Abstract
The regulated equilibrium between mitochondrial fusion and fission is essential to maintain integrity of the organelle. Mechanisms of mitochondrial fusion are largely uncharacterized in mammalian cells. It is unclear whether OPA1, a dynamin-related protein of the inner membrane mutated in autosomal dominant optic atrophy, participates in fusion or fission. OPA1 promoted the formation of a branched network of elongated mitochondria, requiring the integrity of both its GTPase and C-terminal coiled-coil domain. Stable reduction of OPA1 levels by RNA interference resulted in small, fragmented, and scattered mitochondria. Levels of OPA1 did not affect mitochondrial docking, but they correlated with the extent of fusion as measured by polyethylene glycol mitochondrial fusion assays. A genetic analysis proved that OPA1 was unable to tubulate and fuse mitochondria lacking the outer membrane mitofusin 1 but not mitofusin 2. Our data show that OPA1 functionally requires mitofusin 1 to regulate mitochondrial fusion and reveal a specific functional difference between mitofusin 1 and 2.
Mitochondria are crucial organelles for life and death of the cell: they produce most cellular ATP, shape cytosolic Ca2+ transients, and integrate diverse apoptotic stimuli by releasing protein cofactors needed for the efficient activation of effector caspases (1, 2). Such a functional versatility is matched by a complex structural organization. The mitochondrial cristae have been identified as a separate compartment connected to the thin intermembrane space by narrow tubular junctions (3), which may generate gradients of ions and small molecules along the cristae (4) and are responsible for the segregation of cytochrome c in the cristae compartment (5, 6). In the cytosol of certain cell types, mitochondria are organized in a network of individual organelles that dynamically fuse and divide (7, 8), generating functional mitochondrial cables. This organization allows stimuli hitting one end of the mitochondrial wire to be readily transmitted to distal components of the net (9), a useful property in large cells such as cardiomyocytes (10). On the other hand, mitochondria can also behave as individual units in other cell types, such as pancreatic beta cells (11). Mitochondrial shape is not static, because during mitosis, mitochondria divide and partition into daughter cells (12). Major changes of mitochondrial morphology have been described during apoptosis, with fragmentation of the mitochondrial network, cristae fusion, and enlargement of cristae junctions (6, 13).
Dynamic control of mitochondrial structure is performed by a growing set of “mitochondria-shaping” proteins that include both pro-fusion and pro-fission members, several of which have been identified in budding yeast (14). Fission of yeast mitochondria is accomplished by the recruitment of the dynamin-related large GTPase Dnm1p to the outer membrane, where it forms a complex with the adapter Mdv1p and the integral membrane protein Fis1p (15–18). Fusion involves proteins localized at both the outer and the inner membrane. A crucial role has been ascribed to the GTPase Fzo1p (19, 20), which interplays with the adapter Ugo1p in the outer membrane (21) and associates with the inner membrane dynamin-related protein Mgm1p to coordinate the fusion of the four membranes of two juxtaposed mitochondria (22–24).
Mammalian orthologues of DNM1 and FIS1, called dynamin-related protein 1 (Drp1) and hFis1, respectively, have been identified and shown to participate in mitochondrial fission (25–27). Fusion processes seem to be more complex. Two FZO1 homologues have been identified, mitofusin (Mfn) 1 and 2 (28, 29). Both are required for embryonic development, as substantiated by their genetic ablation in the mouse (30), but it is unclear whether Mfn redundancy reflects specific functional differences between MFN1 and MFN2. The mammalian orthologue of mgm1p is the inner membrane dynamin-related protein OPA1. Mutations in Opa1 are associated with autosomal dominant optic atrophy, the leading cause of inherited optic neuropathy (31, 32). It has been proposed that OPA1 participates in a fission/fragmentation pathway (33–35) or in the maintenance of the structural integrity of the mitochondrial reticulum (36), but, until now, function of OPA1 was unclear, as was its interaction with other mitochondria-shaping proteins.
Here we investigated the role of OPA1 in controlling mitochondrial shape. A genetic analysis revealed that OPA1 requires MFN1 but not MFN2 to induce mitochondrial fusion. Our results identify a role for OPA1 in fusion of mammalian mitochondria and reveal a functional difference between MFN1 and MFN2.
Experimental Procedures
Plasmid Construction. Murine OPA1 cDNA (33) (corresponding to human transcript variant 1; a kind gift of Y. Kubo, Tokyo University, Tokyo) was subcloned into the ClaI site of pMSCV (BD-Clontech). K301A and R905stop mutants of OPA1 were generated by using QuikChange (Stratagene) and confirmed by DNA sequencing. Details on other plasmids used can be found in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
RNA Interference (RNAi). Stable RNAi against murine OPA1 nucleotide region 1813–1831 and scrambled control was delivered by using pSilencer2.1-U6/hygro (Ambion, Austin, TX). Details on this and on the generation of stable clones can be found in Supporting Materials and Methods.
Imaging. Details on epifluorescence, real-time, and static confocal imaging systems, parameters, and operations can be found in Supporting Materials and Methods.
Polyethylene Glycol (PEG) Fusion Assay. For PEG fusion assay, 5 × 105 mouse embryonic fibroblasts (MEFs) of the indicated genotype were transfected with mitochondrially targeted yellow fluorescent protein (mtYFP) or with mitochondrially targeted dsRED (mtRFP) alone or contransfected with mtYFP or mtRFP plus the indicated constructs. After 24 h, cells labeled with different fluorescent proteins were coplated at a 1:1 ratio onto 13-mm round coverslips. Fusion was then induced after 24 h by a 60-sec treatment with a 50% (wt/vol) solution of PEG 1500 in PBS (Sigma), followed by extensive washes in DMEM supplemented with 10% FCS. To inhibit de novo synthesis of fluorescent proteins, 30 min before PEG treatment cells were incubated with the protein synthesis inhibitor cyclohexymide (20 μg/ml, Sigma), which was subsequently kept in all solutions and tissue culture media until cells were fixed for 30 min with ice-cold 3.7% (vol/vol) formaldehyde in PBS. After two washes with PBS, coverslips were mounted on slides with Anti-Fade Reagent (Molecular Probes).
