Abstract
Lysine 5,6-aminomutase is an adenosylcobalamin and pyridoxal-5′-phosphate-dependent enzyme that catalyzes a 1,2 rearrangement of the terminal amino group of dl-lysine and of l-β-lysine. We have solved the x-ray structure of a substrate-free form of lysine-5,6-aminomutase from Clostridium sticklandii. In this structure, a Rossmann domain covalently binds pyridoxal-5′-phosphate by means of lysine 144 and positions it into the putative active site of a neighboring triosephosphate isomerase barrel domain, while simultaneously positioning the other cofactor, adenosylcobalamin, ≈25 Å from the active site. In this mode of pyridoxal-5′-phosphate binding, the cofactor acts as an anchor, tethering the separate polypeptide chain of the Rossmann domain to the triosephosphate isomerase barrel domain. Upon substrate binding and transaldimination of the lysine-144 linkage, the Rossmann domain would be free to rotate and bring adenosylcobalamin, pyridoxal-5′-phosphate, and substrate into proximity. Thus, the structure embodies a locking mechanism to keep the adenosylcobalamin out of the active site and prevent radical generation in the absence of substrate.
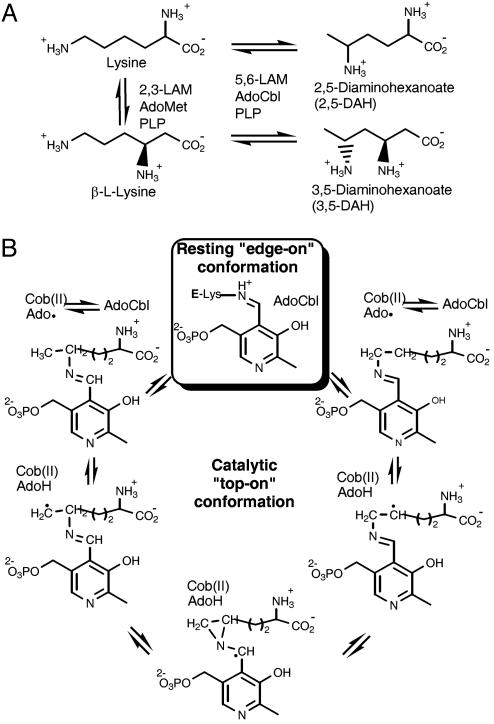
Adenosylcobalamin (AdoCbl; coenzyme B12) is nature's biochemical radical reservoir, capable of catalyzing challenging chemical reactions by way of H atom abstraction and the generation of free-radical intermediates (1–3). AdoCbl-dependent isomerases catalyze 1,2 shifts between an H atom and a functional group such as –OH, –NH3+, –(CO)S-coenzyme A, or other carbon-based groups. The catalytic power of AdoCbl lies in the homolytic cleavage of its weak (≈30 kcal/mol) organometallic C–Co bond, formed between an octahedral Co(III) center with five N coordinations and a 5′-deoxyadenosyl (Ado) axial ligand. C–Co bond homolysis results in the transient formation of cob(II)alamin and 5′-deoxyadenosyl radical (Ado•). Ado• abstracts an H atom from the substrate, forming a substrate radical and 5′-deoxyadenosine (AdoH). To close the catalytic cycle, substrate reabstracts the H atom from AdoH, and recombination of cob(II)alamin and Ado• accompanies product formation. Amazingly, the enzymatic rate of C–Co bond homolysis is enhanced by a factor of ≈1012 over nonenzymatic homolysis (4, 5). AdoCbl-dependent isomerases are often present in catabolic pathways and can serve to rearrange the substrate's carbon skeleton and/or functional groups for further degradation. One such pathway that operates in several bacterial species is the fermentation of lysine to yield acetate. Interestingly, the lysine fermentation pathway contains two analogous enzymes: lysine 5,6-aminomutase (5,6-LAM), which is AdoCbl-dependent (6, 7), and lysine 2,3-aminomutase (2,3-LAM), which is an S-adenosylmethionine (AdoMet or SAM)-dependent iron–sulfur enzyme (8–10). Both enzymes require pyridoxal 5′-phosphate (PLP) (8, 11) in addition to AdoCbl or AdoMet, and both catalyze a 1,2 amino group shift with concomitant H atom migration (Fig. 1A). In 5,6-LAM, AdoCbl is the source of the transient Ado•, whereas, in 2,3-LAM, an “AdoMet radical” or “radical SAM” enzyme, Ado•, is produced from AdoMet coordinated to the unique iron of a [4Fe-4S]2+ cluster and an exogenous electron. The exogenous electron is transferred first into the [4Fe-4S]2+ cluster and then into AdoMet, cleaving AdoMet to give methionine and the transient Ado•. For both 5,6-LAM and 2,3-LAM, radical propagation from Ado• to the substrate–PLP covalent complex (known as the external aldimine) initiates the isomerization, and both reaction mechanisms are likely to involve analogous intermediates. The requirement for PLP in these and one other 1,2-aminomutase (AdoCbl-dependent ornithine aminomutase) is not fully understood. Although computational studies suggest that the role of PLP is to stabilize high-energy radical intermediates by providing a conjugated π-electron system, over which the unpaired electron may be delocalized (12) (Fig. 1B), model studies show that the rearrangement readily takes place at cryogenic temperatures in the simplest model, the aziridylcarbinyl radical, in the absence of any aromatic substituent (13).
