by GFP-RV-, STAT6-VT RV-, or GATA3-RV-transduced STAT6–/– Th1 cells stimulated with PMA/ionomycin for ≈20 h by ELISA. Data shown are the mean ± SD of triplicate readings of two independent cell culture experiments. Comparison of the bars marked with asterisks shows that infection with GATA3-RV consistently induces lower levels of IL-4 production than infection with STAT6-VT RV. (C) (Upper) Model for the actions of STAT6 and GATA3 at the extended RAD50/IL-13/IL-4 locus. STAT6 and GATA3 cooperate functionally to induce optimal cytokine expression but act at disparate regions of the locus. STAT6 up-regulates the expression of GATA3, which in turn autoregulates its own expression. Both STATA6 and GATA3 can induce Th2-specific chromatin changes at cis-regulatory regions surrounding the IL-4 locus, even in differentiating Th1 cells, but Th2-specific chromatin remodeling at the RAD50 locus requires STAT6 and not GATA3. Note that there is evidence for direct actions of STAT5 and STAT6 in the IL-4 locus and that we cannot exclude a late effect of GATA3 at the RAD50 region. (Lower) Summary of the changes in DNase I HS at the extended RAD50/IL-13/IL-4 locus.
In parallel, we compared the ability of STAT6-VT and GATA3 to induce IL-4 expression. Two days after infection with STAT6-VT and GATA3 retroviruses, the transduced GFP+ cells were sorted and allowed to expand for an additional 2 days before restimulation and analysis of cytokine production by ELISA (Fig. 6B). Control GFP-RV-infected STAT6–/– Th1 cells produced high levels of IFN-γ and negligible levels of IL-4, indicating efficient polarization to the Th1 lineage (Fig. 6B Left), whereas cells transduced with either STAT6-VT or GATA3 retroviruses produced lower levels of IFN-γ and higher levels of IL-4, indicating realignment of differentiation toward the Th2 cytokine pattern (Fig. 6B Center and Right). Reproducibly, GATA3-expressing Th1 cells showed 2- to 3-fold lower IL-4 expression compared with STAT6-VT transduced Th1 cells (Fig. 6B, compare right and center bars with asterisks). Similarly, ectopic expression of GATA3 in STAT6-deficient Th1 cells led to lower IL-4 production than observed in wild-type Th1 cells reconstituted with GATA3 (42). This difference most likely reflects the fact that GATA3 is unable to induce early Th2-specific chromatin changes in the RAD50 LCR.
Conclusions
Our analysis predicts that the RAD50 3′ region will differentially regulate IL-4 and IL-13 gene expression in Th1 and Th2 cells. LCR assays are known to be biased toward strong enhancers (23, 24, 43), and it is likely that the LCR function of the RAD50 3′ region (22) is primarily mediated through RAD50-B and -C, which are good candidates for Th2-specific enhancers. In Th1 cells, the same two sites are selectively H3-K9-methylated and so could have an opposing repressive effect on IL-4 and IL-13 gene transcription by recruiting HP-1 and localizing the cytokine genes to heterochromatic regions of the nucleus (44). Alternatively, the RAD50-B HS site, which is basally present in Th2 cells but appears as an inducible HS site in Th1 cells, could be involved in acute repression of the neighboring cytokine genes in activated Th1 cells. The RAD50-A and -O HS sites could function as insulator or enhancer blocking elements, which separate the IL-4 and IL-13 cytokine promoters from the RAD50 promoter. The sites develop in both Th1 and Th2 cells, and they are located at the 5′ end of the LCR region, away from their target cytokine genes, which are 3′ of the LCR. This spatial relationship is similar to that of the chicken β-globin locus, where the insulator element, HS4, is also found 5′ of the other HS sites in the LCR at the opposite end from the globin target genes (45).
The appearance of site RAD50-B in stimulated naïve cells is not dependent on STAT6 or IL-4 (Fig. 4 A and B). However, a Th2-differentiative program is required for RAD50-B to become a constitutive HS site under basal resting conditions. The inducible appearance of this site in stimulated Th1 cells may reflect activation of another STAT family member, such as STAT1 (downstream of IFN-γ). Indeed it is reported that STAT1 and STAT6 often use similar DNA-binding elements (46).