Results
OPA1 Induces Mitochondrial Tubulation. We addressed whether expression of OPA1 affected mitochondrial morphology. Transient transfection of WT MEFs from 129/SvEv background (30) with OPA1 (33) resulted in a several-fold elevation of OPA1 levels after 24 h, as judged by immunoblotting (Fig. 5, which is published as supporting information on the PNAS web site). Imaging of WT MEFs transfected with a mitochondrially targeted cyan fluorescent protein (mtCFP) showed mitochondria as individual, rod-shaped, or round-shaped organelles, with an average length of 3 ± 0.34 μm (n = 100 cells in five different experiments) along their major axis (Fig. 1A). Morphometric analysis confirmed that only 23% of the analyzed cells displayed elongated mitochondria, i.e., cells with axial length >5 μm and roundness index <0.5 in >50% of mitochondria (Fig. 1I). When mtCFP was cotransfected with OPA1, mitochondria appeared tubular and interconnected in a branched network, with axial lengths of up to 18 μm (Fig. 1B). More than 50% of the transfected cells displayed elongated mitochondria (Fig. 1I). Strong overexpression of OPA1 in HeLa (35) and COS-7 (33) cells causes clustering of small mitochondria, whereas in 3T3 fibroblasts it promotes tubulation (37). We tested whether under a strong CMV promoter OPA1 caused mitochondrial fragmentation in MEFs. Levels of OPA1 in MEFs transfected with pCDNA3.1-OPA1 were >3-fold higher than those obtained by transfection with pMSCV-OPA1. Nevertheless, pCDNA3.1-OPA1 also promoted mitochondrial elongation in MEFs (data not shown). This finding suggests that overexpression of OPA1 result in mitochondrial fragmentation only in cells where mitochondria are already organized in a network of interconnected tubuli, such as HeLa cells.
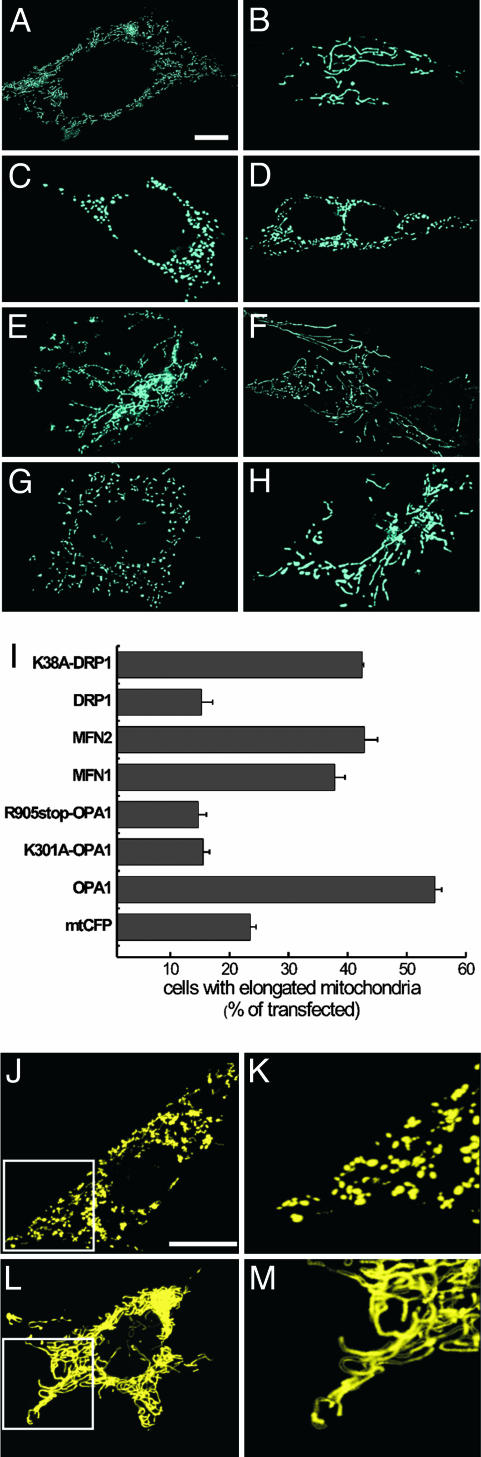
Fig. 1.
Overexpression of OPA1 promotes mitochondrial elongation. (A–H) Effect of mutations in the GTPase and coiled-coil domains. Mitochondrial shape in MEFs transfected with mitochondria-shaping proteins is shown. WT MEFs grown on coverslips were cotransfected with mtCFP and empty vector (A), WT OPA1 (B), K301A OPA1 (C), R905stop OPA1 (D), Mfn1 (E), Mfn2 (F), WT DRP1 (G), or K38A DRP1 (H). After 24 h, images of mtCFP fluorescence from randomly selected cells were acquired, deconvoluted, and stored. (Scale bar, 15 μm.) (I) Morphometric analysis of mitochondrial elongation. WT MEFs grown on coverslips were cotransfected with mtCFP and empty vector or the indicated mitochondria-shaping protein. After 24 h, 20 randomly selected images of mtCFP fluorescence were acquired, deconvoluted, stored, and subsequently classified as described in Supporting Materials and Methods. Data represent mean ± SE of 14 different experiments. (J–M) Volume-rendered 3D reconstructions of mitochondrial network in MEFs. MEFs grown on coverslips were cotransfected with mtYFP and empty vector (J) or OPA1 (L), and, after 24 h, randomly selected confocal z-axis stacks of mtYFP fluorescence were acquired, stored, reconstructed, and volume-rendered as described in Supporting Materials and Methods. K and M represent 3× magnification of the boxed area in J and L, respectively. The depth of stacks was 20 μm. (Scale bar, 15 μm.)