Fig. 1.
Aminomutases in the bacterial lysine fermentation pathway. (A) 5,6-LAM and 2,3-LAM catalyze similar reactions and act on similar substrates. Both enzymes require PLP, but 5,6-LAM is AdoCbl-dependent, whereas 2,3-LAM is an AdoMet-dependent iron-sulfur enzyme. The natural substrates of 5,6-LAM include dl-lysine and β-l-lysine. 2,3-LAM acts on l-lysine and does not accept d-lysine as a substrate. (B) Proposed mechanism of 5,6-LAM, modified from ref. 38. The boxed step represents the state of the enzyme observed in this study. The unboxed steps are proposed to occur while 5,6-LAM is in the hypothetical top-on conformation (see the Introduction).
All radical AdoMet- or AdoCbl-dependent enzymes rely on Ado• for catalysis, yet the formation of this highly oxidative intermediate must be controlled to prevent aberrant reactions. C–Co bond homolysis and the transient formation of Ado• is triggered by substrate binding in the AdoCbl-dependent enzymes methylmalonyl-coenzyme A mutase (MCM) (14), glutamate mutase (GM) (15, 16), and diol dehydratase (17), whereas effector binding triggers the C–Co bond homolysis in the AdoCbl-dependent ribonucleotide reductase (18). How substrates or effectors afford such enormous Co–C bond cleavage rate accelerations in AdoCbl-dependent enzymes is a question that has intrigued scientists for decades.
We present the crystal structure of the substrate-free holoenzyme form of 5,6-LAM from Clostridium sticklandii, which is the first structure of an enzyme that utilizes both AdoCbl and cofactors. The structure reveals that AdoCbl is bound by a Rossmann domain and that PLP, which is tethered to the B12-binding domain by means of its imine linkage, is bound in the putative active site, at the top of a triosephosphate isomerase (TIM) barrel domain. Thus, 5,6-LAM joins a group of three other AdoCbl-dependent radical enzymes (GM, MCM, and diol dehydratase) (19–21) and one AdoMet-dependent radical enzyme (biotin synthase) (22) in using the TIM barrel fold to sequester substrates that form free-radical intermediates. Our structure is distinctive among all PLP-dependent enzymes in that the imine linkage is provided by a Rossmann domain. Strikingly, in the precatalytic, substrate-free conformation captured in our structure, the PLP and AdoCbl cofactors are separated by a distance of ≈25 Å, suggesting that a gross conformational change occurs upon substrate binding.
Materials and Methods
Enzyme Preparation. Selenomethionine (SeMet)-incorporated 5,6-LAM from C. sticklandii was purified and reconstituted with PLP as follows. A plasmid KamDE containing the genes encoding both β and α subunits of 5,6-LAM was used to transform Escherichia coli B834(DE3) cells. A 1% overnight culture was used to inoculate 1 liter of SeMet media, prepared according to Budisa et al. (23) with the omission of l-cysteine and addition of 5 μM pyridoxal hydrochloride. The culture was grown for 16 h at 37°C. The yield was 25 g of wet cells from 10 liters of minimal media. The SeMet recombinant protein was purified by the procedure of Chang and Frey (24) without the gel filtration chromatography step.