We have shown that STAT6 and GATA3 act synergistically at widely separated regions to induce chromatin structural changes throughout the RAD50/IL-13/IL-4 locus (Fig. 6C). To our knowledge, a role for STAT6 in chromatin remodeling that is independent of GATA3 has not been reported previously, and also the key regulatory region in the cytokine locus that responds to Th2-differentiative signals in a STAT6-dependent, GATA3-independent manner has not been previously identified. Although GATA3 overexpression partially bypasses the requirement for STAT6 in Th2 differentiation and chromatin remodeling near the IL-4 gene (42, 44, 47), an additional input from STAT6 is needed for chromatin changes at the RAD50 locus and optimal IL-4 expression.
Supplementary Material
Acknowledgments
We thank Drs. W. E. Paul (National Institutes of Health, Bethesda) and K. Murphy (Washington University, St. Louis, MO) for the STAT6-VT and GATA3 retroviral constructs, respectively; Dr. Julie Nardone for assistance with vista and analysis of transcription factor binding sites in the RAD50 gene; and members of the laboratory for helpful comments and discussion. This research was supported by National Institutes of Health Grants R01 AI44432 and P01 HL67664 and by a grant from the Sandler Program for Asthma Research (to A.R.). D.U.L. is an Albert J. Ryan Foundation Fellow.
Author contirbutions: D.U.L. and A.R. designed research; D.U.L. performed research; D.U.L. and A.R. analyzed data; and D.U.L. and A.R. wrote the paper.
Abbreviations: ChIP, chromatin immunoprecipitation; HS, hypersensitivity; LCR, locus control region; PMA, phorbol 12-myristate 13-acetate; STAT, signal transducers and activators of transcription; TCR, T cell antigen receptor; Th, T helper.
Note Added in Proof. While this manuscript was under review, Spilianakis and Flavell (48) reported on the spatial organization of chromatin in the IL-4/IL-13/RAD50/IL-5 locus. By using the chromosome conformation capture assay, they showed that the RAD50 LCR is in close apposition to the promoters and other cis-regulatory regions (DNase I HS sites) of the cytokine genes in naïve, Th1, and Th2 cells, but not in fibroblasts that do not express the cytokine genes. Their results support a DNA looping model as previously suggested for the β-globin locus, with the surprising finding that Th1 and Th2 cells show almost identical configurations of the DNA loops that form the chromatin hub. Thus, although the gross associations between regulatory elements appear indistinguishable in Th1 vs. Th2 cells, our analyses of DNase I HS and transcription factor binding point to striking differences in the chromatin accessibility of the LCR HS sites in these two differentiated cell types.
References
- 1.Avni, O. & Rao, A. (2000) Curr. Opin. Immunol. 12, 654–659. [DOI] [PubMed] [Google Scholar]
- 2.Grogan, J. L. & Locksley, R. M. (2002) Curr. Opin. Immunol. 14, 366–372. [DOI] [PubMed] [Google Scholar]