We compared the observed effects of OPA1 on mitochondrial morphology with those induced by other mitochondria-shaping proteins. After expression of MFN1 or MFN2, clusters of rod-shaped mitochondria coexisted with elongated organelles (28, 29, 38) (Fig. 1 E and F). Quantification revealed a positive effect of these proteins on mitochondrial tubulation in MEFs, albeit lower than that induced by OPA1 (Fig. 1I). Expression of the fission protein DRP1 resulted in the appearance of globular and dispersed mitochondria (Fig. 1 G and I), whereas its dominant negative K38A mutant (26) promoted mitochondrial elongation and formation of tubuli of interconnected mitochondria (Fig. 1 H and I). MEFs appear to be a valid model cell line to study mitochondria-shaping proteins.
Tubular structures that move in and out of the focal plane can be easily mistaken for individual rod or spherical organelles in conventional imaging. Therefore, we acquired stacks of mitochondrial images along the z-axis of the entire cell, followed by 3D-image reconstruction and volumetric rendering. In mock-transfected MEFs, mitochondria were separated and dispersed throughout the cytosol, with clusters of individual organelles visible at high-power magnification (Fig. 1 J and K). Cells transfected with OPA1 showed an intricate network of highly interconnected mitochondria that spanned the entire volume of the cytoplasm (Fig. 1 L and M). Thus, overexpression of OPA1 in MEFs results in efficient formation of a network of elongated mitochondria.
Function and Levels of OPA1 Determine Shape of the Mitochondrial Reticulum. OPA1 is affected in dominant optic atrophy, its mutations clustering in the GTPase, and in the C-terminal coiled-coil domain (39). Because the disease is transmitted as a dominant trait, it has been suggested that these mutations either act as dominant negative or induce a condition of haploinsufficiency leading to the clinical phenotype (36, 40). Therefore, we tested whether pathogenetic mutants of OPA1 or reduction of OPA1 levels modified mitochondrial morphology. We generated mutants of OPA1 GTPase and coiled-coil domains by substituting Lys-301 with Ala (K301A OPA1) and Arg-905 with a stop codon (R905stop OPA1), respectively. The K301A mutation has been shown to reduce the GTPase activity of OPA1 by >80%, whereas the R905stop mutant lacks the C-terminal coiled-coil domain thought to participate in protein–protein interactions (35). Transfection with K301A or R905stop OPA1 resulted in OPA1 levels comparable with those observed after transfection with WT OPA1 (Fig. 5), suggesting that these mutations do not affect protein stability. Mitochondria of cells transfected with K301A (Fig. 1C) or R905stop OPA1 (Fig. 1D) appeared globular. The reduction in mitochondrial elongation induced by these two mutants was comparable with that observed after transfection with the fission protein DRP1 (Fig. 1I).
To address the effect of reduced OPA1 levels on mitochondrial morphology, we turned to stable, plasmid-generated RNAi. We generated clones of MEFs from a mixed 129/CD1 background (41) stably expressing scrambled (control) or OPA1-targeted RNAi. In the OPA1-ablated clone A7, OPA1 levels were ≈30% compared with those of the control clone H4 (Fig. 2A). Other mitochondria-shaping proteins, such as DRP1, hFIS1, and MFN2, were not affected (data not shown). Clone A7 mitochondria appeared globular and fragmented as opposed to the rod, elongated organelles of clone H4 (Fig. 2B; see morphometric analysis in Fig. 2C). Fragmentation of mitochondria could be achieved by transient RNAi against OPA1 with 19-mer duplexes RNA in clone H4, suggesting that the mitochondrial morphology observed in clone A7 does not result from long-term adaptation of this particular clone (data not shown). In other screened clones, the degree of mitochondrial fragmentation correlated with levels of expressed OPA1 (data not shown). Taken together, these data demonstrate that levels of OPA1, as well as integrity of GTPase and coiled-coil domains of OPA1, determine mitochondrial morphology.
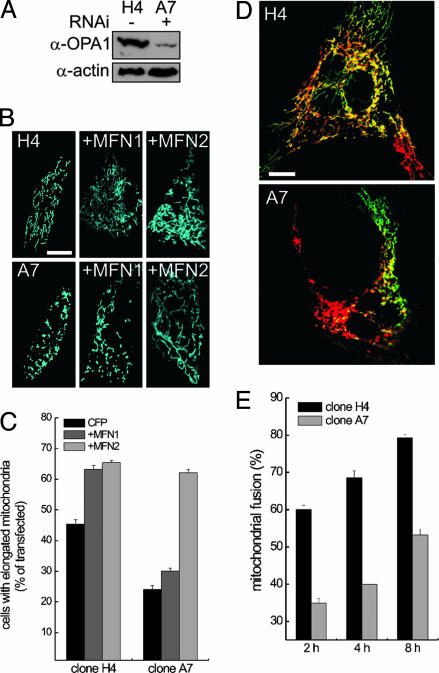
Fig. 2.