Crystallization, Structure Determination, and Refinement. Crystals were grown by using hanging-drop vapor diffusion techniques at room temperature in a dark room, and all solutions and crystals were manipulated under red light until after cryocooling. Protein solution (12 mg/ml 5,6-LAM/0.5 mM 2-mercaptoethanol/10 mM triethanolamine, pH 7.2/4.5 mM AdoCbl) was mixed in a 1:1 ratio with precipitant solution [(0.1 M Tris·HCl, pH 8.0/0.2 M sodium acetate/24% (wt/vol) polyethylene glycol 2000 monomethyl ether] and equilibrated over 0.5 ml of precipitant solution. Crystals appeared within 24 h and were immediately cryoprotected by quickly soaking in precipitant solution with 20% glycerol added and plunging into liquid nitrogen. Crystals belong to space group P3121, with one αβ heterodimer per asymmetric unit, and a = b = 99.7 Å, c = 168.8 Å (see Table 1). Data were collected at the Argonne National Laboratory beamline NE-CAT 8-BM, equipped with a charge-coupled device detector (Area Detector Systems, Poway, CA). Data were processed and scaled with denzo and scalepack (25). A single set of data, collected at the Se absorption peak wavelength (0.97918 Å), was used to solve the structure. Twenty-nine Se atoms and the Co atom of AdoCbl were located and refined with solve (26), with a mean figure of merit of 0.40–3.0 Å resolution. The experimental electron density map was subjected to solvent-flattening with resolve (26), resulting in a good-quality map that allowed us to begin model building. Iterative rounds of model building in xfit (27) and refinement in cns (28) resulted in the final model at 2.8-Å resolution. The refined structure contains all 516 residues of the α subunit (chain A), residues 24–84 and 102–261 of the β subunit (chain B), one AdoCbl molecule, and one PLP molecule (as the imine adduct to Lys-144β). A simulated annealing composite omit map was used to validate the final structure. There is no electron density for the histidine tag, residues 1–23β, 85–101β, or 262β. The final model has all residues residing in the allowed regions of the Ramachandran plot (87.7% in the most-favored regions, 11% in additionally allowed regions, and 1.3% in generously allowed regions), as calculated by procheck (29). The average B factors are as follows: TIM barrel and “accessory clamp,” 31.3 Å2 (main chain) and 36.1 Å2 (side chain); dimerization domain, 62.9 Å2 (main chain) and 67.7 Å2 (side chain); Rossmann domain, 31.6 Å2 (main chain) and 38.1 Å2 (side chain); cobalamin (Cbl), 34.8 Å2; AdoH, 70.1 Å2; and PLP, 24.9 Å2.
Table 1. Data collection and refinement statistics.
| Wavelength, Å | 0.97918 |
| Resolution range, Å | 50.0-2.8 |
| l/σ(l) | 18.6 (6.6) |
| Unique reflections* | 45,836 |
| Rsym† | 9.3 (28.4) |
| Redundancy | 6.8 |
| Completeness, % | 99.8 (99.8) |
| Rcryst‡/Rfree, % | 21.9/26.9 |
| Non-hydrogen atoms in ASU§ | 5,864 |
| No. of reflections | |
| Working set | 24,098 |
| Test set | 2,378 |
| RMSD¶ | |
| Protein bonds, Å | 0.009 |
| Protein angles, ° | 1.4 |
Values in parentheses are for the highest-resolution shell.
For the purpose of phasing, Friedel pairs were not merged, and this is accounted for in the number of unique reflections.
Rsym = [ΣhklΣi|li(hkl) - 〈l(hkl)〉|]/ΣhklΣili(hkl) for hkl independent reflections and i observations of a given reflection. 〈l(hkl)〉 is the mean intensity of the Miller index (hkl).
Rcryst = Σhkl|Fo(hkl)| - |Fc(hkl)|/Σhkl|Fo(hkl)|. Rfree = Rcryst for a test set of reflections (9.9% of all reflections) not included in refinement. No sigma cutoff was used in the refinement.
Asymmetric unit.
Root mean-square deviation.