- 3.Ansel, K. M., Lee, D. U. & Rao, A. (2003) Nat. Immunol. 4, 616–623. [DOI] [PubMed] [Google Scholar]
- 4.O'Garra, A. (1998) Immunity 8, 275–283. [DOI] [PubMed] [Google Scholar]
- 5.Murphy, K. M., Ouyang, W., Farrar, J. D., Yang, J., Ranganath, S., Asnagli, H., Afkarian, M. & Murphy, T. L. (2000) Annu. Rev. Immunol. 18, 451–494. [DOI] [PubMed] [Google Scholar]
- 6.Seder, R. A. & Paul, W. E. (1994) Annu. Rev. Immunol. 12, 635–673. [DOI] [PubMed] [Google Scholar]
- 7.Abbas, A. K., Murphy, K. M. & Sher, A. (1996) Nature 383, 787–793. [DOI] [PubMed] [Google Scholar]
- 8.Wurster, A. L., Tanaka, T. & Grusby, M. J. (2000) Oncogene 19, 2577–2584. [DOI] [PubMed] [Google Scholar]
- 9.Loots, G. G., Locksley, R. M., Blankespoor, C. M., Wang, Z. E., Miller, W., Rubin, E. M. & Frazer, K. A. (2000) Science 288, 136–140. [DOI] [PubMed] [Google Scholar]
- 10.Frazer, K. A., Ueda, Y., Zhu, Y., Gifford, V. R., Garofalo, M. R., Mohandas, N., Martin, C. H., Palazzolo, M. J., Cheng, J. F. & Rubin, E. M. (1997) Genome Res. 7, 495–512. [DOI] [PubMed] [Google Scholar]
- 11.Agarwal, S. & Rao, A. (1998) Immunity 9, 765–775. [DOI] [PubMed] [Google Scholar]
- 12.Agarwal, S., Avni, O. & Rao, A. (2000) Immunity 12, 643–652. [DOI] [PubMed] [Google Scholar]
- 13.Takemoto, N., Koyano-Nakagawa, N., Yokota, T., Arai, N., Miyatake, S. & Arai, K. (1998) Int. Immunol. 10, 1981–1985. [DOI] [PubMed] [Google Scholar]
- 14.Lee, D. U., Agarwal, S. & Rao, A. (2002) Immunity 16, 649–660. [DOI] [PubMed] [Google Scholar]
- 15.Avni, O., Lee, D., Macian, F., Szabo, S. J., Glimcher, L. H. & Rao, A. (2002) Nat. Immunol. 3, 643–651. [DOI] [PubMed] [Google Scholar]
- 16.Fields, P. E., Kim, S. T. & Flavell, R. A. (2002) J. Immunol. 169, 647–650. [DOI] [PubMed] [Google Scholar]
- 17.Yamashita, M., Ukai-Tadenuma, M., Kimura, M., Omori, M., Inami, M., Taniguchi, M. & Nakayama, T. (2002) J. Biol. Chem. 277, 42399–42408. [DOI] [PubMed] [Google Scholar]
- 18.Smale, S. T. & Fisher, A. G. (2002) Annu. Rev. Immunol. 20, 427–462. [DOI] [PubMed] [Google Scholar]
- 19.Mohrs, M., Blankespoor, C. M., Wang, Z. E., Loots, G. G., Afzal, V., Hadeiba, H., Shinkai, K., Rubin, E. M. & Locksley, R. M. (2001) Nat. Immunol. 2, 842–847. [DOI] [PubMed] [Google Scholar]
- 20.Solymar, D. C., Agarwal, S., Bassing, C. H., Alt, F. W. & Rao, A. (2002) Immunity 17, 41–50. [DOI] [PubMed] [Google Scholar]
- 21.Lee, G. R., Fields, P. E. & Flavell, R. A. (2001) Immunity 14, 447–459. [DOI] [PubMed] [Google Scholar]
- 22.Lee, G. R., Fields, P. E., Griffin, T. J. & Flavell, R. A. (2003) Immunity 19, 145–153. [DOI] [PubMed] [Google Scholar]
- 23.Engel, J. D. & Tanimoto, K. (2000) Cell 100, 499–502. [DOI] [PubMed] [Google Scholar]
- 24.Festenstein, R. & Kioussis, D. (2000) Curr. Opin. Genet. Dev. 10, 199–203. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan, M. H., Schindler, U., Smiley, S. T. & Grusby, M. J. (1996) Immunity 4, 313–319. [DOI] [PubMed] [Google Scholar]
- 26.Kuhn, R., Rajewsky, K. & Muller, W. (1991) Science 254, 707–710. [DOI] [PubMed] [Google Scholar]