Mitochondrial shape, mitochondrial fusion, and response to mitofusins after RNAi against OPA1. (A) Expression levels of OPA1 in MEF clones. Cells (5 × 106) from clone H4 carrying pSilencer-control and clone A7 carrying pSilencer-OPA1 were lysed, and equal amounts of protein were separated and immunoblotted with anti-OPA1 (1:2,000) antiserum and an anti-actin monoclonal antibody (1:2,000, Chemicon). (B) Mitochondrial shape and response to mitofusins after OPA1 ablation. MEFs from clones H4 and A7 grown on coverslips were transfected with mtCFP or cotransfected with the indicated mitofusins. After 24 h, images of mtCFP fluorescence were acquired exactly as in Fig. 1 A–H. (Scale bar, 15 μm.) (C) Morphometric analysis. H4 and A7 MEFs grown on coverslips were transfected as indicated. Experiments were performed exactly as in Fig. 1I. Data represent mean ± SE of four different experiments. (D) Representative heteropolykaryons from H4 and A7 clones. MEFs from the indicated clones were transfected with mtYFP or mtRFP, coplated on glass coverslips, fused as described in Experimental Procedures, and fixed after 4 h. Confocal images of representative polykaryons are shown. (Scale bar, 20 μm.) (E) Quantitation of the effects of ablation of OPA1 on a mitochondrial PEG fusion assay. Experiments were carried out as described in D, except that cells were fixed at the indicated times. Mitochondrial fusion was evaluated as described in Experimental Procedures from 30 randomly selected polykaryons. Data represent mean ± SE of three different experiments.
OPA1 Participates in Mitochondrial Fusion. Mitochondrial shape is the result of the equilibrium between fusion and fission events. In principle, a shift from a rod-globular to a tubular mitochondrial shape could result from increased fusion as well as decreased fission. We addressed which direction of the fusion/fission equilibrium was controlled by OPA1. We performed PEG fusion assays on clones H4 and A7. MEFs transfected with mtYFP or mtRFP were coplated and triggered to fuse by treatment with PEG. Differential interference contrast light microscopy revealed cytoplasmic fusion as early as 30 min after treatment, with numerous polykaryons becoming apparent after 2 h. We quantified fusion events in the heteropolykaryons by measuring the fraction of mitochondria simultaneously positive for both mtYFP and mtRFP, which cannot be exchanged between mitochondria because the targeting sequences are cleaved after mitochondrial import. Double labeling thus reports only fusion of mitochondria with mixing of matricial content. Four hours after PEG treatment, polykaryons derived from control clone (H4) displayed ≈65% of mitochondria positive for both yellow fluorescent protein and red fluorescent protein (Fig. 2 D and E). Mitochondrial fusion increased with time, reaching 78% after 8 h. Ablation of OPA1 in clone A7 impaired fusion, reducing it to ≈52% even after 8 h (Fig. 2 D and E). We turned to WT MEFs to address whether increased OPA1 levels enhanced mitochondrial fusion. The number of mitochondria that were positive for both yellow fluorescent protein and red fluorescent protein increased linearly in WT MEFs after PEG treatment, reaching ≈55% after 8 h (Fig. 3 C and D, open squares). OPA1 increased fusion to ≈50% after 2 h and to ≈80% after 8 h (Fig. 3 C and D, filled squares). On the other hand, K301A-OPA1 impaired mitochondrial fusion, reducing it to 31 ± 3.4% after 8 h (data not shown). These results show that levels and function of OPA1 regulate mitochondrial shape by impinging on organellar fusion.
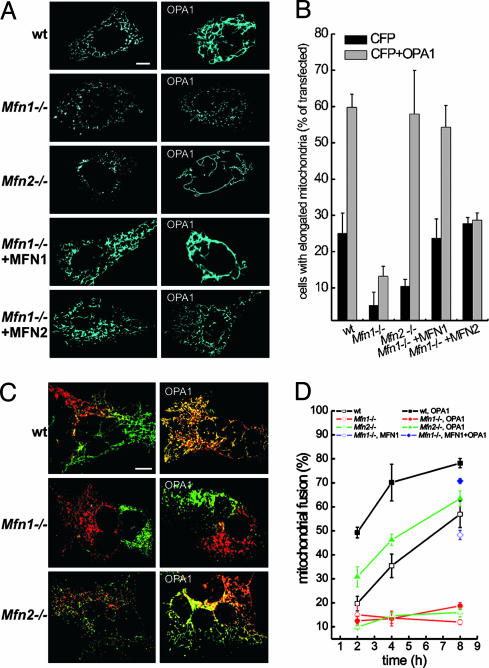
Fig. 3.
OPA1 changes the shape of the mitochondrial reticulum by promoting MFN1-dependent mitochondrial fusion. (A) Effect of OPA1 on the shape of the mitochondrial network in WT, Mfn1–/–, and Mfn2–/– MEFs. MEFs of the indicated genotype grown on coverslips were cotransfected with mtCFP and empty vector (Left) or OPA1 (Right). When indicated, MFN1 or MFN2 were cotransfected with mtCFP in Mfn1–/– MEFs. After 24 h, images of mtCFP fluorescence were acquired exactly as in Fig. 1 A–H.(B) Morphometric analysis of WT, Mfn1–/–, and Mfn2–/– mitochondria. MEFs of the indicated genotype grown on coverslips were cotransfected with mtCFP and empty vectors (black bars) or with OPA1 (gray bars). Where indicated, Mfn1–/– cells were complemented by cotransfection with MFN1 or MFN2. Experiments were performed exactly as in Fig. 1I. Data represent mean ± SE of nine different experiments. (C) MEFs of the indicated genotype were transfected with mtYFP or mtRFP (Left) or cotransfected with mtYFP or mtRFP plus OPA1 (Right), coplated on glass coverslips, fused with PEG as described in Experimental Procedures, and fixed after 4 h. Confocal images of representative polykaryons are shown. (Scale bar, 20 μm.) (D) Quantification of the effect of OPA1 on fusion of WT, Mfn1–/–, and Mfn2–/– mitochondria. The experiments were conducted as described in C, except that heteropolykaryons were fixed at the indicated times. In the experiments depicted with diamonds, Mfn1–/– cells were cotransfected with mtYFP or mtRFP plus MFN1 (⋄) and with mtYFP or mtRFP plus MFN1 and OPA1 (♦). Mitochondrial fusion was evaluated as described in Experimental Procedures from 30 randomly selected polykaryons. Data represent mean ± SE of four different experiments.