Results
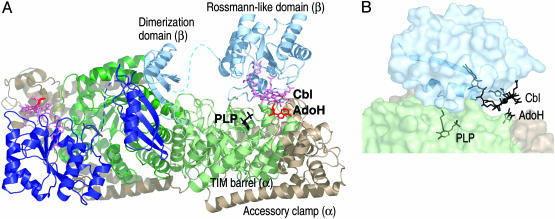
Overall Structure. 5,6-LAM is an α2β2 tetramer (30) that can be thought of as a dimer of αβ units (Fig. 2A). In the crystal, the asymmetric unit contains one αβ dimer, and crystallographic symmetry produces a likely physiological tetramer that buries 5,736 Å2 (19%) of the αβ heterodimer surface area. The large α subunit (538 residues) is composed of the PLP-binding TIM barrel domain and several additional α-helices and β-strands at the N and C termini (for topology diagram, see Fig. 6, which is published as supporting information on the PNAS web site). These helices and strands form an intertwined accessory clamp structure that wraps around the sides of the TIM barrel and extends up toward the Ado ligand of the Cbl cofactor (Fig. 2). This accessory clamp provides most of the interactions observed between the protein and the Ado ligand of the Cbl, suggesting that its role is mainly in stabilizing AdoCbl in the precatalytic resting state. The small β subunit (262 residues) comprises two domains: the N-terminal dimerization domain, which has the same fold as the Cu-binding domain of the Alzheimer's disease amyloid precursor protein (Protein Data Bank ID code 1OWT), and the AdoCbl-binding Rossmann domain, which also provides the imine bond to PLP. The Rossmann domain interacts with the C terminus of the TIM barrel, placing PLP into the top of the barrel while projecting AdoCbl to the edge of the barrel, far from the PLP-binding site (Fig. 2B). The dimerization domain of β forms a continuous β-sheet with the dimerization domain of the second αβ unit and buries an extensive surface of hydrophobic residues. In addition, this dimerization domain forms hydrogen-bonding and hydrophobic contacts with the TIM barrel domain of the α subunit, whereas no contacts are observed with the Rossmann domain, although these domains are linked by a 16-residue-long disordered loop that we could not model. The average B factor of the dimerization domain is approximately twice that of either the Rossmann domain or the TIM barrel and accessory clamp, suggesting a higher degree of mobility for the dimerization domain. The α2β2 tetramer is arranged such that there is no direct interaction between the active sites of either αβ pair. The presence of a TIM barrel and a Rossmann domain in 5,6-LAM is consistent with the domain usage in other Cbl-dependent base-off enzymes, such as GM (19), MCM (20), and methionine synthase (31, 32), but the orientation of the Rossmann domain relative to the TIM barrel is markedly different and is likely to be mechanistically relevant (discussed below). It is also interesting that both subunits play a role in binding both cofactors; AdoCbl is bound at the C terminus of the Rossmann domain of β and by the accessory clamp at the edge of the α subunit, whereas PLP is bound at the C terminus of the TIM domain of α and by a lysine residue at the edge of the β subunit (Fig. 2B).
Fig. 2.
Overall structure of 5,6-LAM and location of cofactors. (A) Ribbon diagram of the α2β2 5,6-LAM tetramer with the accessory clamp of α in brown, the TIM barrel of α in green, and the Rossmann domain and the dimerization domain of β in blue. The dashed line represents the disordered loop connecting the two domains of β. The second αβ unit is represented in darker colors. AdoH is shown in red sticks, Cbl in pink sticks and sphere, and PLP in black sticks. With the exception of Figs. 1, 2, and 3B, all figures were prepared by using pymol (42). (B) Relative positions of PLP and AdoCbl. PLP is inserted into the C terminus of the TIM barrel by Lys-144β, which anchors the Rossmann domain in an off-center conformation on the top corner of the TIM barrel. His-133β (the lower axial ligand to Co in Cbl), Lys-144β (which forms the imine bond to PLP), PLP, Cbl, and AdoH are all depicted in black sticks. The secondary structural elements of the Rossmann domain that contain His-133β and Lys-144β are shown in a ribbon representation. Opaque domain surfaces are shown and are colored as in Fig. 1 A.
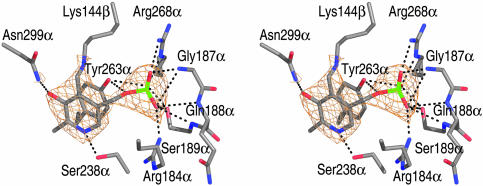
PLP–Protein Interactions. PLP-dependent enzymes of known structure can be sorted into families based on their respective folds. Enzymes of fold types I, II, III, IV, and V are represented by aspartate aminotransferase, tryptophan synthase, alanine racemase, d-amino acid aminotransferase, and glycogen phosphorylase, respectively (33). 5,6-LAM cannot be placed into any of these five PLP enzyme families, although it does share features with fold types II, III, and IV. As with fold type III, PLP is bound at the top of the TIM barrel pore, but the imine linkage is formed to the Rossmann domain rather than to a lysine of the TIM barrel, and a least-squares superposition of 5,6-LAM and alanine racemase reveals that PLP does not occupy analogous positions at the C termini of the TIM barrels. In addition, the second domain of fold type III is composed mainly of β-strands rather than having a Rossmann fold. The similarity to fold type II proteins is more limited and focuses on the use of a serine residue (Ser-238α in 5,6-LAM), rather than the more typical aspartic acid, to hydrogen-bond with the pyridine nitrogen. In agreement with fold types III and IV, the si face of PLP is solvent-exposed (33).