- 27.Santoso, B., Ortiz, B. D. & Winoto, A. (2000) J. Biol. Chem. 275, 1952–1958. [DOI] [PubMed] [Google Scholar]
- 28.Lee, D. U., Avni, O., Chen, L. & Rao, A. (2004) J. Biol. Chem. 279, 4802–4810. [DOI] [PubMed] [Google Scholar]
- 29.Ouyang, W., Ranganath, S. H., Weindel, K., Bhattacharya, D., Murphy, T. L., Sha, W. C. & Murphy, K. M. (1998) Immunity 9, 745–755. [DOI] [PubMed] [Google Scholar]
- 30.Mikita, T., Daniel, C., Wu, P. & Schindler, U. (1998) J. Biol. Chem. 273, 17634–17642. [DOI] [PubMed] [Google Scholar]
- 31.Nardone, J., Lee, D. U., Ansel, K. M. & Rao, A. (2004) Nat. Immunol. 5, 768–774. [DOI] [PubMed] [Google Scholar]
- 32.Duret, L. & Bucher, P. (1997) Curr. Opin. Struct. Biol. 7, 399–406. [DOI] [PubMed] [Google Scholar]
- 33.Wei, L., Liu, Y., Dubchak, I., Shon, J. & Park, J. (2002) J. Biomed. Inform. 35, 142–150. [DOI] [PubMed] [Google Scholar]
- 34.Pennacchio, L. A. & Rubin, E. M. (2001) Nat. Rev. Genet. 2, 100–109. [DOI] [PubMed] [Google Scholar]
- 35.Leighton, P. A., Ingram, R. S., Eggenschwiler, J., Efstratiadis, A. & Tilghman, S. M. (1995) Nature 375, 34–39. [DOI] [PubMed] [Google Scholar]
- 36.Adlam, M. & Siu, G. (2003) Immunity 18, 173–184. [DOI] [PubMed] [Google Scholar]
- 37.Santos-Rosa, H., Schneider, R., Bannister, A. J., Sherriff, J., Bernstein, B. E., Emre, N. C., Schreiber, S. L., Mellor, J. & Kouzarides, T. (2002) Nature 419, 407–411. [DOI] [PubMed] [Google Scholar]
- 38.Lachner, M. & Jenuwein, T. (2002) Curr. Opin. Cell Biol. 14, 286–298. [DOI] [PubMed] [Google Scholar]
- 39.Daniel, C., Salvekar, A. & Schindler, U. (2000) J. Biol. Chem. 275, 14255–14259. [DOI] [PubMed] [Google Scholar]
- 40.Lee, H. J., Takemoto, N., Kurata, H., Kamogawa, Y., Miyatake, S., O'Garra, A. & Arai, N. (2000) J. Exp. Med. 192, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurata, H., Lee, H. J., O'Garra, A. & Arai, N. (1999) Immunity 11, 677–688. [DOI] [PubMed] [Google Scholar]
- 42.Ouyang, W., Lohning, M., Gao, Z., Assenmacher, M., Ranganath, S., Radbruch, A. & Murphy, K. M. (2000) Immunity 12, 27–37. [DOI] [PubMed] [Google Scholar]
- 43.Li, Q., Peterson, K. R., Fang, X. & Stamatoyannopoulos, G. (2002) Blood 100, 3077–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grogan, J. L., Mohrs, M., Harmon, B., Lacy, D. A., Sedat, J. W. & Locksley, R. M. (2001) Immunity 14, 205–215. [DOI] [PubMed] [Google Scholar]
- 45.Gerasimova, T. I. & Corces, V. G. (2001) Annu. Rev. Genet. 35, 193–208. [DOI] [PubMed] [Google Scholar]
- 46.Ehret, G. B., Reichenbach, P., Schindler, U., Horvath, C. M., Fritz, S., Nabholz, M. & Bucher, P. (2001) J. Biol. Chem. 276, 6675–6688. [DOI] [PubMed] [Google Scholar]
- 47.Farrar, J. D., Ouyang, W., Lohning, M., Assenmacher, M., Radbruch, A., Kanagawa, O. & Murphy, K. M. (2001) J. Exp. Med. 193, 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spilianakis, C. G. & Flavell, R. A. (2004) Nat. Immunol. 5, 1017–1027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.