Fusion by OPA1 Depends on the Outer Membrane Protein Mitofusin 1. We next wished to ascertain whether OPA1 required other mitochondria-shaping proteins to control mitochondrial morphology. We turned to a genetic approach, testing the ability of overexpressed OPA1 to promote mitochondrial tubulation in MEFs deficient for either Mfn1 or Mfn2, the orthologues of yeast fzo1p. In Mfn1–/– MEFs, most mitochondria appeared globular, whereas in Mfn2–/– MEFs, the shape of mitochondria ranged from round dots to short rods (Fig. 3A) (30). Of note, levels and cleavage pattern of OPA1 were comparable in WT, Mfn1–/–, and Mfn2–/– MEFs (Fig. 6, which is published as supporting information on the PNAS web site), and transfection with pMSCV-OPA1 yielded similar levels of mitochondrial OPA1 in the MEFs of the different genotypes (Fig. 5). Expression of OPA1 induced mitochondrial tubulation in WT and Mfn2–/– but not in Mfn1–/– cells (Fig. 3A). Morphometric analysis confirmed that OPA1 can efficiently tubulate mitochondria in the absence of MFN2 but not MFN1 (Fig. 3B).
To address why Mfn1–/– cells did not respond to OPA1, we performed PEG fusion assays. In polykaryons derived from Mfn1–/– MEFs, mitochondria labeled with yellow fluorescent protein and red fluorescent protein sometimes segregated at the opposite sides of the cell with extremely low levels of fusion (Fig. 3C) that did not increase in time (Fig. 3D). In keeping with previous findings (30), this segregation occurred in only 20% of Mfn1–/– polykaryons. Mixing of differently labeled mitochondria was a constant feature of Mfn2–/– polykaryons, but mitochondria were still unable to fuse (Fig. 3 C and D) (30). Mitochondrial fusion in Mfn2–/– MEFs transfected with OPA1 was higher than in untransfected WT MEFs, whereas overexpression of OPA1 had no effect on mitochondrial fusion in polykaryons from Mfn1–/– cells (Fig. 3 C and D). These results show that MFN1 is required for OPA1-dependent mitochondrial fusion. Thus, OPA1, like mgm1p in Saccharomyces cerevisiae, requires outer membrane protein(s) to fuse mitochondria. Furthermore, our results reveal a functional difference between MFN1 and MFN2, at least in respect to OPA1-driven mitochondrial tubulation.
Specific Functional Interdependence Between MFN1 and OPA1. These results raised two crucial questions. Is the defect of Mfn1–/– mitochondria specific? If so, which step is affected by lack of MFN1, mitochondrial docking or the actual fusion process?
We first addressed whether the defect of Mfn1–/– mitochondria was specific. We checked whether reintroduction of MFN1 in Mfn1–/– cells corrected OPA1-induced mitochondrial elongation and fusion. Reexpression of MFN1 in Mfn1–/– MEFs restored mitochondrial shape (Fig. 3 A and B). Coexpression of MFN1 with OPA1 in Mfn1–/– cells resulted in tubular and elongated mitochondria (Fig. 3 A and B). Reintroduction of MFN1 corrected the defect of Mfn1–/– MEFs in mitochondrial PEG fusion assays (Fig. 3D, compare open diamonds with open squares) and enabled the pro-fusion effect of OPA1 (Fig. 3D, compare filled diamonds with filled squares). Because the mitochondrial defect of Mfn1–/– MEFs is complemented by MFN2 (30), we assessed whether introduction of MFN2 could also correct OPA1-driven mitochondrial tubulation. Expression of MFN2 in Mfn1–/– cells restored mitochondrial shape but did not allow OPA1-driven mitochondrial elongation (Fig. 3 A and B). To further test functional interdependence between OPA1 and mitofusins, we turned to OPA1 knockdown MEFs. Although levels of OPA1 did not influence mitochondrial elongation induced by MFN2, they proved essential for MFN1, which caused tubulation only in control H4 cells (Fig. 2 B and C). In complex, failure of Mfn1–/– mitochondria to fuse in response to OPA1 is a specific consequence of MFN1 deficiency, which cannot be complemented by MFN2. Moreover, MFN1 but not MFN2 requires adequate levels of OPA1 to promote mitochondrial elongation, further substantiating a functional interdependence between OPA1 and MFN1.