Biochemical, mutagenesis, and mass-spectrometry studies on 5,6-LAM from Porphyromonas gingivalis (34), combined with sequence alignment against the C. sticklandii enzyme (≈67% identity), support our assignment of Lys-144β as the Lys residue that makes an imine linkage to PLP (Fig. 3). Lys-144β resides at the N terminus of a short glycine-rich loop (144β-KGYAGHYG-151β) that is highly conserved across all 5,6-LAM sequences. This loop interrupts the second helix of the Rossmann domain one turn before the end of the helix (Fig. 2B). As was previously recognized (34), this represents a distinctive PLP-binding motif. The terminal amino group of Lys-144β does not seem to be coplanar with the pyridine ring of PLP, leading us to believe that the internal aldimine was photoreduced in the x-ray beam. Significantly, all contacts to the PLP cofactor, except for the imine linkage, are made from residues of the TIM barrel (Fig. 3) and are similar to PLP–protein interactions observed in other PLP-dependent enzymes. These interactions include π-stacking, electrostatic interactions with the phosphate, and hydrogen bonding. Tyr-263α forms π-electron stacking interactions with the pyridine ring and also hydrogen-bonds to the phosphate moiety. The phenolic oxygen of PLP, proposed to be important for intermediate stabilization in PLP- and AdoCbl-dependent 1,2-aminomutases (12), is observed to hydrogen-bond to the side chain of Asn-299α. The phosphate group interacts with two Arg side chains (Arg-184α and Arg-268α), the side chain of Ser-189α, and a number of main-chain amides (Gly-187α, Gln-188α, and Ser-189α). As in all PLP enzymes of known structure, the phosphate moiety of PLP is bound near the N terminus of an α-helix (in this case, a short helical turn, composed of 188α-QSL-190α).
Fig. 3.
PLP bound in the putative active-site of 5,6-LAM. Stereoview of the putative active site of 5,6-LAM. Lys-144β forms an imine bond to PLP; all other protein–PLP interactions are made by residues of the TIM barrel. A simulated annealing composite 2 Fo – Fc omit electron density map (orange mesh), contoured at 1.5 σ, is shown around the PLP. Unless otherwise noted, the coloring scheme for all stick or ball-and-stick diagrams is as follows: gray, C; red, O; blue, N; green, P.
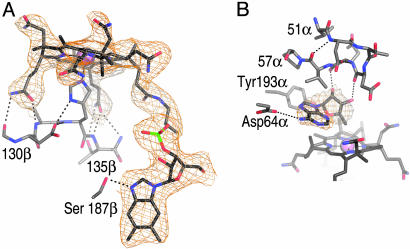
AdoCbl–Protein Interactions. AdoCbl binds to 5,6-LAM with 6.6 μM affinity and stabilizes the enzyme against thermal lability (24). AdoCbl was added to 5,6-LAM immediately before crystallization, and we observed electron density that is consistent with the cofactor (Fig. 4). 5,6-LAM contains a “base-off” AdoCbl-binding sequence (131β-DxHxxG... Sxl... GG-222β) (24) and binds AdoCbl in the base-off conformation, with His-133β replacing the intrinsic dimethylbenzimidazole (DMB) substituent of the cofactor as the lower axial ligand to the cobalt. Binding of AdoCbl by the Rossmann domain of 5,6-LAM is similar to Cbl binding in methionine synthase (MS) (31), MCM (20), and GM (19). The binding determinants for Cbl include several residues that hydrogen-bond to the propionamide side chains of the corrin ring (Thr-130β, Ala-132β, Thr-134β, Val-135β, Thr-191β, and Gln-192β), and, as in the case of MS, MCM, and GM, a serine residue (Ser-187β), which hydrogen-bonds to the DMB N atom (Fig. 4A). One potential hydrogen bond between the ribose moiety of the DMB tail and the protein is observed: the main-chain carbonyl O of Arg-243β is 2.8 Å away from the 2′-OH. The DMB moiety is bound in a largely hydrophobic cavity and is in van der Waals contact with Val-248β, Leu-185β, and Phe-239β, as well as Gly-221β and Gly-222β, two residues of the base-off Cbl-binding sequence motif. The phosphate moiety of the DMB tail, which is not observed to contact the protein directly, is bound near the surface of the Rossmann domain. Based on the structures of GM and MCM, we assume that the phosphate interacts with solvent, but, due to the moderate resolution of our structure, we did not model water molecules.
Fig. 4.
The AdoCbl-binding site. (A) View of Cbl within a 1.8-Å radius simulated annealing composite 2 Fo – Fc omit electron density map contoured at 1.0 σ. Hydrogen bonds between a propionamide side chain of the corrin ring and the backbone N and side chain of Thr-191β and the backbone N of Gln-192β are omitted for clarity. (B) View of AdoH within a 1.8-Å radius simulated annealing 2 Fo – Fc omit electron density map contoured at 1.0 σ. Tyr-193α is part of the TIM barrel domain; otherwise, all protein-AdoH contacts are made by residues of the accessory clamp.
All solutions and crystals containing AdoCbl were handled under red light until after cryocooling. Despite our stringent efforts to prevent the cleavage of the photolabile C–Co bond, the simulated annealing composite omit map suggests that this bond is mostly cleaved (Fig. 4), and we have modeled the Ado moiety as AdoH. We believe that the cleavage of AdoCbl is nonenzymatic and is due to photoreduction of the C–Co bond in the x-ray beam, a phenomenon reported for other AdoCbl enzymes (35). In the structure of 5,6-LAM, AdoH adopts a syn conformation about the glycosidic bond, in contrast to GM (36) and MCM (37), where the adenine ring of Ado is anti to the ribose ring. Here, AdoH is forced to adopt the syn conformation because of a steric clash between the adenine ring and Tyr-193α that would arise if AdoH were in the anti conformation. AdoH interacts with residues of the accessory clamp: the 2′ and 3′ –OH groups of the ribose moiety of AdoH hydrogen-bond to the main-chain carbonyls of Glu-55α and Asp-54α, respectively, and the exocyclic amino group of the adenine ring hydrogen-bonds to Asp-64α (Fig. 4B). Tyr-193α and Val-56α are positioned for hydrophobic interaction with the adenine moiety of AdoH. Interestingly, the AdoH-binding loop, composed of residues 51α–57α, forms a distorted β-hairpin structure that allows for two adjacent main-chain carbonyls to hydrogen-bond with the 2′ and 3′ –OH groups of AdoH (Fig. 4B).
Discussion
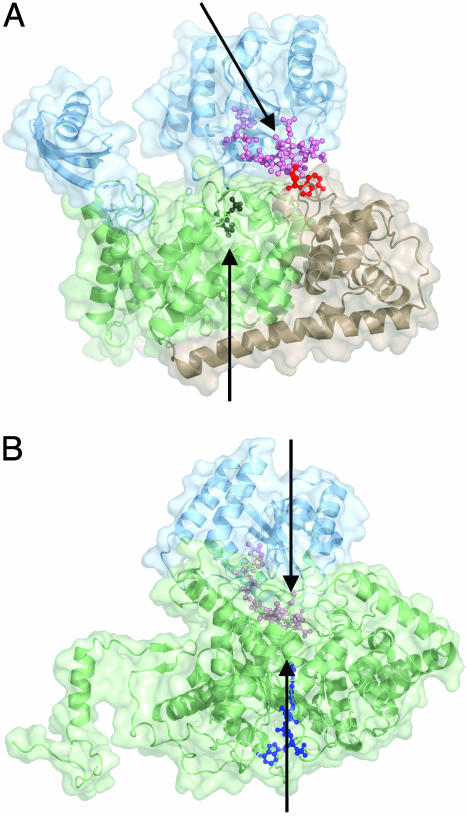
One fascinating feature of this structure is that the relative orientation of the Rossmann domain to the TIM barrel in 5,6-LAM is markedly different from that observed in either GM or MCM. Although in 5,6-LAM the Rossmann domain is bound off-center on top of the TIM barrel, resulting in the ≈25-Å separation of the AdoCbl cofactor from the active-site PLP, in the enzyme-substrate complex of both MCM and GM, the Rossmann domain is docked directly over the center of the TIM barrel, thereby placing the Cbl cofactor in the active site and sequestering the active site from bulk solvent (19, 20) (Fig. 5). MCM buries ≈15% and ≈60% of the TIM-barrel and Rossmann domain surface areas, respectively, at the TIM barrel–Rossmann interface. Likewise, GM buries ≈17.5% of the TIM barrel surface area and ≈47% of the Rossmann domain surface area. In contrast, 5,6-LAM buries only ≈6.6% of the TIM barrel domain and 17% of the Rossmann domain at the TIM barrel–Rossmann interface. Therefore, we describe this structure of 5,6-LAM, in which the active site is solvent-accessible and the AdoCbl cofactor is not positioned for catalysis, as having the resting state “edge-on” conformation (Fig. 5A), in contrast to the hypothetical catalytic “top-on” conformation that is analogous to the domain arrangement in the substrate-bound forms of MCM (Fig. 5B) and GM.
Fig. 5.