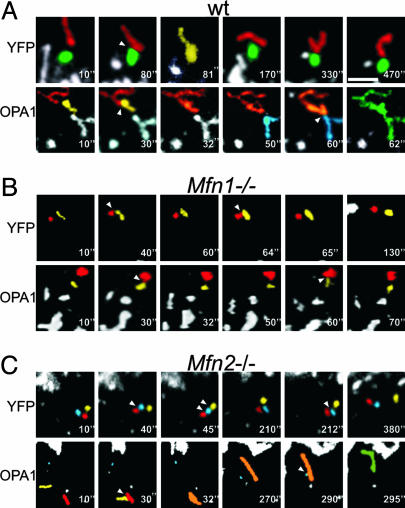
Number of Intermitochondrial Contacts Is Not Affected by Levels of Mitofusins and OPA1. Mfn1–/– mitochondria are characterized by motion defects resulting in disordered, Brownian-like wandering of the organelles around the cytoplasm (30). If this phenotype resulted in fewer contacts between mitochondria, it might cause the inability of OPA1 to promote fusion in Mfn1–/– MEFs. To address whether contacts between Mfn1–/– mitochondria were impaired, we performed 4D imaging of mitochondria, i.e., time series of z-stacks of mitochondrial images. Yellow fluorescent protein-expressing WT mitochondria juxtaposed and merged in tubuli in which individual mitochondria were not recognizable (Fig. 4A Upper). These tubuli reverted then to the original elemental mitochondria, probably by fission (Fig. 4A Upper). Mfn1–/– (Fig. 4B) and Mfn2–/– mitochondria (Fig. 4C) juxtaposed closely, remaining in contact for seconds, even minutes, but did not fuse to constitute tubuli in any of the acquired planes. We cannot exclude the formation of tubuli that rapidly reverted by fission, albeit we never observed fission events completing in <500 msec (the acquisition time of one stack). We quantified contacts by following randomly chosen mitochondria in the volume of the z-stack for the entire acquisition sequence. The number of mitochondrial contacts was similar in WT, Mfn1–/–, and Mfn2–/– MEFs (8.5 ± 1.5 contacts in WT, 7 ± 0.9 in Mfn1–/–, and 7.5 ± 0.8 in Mfn2–/– MEFs; n = 16 in four different experiments). However, differences existed in the number of “productive” contacts, leading to the formation of mitochondrial tubuli, which were 2.3 ± 0.4 in WT, 0.7 ± 0.3 in Mfn2–/–, and 0.4 ± 0.3 in Mfn1–/– cells (n = 16 in four different experiments). Although OPA1 overexpression did not change the total number of contacts, in WT MEFs it caused a 2- to 3-fold increase in tubulation events (5.25 ± 1.25 productive events in a total of 9.75 ± 2.25 contacts; n = 16 in four different experiments) (Fig. 4A). A similar pattern was observed in Mfn2–/– MEFs (Fig. 4C), with 4.2 ± 1.1 productive events in a total of 8.1 ± 0.8 contacts (n = 16 in four different experiments). Mfn1–/– mitochondria also were unable to merge into tubuli after OPA1 overexpression (Fig. 4B). In this case, contacts were 7.25 ± 2.6, but only 0.5 ± 0.3 were productive. In summary, the total number of contacts between mitochondria is not affected by OPA1 overexpression or by MFN deficiency. OPA1 facilitates fusion after contacts between WT and Mfn2–/– but not Mfn1–/– mitochondria. Taken together, our results suggest that OPA1 requires MFN1 to fuse the membranes of two juxtaposed mitochondria and not to produce intermitochondrial contacts.
Fig. 4.
The lack of MFN1 does not affect mitochondrial juxtaposition but blocks OPA1-induced mitochondrial tubulation. WT (A), Mfn1–/– (B), and Mfn2–/– (C) MEFs were cotransfected with mtYFP and empty vector (Upper) or with mtYFP and OPA1 (Lower), and, after 24 h, confocal z-stacks were acquired as described in Supporting Materials and Methods. Stacks were reconstructed and volume-rendered, and movements of individual mitochondria were tracked. Five-fold magnified portions of single z-planes corresponding to the indicated frames are shown. For the sake of clarity, mitochondria were individually labeled in different colors. Arrowheads indicate sites of mitochondrial contact. Colors of tubular mitochondria resulted from the merging of the colors of the individual mitochondria from which they originated. (Scale bar, 4 μm.)
Discussion
OPA1 is so far the only mammalian mitochondria-shaping protein localized in the inner mitochondrial membrane (34, 37). Albeit it shares 30% homology with S. cerevisiae mgm1p, which in budding yeast is crucial to maintain fusion-competent mitochondria (22, 23), its biological function remains unclear. Overexpression of OPA1 has been reported to promote fission and perinuclear clustering of mitochondria (33–35). Conversely, ablation of OPA1 resulted in mitochondrial depolarization, derangement of the mitochondrial network, and release of cytochrome c (36), or in localized constrictions, followed by fragmentation (35). It is difficult to reconcile the proposed pro-fission role of OPA1 with the known function in mitochondrial fusion of its yeast orthologue mgm1p. To define the biological function of OPA1, we have used a combination of genetics and imaging. Overexpression of OPA1 in MEFs induces mitochondrial elongation and tubulation, in keeping with what was observed in 3T3 fibroblasts (37), but at a variance from the results obtained in HeLa and COS7 cells (33–35). OPA1 might cause fragmentation only when it is expressed at very high levels in cells such as HeLa and COS7, where mitochondria are already elongated. This interpretation is also consistent with the finding that in COS7 cells the inactive K301A mutant of OPA1, which does not hydrolyze GTP (35), also causes fragmentation (33). Stable reduction of OPA1 levels by RNAi results in mitochondrial fragmentation, often associated with clustering. A similar mitochondrial phenotype is observed in HeLa cells after transient RNAi (35, 36). Assays of mitochondrial fusion show that OPA1 controls the complex process of mitochondrial fusion, a direct correlation being observed between levels of OPA1 and fusion efficiency. PEG fusion assays are limited in that they measure not only mitochondrial fusion per se, but also the preparatory events of mitochondrial juxtaposition and docking, which therefore need to be measured. Real-time imaging experiments show that OPA1 does not promote mitochondrial docking. Taken together, our data suggest that OPA1 impinges on the fusion step to regulate mitochondrial morphology.