Edge-on vs. top-on enzyme conformations. (A) Structure of the substrate-free form of 5,6-LAM with the Rossmann domain in an edge-on conformation above the TIM barrel. Protein domains and cofactors are colored as in Fig. 2 A. Arrows represent the axes of the TIM barrel and Rossmann domains. (B) Structure of substrate-bound MCM (Protein Data Bank ID code 1REQ) with the Rossmann domain sitting directly on top of the TIM barrel (top-on). The substrate fragment, desulfo-coenzyme A (dark blue), threads through the TIM barrel domain, effecting the closure of the TIM barrel to the more compact structure shown. The Ado moiety of AdoCbl was not observed. We propose that the substrate-bound 5,6-LAM adopts a subunit arrangement like that of substrate-bound MCM, with the Rossmann domain and AdoCbl docked directly onto the center of the TIM barrel (see Results).
A key feature that is distinctive to PLP-binding in the resting-state structure of 5,6-LAM is the intersubunit nature of the imine linkage (Fig. 2B). In this mode of PLP-binding, the cofactor acts as an anchor, tethering the separate polypeptide chain of the Rossmann domain to the TIM barrel domain through PLP. We propose that the anchoring role of PLP, which forces the unusual orientation of the Rossmann domain with respect to the TIM barrel, is important for positioning AdoCbl outside of the putative active site and for regulating the formation of the highly oxidizing Ado• in the absence of substrate. In the resting state, a large cleft separates the dimerization domain from the Rossmann domain (Figs. 2 A and 5A). This cleft, which leads to the top of the TIM barrel and the PLP cofactor, is presumably the path that the substrate must follow to arrive at the active site. Introduction of the substrate and transaldimination would then release Lys-144β and effectively break the PLP-mediated anchoring interactions between the Rossmann and TIM barrel domains. With the Rossmann domain freed from its constrained position, a large-scale conformational change could occur. We suggest that such a conformational change must occur and must result in the docking of the Rossmann domain directly atop the center of the TIM barrel domain, filling the cleft between the Rossmann domain and the dimerization domain. This catalytic “top-on” conformation would effectively sequester the active site and position the Ado group of AdoCbl near the substrate–PLP complex, allowing for substrate radical generation and catalysis. Such a conformational change could provide the energy that is required for the large rate acceleration of AdoCbl C–Co bond homolysis that characterizes this enzyme family. 5,6-LAM is not exceptionally fast [kcat = 750 ± 44 min–1 for d-lysine (38)] and would not require a very fast conformational change for catalysis. The hypothetical conformational change that accompanies formation of the catalytic state of 5,6-LAM is not rate-limiting, because a primary deuterium kinetic isotope effect is observed in the reaction of 5,6-LAM with deuterium-labeled substrates.
There is precedent in the Cbl enzyme literature for a large-scale conformational change upon substrate binding (32, 37, 39). For methylcobalamin-dependent methionine synthase, the enzyme exists as an ensemble of conformational states that interconvert upon substrate or product binding (39). X-ray analysis reveals that the two active sites that alternatively methylate and demethylate the Cbl cofactor are ≈50 Å apart (32), requiring large conformational rearrangements of the enzyme during each catalytic cycle. For AdoCbl-bound MCM, the presence of substrate has a dramatic effect on the structure of the TIM barrel (37). In the x-ray structure of the substrate-free form of MCM, the TIM barrel is pried apart, leaving a large gap in the center of the barrel (37). Substrate binds in this gap, threading through the N terminus of the TIM barrel to the Cbl cofactor, which is located at the C terminus of the barrel (20). Binding of substrate seems to trigger the barrel closure (Fig. 5B), which, in turn, causes Tyr-89 to swing toward the top of the AdoCbl corrin ring, presumably facilitating the homolytic cleavage of the Ado moiety to give Ado• (37, 40). Thus, whereas MCM and 5,6-LAM have very similar structures, the way in which conformational changes are linked to substrate binding is different and takes advantage of the unique properties of the substrates. The ≈23-Å-long methylmalonyl-coenzyme A can act as a thread to sew together the TIM barrel, whereas a smaller lysine substrate can release an enzyme-bound Lys from the PLP, freeing the Rossmann domain to rotate.
Several open questions remain concerning the hypothetical conformational change that we propose is necessary for catalysis in 5,6-LAM. What role, if any, does the dimerization domain play in the conformational change? Could the accessory clamp swing up and down to facilitate the edge-to-top rotation of the Rossmann domain? What drives the reformation of the imine linkage with Lys-144β that would result in reversion to the edge-on conformation and product release? What is the structural rationale for the observed suicide inactivation of 5,6-LAM (41)? Although many interesting details remain to be discovered, our structural analysis suggests a distinctive mechanism for substrate-mediated control of radical generation. The structure of 5,6-LAM shows that AdoCbl-dependent enzymes can control radical generation by using a covalent bond that must be broken when substrate binds, effectively locking AdoCbl into a noncatalytic position in the absence of substrate.