Function of OPA1 requires an intact GTPase and C-terminal coiled-coil domain, two hot spots for OPA1 mutations in dominant optic atrophy (31, 32, 39). Overexpression of the K301A mutant of OPA1 impairs mitochondrial fusion; interestingly, monocytes from dominant optic atrophy patients carrying mutations in OPA1 GTPase domain display fragmented mitochondria (32). The C-terminal coiled-coil domain is also necessary for mitochondrial fusion. Coiled-coil regions often participate in protein–protein interaction (42), and integrity of the C-terminal domain of the S. cerevisiae orthologue of OPA1, mgm1p, is required for its activity (23). Recent evidence supports a model in which mgm1p mediates mitochondrial fusion through a multiprotein complex with the outer membrane proteins fzo1p and ugo1p (23). Although the mammalian homologue of ugo1p is not yet identified, fzo1p orthologues have been characterized in MFN1 and MFN2. Their ablation results in embryonic lethality, mitochondrial fragmentation, and heterogeneous mitochondrial dysfunction (30). We reasoned that OPA1 function might depend on other mitochondria shaping proteins, such as the outer membrane MFNs. In cells deficient in MFN1, OPA1 cannot promote mitochondrial fusion. This defect is complemented by reintroduction of MFN1 but not MFN2, unequivocally identifying outer membrane MFN1 as an essential functional partner of OPA1. Moreover, MFN1 is unable to promote mitochondrial elongation if OPA1 is ablated. Thus, OPA1 and MFN1 appear to functionally depend on each other.
A major dilemma is whether the two fzo1p homologues MFN1 and MFN2 are functionally redundant. Our genetic analysis provides evidence of a functional diversity between MFN1 and MFN2, suggesting a functional axis between OPA1 and MFN1. This finding might explain the morphological difference between mitochondria of Mfn1–/– and Mfn2–/– cells. In the absence of MFN2 the inner–outer membrane fusion machinery composed by OPA1 and MFN1 is still intact and can provide a low degree of fusion resulting in the few tubular mitochondria of Mfn2–/– MEFs. Because MFN1 appears to allow fusion of the inner and outer membrane, it will be interesting to assess whether during evolution MFN2 acquired other specialized functions like regulation of mitochondrial positioning and/or communication with other cellular structures.
It has been suggested that OPA1 could maintain cristae structure (36). During cell death mitochondrial fusion is inhibited (43); inactivation of OPA1 might therefore be a crucial step in mitochondrial remodeling during apoptosis. Because OPA1 is unable to promote fusion in the absence of MFN1, our results provide a framework to dissect specific effects of OPA1 during apoptosis on cristae structure vs. preservation of mitochondrial fusion.
Supplementary Material
Acknowledgments
We thank D. Chan (California Institute of Technology, Pasadena, CA) for the kind gift of WT, mfn1–/–, and mfn2–/– MEFs and for critical reading of the manuscript; V. Petronilli for helpful discussions; and P. Bernardi and M. Zaccolo for critical reading of the manuscript. O.M.d.B. is the recipient of a Bolsa de Doutoramento of the Fundação para a Ciência e Tecnologia (Lisbon). L.S. is an assistant Telethon scientist of the Dulbecco–Telethon Institute. This work was supported by Telethon Italy, the Glaucoma Research Foundation, and the Human Frontier Science Program Organization.
Author contributions: L.S. designed research; S.C., O.M.d.B., and B.D.Z. performed research; S.C. analyzed data; O.M.d.B. contributed new reagents/analytic tools; and L.S. wrote the paper.
Abbreviations: DRP1, dynamin-related protein 1; MEF, mouse embryonic fibroblast; mtCFP, mitochondrially targeted cyan fluorescent protein; mtYFP, mitochondrially targeted yellow fluorescent protein; mtRFP, mitochondrially targeted dsRED; MFN, mitofusin; PEG, polyethylene glycol; RNAi, RNA interference.
References
- 1.Jouaville, L. S., Ichas, F., Holmuhamedov, E. L., Camacho, P. & Lechleiter, J. D. (1995) Nature 377, 438–441. [DOI] [PubMed] [Google Scholar]
- 2.Wang, X. (2001) Genes Dev. 15, 2922–2933. [PubMed] [Google Scholar]
- 3.Frey, T. G. & Mannella, C. A. (2000) Trends Biochem. Sci. 25, 319–324. [DOI] [PubMed] [Google Scholar]
- 4.Mannella, C. A., Pfeiffer, D. R., Bradshaw, P. C., Moraru, I. I., Slepchenko, B., Loew, L. M., Hsieh, C. E., Buttle, K. & Marko, M. (2001) IUBMB Life 52, 93–100. [DOI] [PubMed] [Google Scholar]
- 5.Bernardi, P. & Azzone, G. F. (1981) J. Biol. Chem. 256, 7187–7192. [PubMed] [Google Scholar]
- 6.Scorrano, L., Ashiya, M., Buttle, K., Weiler, S., Oakes, S. A., Mannella, C. A. & Korsmeyer, S. J. (2002) Dev. Cell 2, 55–67. [DOI] [PubMed] [Google Scholar]
- 7.Griparic, L. & van der Bliek, A. M. (2001) Traffic 2, 235–244. [DOI] [PubMed] [Google Scholar]
- 8.Legros, F., Lombes, A., Frachon, P. & Rojo, M. (2002) Mol. Biol. Cell 13, 4343–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amchenkova, A. A., Bakeeva, L. E., Chentsov, Y. S., Skulachev, V. P. & Zorov, D. B. (1988) J. Cell Biol. 107, 481–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pacher, P. & Hajnoczky, G. (2001) EMBO J. 20, 4107–4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, T. J., Berridge, M. J., Lipp, P. & Bootman, M. D. (2002) EMBO J. 21, 1616–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Catlett, N. L. & Weisman, L. S. (2000) Curr. Opin. Cell Biol. 12, 509–516. [DOI] [PubMed] [Google Scholar]
- 13.Frank, S., Gaume, B., Bergmann-Leitner, E. S., Leitner, W. W., Robert, E. G., Catez, F., Smith, C. L. & Youle, R. J. (2001) Dev. Cell 1, 515–525. [DOI] [PubMed] [Google Scholar]
- 14.Shaw, J. M. & Nunnari, J. (2002) Trends Cell Biol. 12, 178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bleazard, W., McCaffery, J. M., King, E. J., Bale, S., Mozdy, A., Tieu, Q., Nunnari, J. & Shaw, J. M. (1999) Nat. Cell Biol. 1, 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sesaki, H. & Jensen, R. E. (1999) J. Cell Biol. 147, 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mozdy, A. D., McCaffery, J. M. & Shaw, J. M. (2000) J. Cell Biol. 151, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tieu, Q., Okreglak, V., Naylor, K. & Nunnari, J. (2002) J. Cell Biol. 158, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermann, G. J., Thatcher, J. W., Mills, J. P., Hales, K. G., Fuller, M. T., Nunnari, J. & Shaw, J. M. (1998) J. Cell Biol. 143, 359–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rapaport, D., Brunner, M., Neupert, W. & Westermann, B. (1998) J. Biol. Chem. 273, 20150–20155. [DOI] [PubMed] [Google Scholar]
- 21.Sesaki, H. & Jensen, R. E. (2001) J. Cell Biol. 152, 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong, E. D., Wagner, J. A., Gorsich, S. W., McCaffery, J. M., Shaw, J. M. & Nunnari, J. (2000) J. Cell Biol. 151, 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong, E. D., Wagner, J. A., Scott, S. V., Okreglak, V., Holewinske, T. J., Cassidy-Stone, A. & Nunnari, J. (2003) J. Cell Biol. 160, 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sesaki, H., Southard, S. M., Yaffe, M. P. & Jensen, R. E. (2003) Mol. Biol. Cell 14, 2342–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labrousse, A. M., Zappaterra, M. D., Rube, D. A. & van der Bliek, A. M. (1999) Mol. Cell 4, 815–826. [DOI] [PubMed] [Google Scholar]
- 26.Smirnova, E., Griparic, L., Shurland, D. L. & van der Bliek, A. M. (2001) Mol. Biol. Cell 12, 2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James, D. I., Parone, P. A., Mattenberger, Y. & Martinou, J. C. (2003) J. Biol. Chem. 278, 36373–36379. [DOI] [PubMed] [Google Scholar]
- 28.Santel, A. & Fuller, M. T. (2001) J. Cell Sci. 114, 867–874. [DOI] [PubMed] [Google Scholar]
- 29.Rojo, M., Legros, F., Chateau, D. & Lombes, A. (2002) J. Cell Sci. 115, 1663–1674. [DOI] [PubMed] [Google Scholar]
- 30.Chen, H., Detmer, S. A., Ewald, A. J., Griffin, E. E., Fraser, S. E. & Chan, D. C. (2003) J. Cell Biol. 160, 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexander, C., Votruba, M., Pesch, U. E., Thiselton, D. L., Mayer, S., Moore, A., Rodriguez, M., Kellner, U., Leo-Kottler, B., Auburger, G., et al. (2000) Nat. Genet. 26, 211–215. [DOI] [PubMed] [Google Scholar]
- 32.Delettre, C., Lenaers, G., Griffoin, J. M., Gigarel, N., Lorenzo, C., Belenguer, P., Pelloquin, L., Grosgeorge, J., Turc-Carel, C., Perret, E., et al. (2000) Nat. Genet. 26, 207–210. [DOI] [PubMed] [Google Scholar]
- 33.Misaka, T., Miyashita, T. & Kubo, Y. (2002) J. Biol. Chem. 277, 15834–15842. [DOI] [PubMed] [Google Scholar]
- 34.Satoh, M., Hamamoto, T., Seo, N., Kagawa, Y. & Endo, H. (2003) Biochem. Biophys. Res. Commun. 300, 482–493. [DOI] [PubMed] [Google Scholar]
- 35.Griparic, L., van der Wel, N. N., Orozco, I. J., Peters, P. J. & van der Bliek, A. M. (2004) J. Biol. Chem. 279, 18792–18798. [DOI] [PubMed] [Google Scholar]
- 36.Olichon, A., Baricault, L., Gas, N., Guillou, E., Valette, A., Belenguer, P. & Lenaers, G. (2003) J. Biol. Chem. 278, 7743–7746. [DOI] [PubMed] [Google Scholar]
- 37.Olichon, A., Emorine, L. J., Descoins, E., Pelloquin, L., Brichese, L., Gas, N., Guillou, E., Delettre, C., Valette, A., Hamel, C. P., et al. (2002) FEBS Lett. 523, 171–176. [DOI] [PubMed] [Google Scholar]
- 38.Santel, A., Frank, S., Gaume, B., Herrler, M., Youle, R. J. & Fuller, M. T. (2003) J. Cell Sci. 116, 2763–2774. [DOI] [PubMed] [Google Scholar]
- 39.Delettre, C., Griffoin, J. M., Kaplan, J., Dollfus, H., Lorenz, B., Faivre, L., Lenaers, G., Belenguer, P. & Hamel, C. P. (2001) Hum. Genet. 109, 584–591. [DOI] [PubMed] [Google Scholar]
- 40.Delettre, C., Lenaers, G., Pelloquin, L., Belenguer, P. & Hamel, C. P. (2002) Mol. Genet. Metab. 75, 97–107. [DOI] [PubMed] [Google Scholar]
- 41.Wei, M. C., Zong, W. X., Cheng, E. H., Lindsten, T., Panoutsakopoulou, V., Ross, A. J., Roth, K. A., MacGregor, G. R., Thompson, C. B. & Korsmeyer, S. J. (2001) Science 292, 727–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lupas, A. (1996) Trends Biochem. Sci. 21, 375–382. [PubMed] [Google Scholar]
- 43.Karbowski, M., Arnoult, D., Chen, H., Chan, D. C., Smith, C. L. & Youle, R. J. (2004) J. Cell Biol. 164, 493–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.