Supplementary Material
Acknowledgments
We thank C. Ogata at the Argonne National Laboratory for technical assistance during data collection. This research was supported in part by the National Institutes of Health (NIH) (C.L.D.), National Institute of Diabetes and Digestive Diseases Grant DK28607 (to P.A.F.), the Searle Scholars Program (C.L.D.), the Alfred P. Sloan Foundation (C.L.D.), a Lester Wolfe Predoctoral Fellowship (to F.B.), and the Massachusetts Institute of Technology Undergraduate Research Opportunities Program John Reed Fund (E.A.E.). Synchrotron facilities are funded by NIH National Center of Research Resources Grant APS NE-CAT 8BM.
Author contributions: E.B., P.A.F., and C.L.D. designed research; F.B. and E.A.E. performed research; E.B., K.-H.T., and P.A.F. contributed new reagents/analytic tools; F.B. and C.L.D. analyzed data; and F.B. and C.L.D. wrote the paper.
Abbreviations: AdoCbl, adenosylcobalamin or coenzyme B12; Ado, adenosyl group; Ado•, 5′-deoxyadenosyl radical; AdoH, 5′-deoxyadenosine; AdoMet, S-adenosylmethionine; Cbl, cobalamin; DMB, dimethylbenzimidazole; GM, glutamate mutase; 5,6-LAM, lysine 5,6-aminomutase; 2,3-LAM, lysine 2,3-aminomutase; MCM, methylmalonyl-coenzyme A mutase; PLP, pyridoxal 5′-phosphate; TIM, triosephosphate isomerase.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1XRS).
References
- 1.Toraya, T. (2003) Chem. Rev. 103, 2095–2127. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee, R. (2003) Chem. Rev. 103, 2083–2094. [DOI] [PubMed] [Google Scholar]
- 3.Frey, P. A. & Abeles, R. H. (1966) J. Biol. Chem. 241, 2732–2733. [PubMed] [Google Scholar]
- 4.Hay, B. P. & Finke, R. G. (1988) Polyhedron 7, 1469–1481. [Google Scholar]
- 5.Hay, B. P. & Finke, R. G. (1987) J. Am. Chem. Soc. 109, 8012–8018. [Google Scholar]
- 6.Dekker, E. E. & Barker, H. A. (1968) J. Biol. Chem. 243, 3232–3237. [PubMed] [Google Scholar]
- 7.Morley, C. G. D. & Stadtman, T. C. (1970) Biochemistry 9, 4890–4900. [DOI] [PubMed] [Google Scholar]
- 8.Chirpich, T. P., Zappia, V., Costilow, R. N. & Barker, H. A. (1970) J. Biol. Chem. 245, 1778–1789. [PubMed] [Google Scholar]
- 9.Petrovich, R. M., Ruzicka, F. J., Reed, G. H. & Frey, P. A. (1991) J. Biol. Chem. 266, 7656–7660. [PubMed] [Google Scholar]
- 10.Moss, M. & Frey, P. A. (1987) J. Biol. Chem. 262, 14859–14862. [PubMed] [Google Scholar]
- 11.Morley, C. G. D. & Stadtman, T. C. (1972) Biochemistry 11, 600–605. [DOI] [PubMed] [Google Scholar]
- 12.Wetmore, S. D., Smith, D. M. & Random, L. (2001) J. Am. Chem. Soc. 123, 8678–8689. [DOI] [PubMed] [Google Scholar]
- 13.Danen, W. C. & West, C. T. (1973) J. Am. Chem. Soc. 96, 2447–2453. [Google Scholar]
- 14.Zhao, Y., Such, P. & Rétey, J. (1992) Angew. Chem. Int. Ed. Engl. 31, 215–216. [Google Scholar]
- 15.Marsh, E. N. & Ballou, D. P. (1998) Biochemistry 37, 11864–11872. [DOI] [PubMed] [Google Scholar]
- 16.Zelder, O., Beatrix, B., Leutbecher, U. & Buckel, W. (1994) Eur. J. Biochem. 226, 577–585. [DOI] [PubMed] [Google Scholar]
- 17.Finlay, T. H., Valinsky, J., Mildvan, A. S. & Abeles, R. H. (1973) J. Biol. Chem. 248, 1285–1290. [PubMed] [Google Scholar]
- 18.Tamao, Y. & Blakley, R. L. (1973) Biochemistry 12, 24–34. [DOI] [PubMed] [Google Scholar]
- 19.Reitzer, R., Gruber, K., Jogl, G., Wagner, U. G., Bothe, H., Buckel, W. & Kratky, G. (1999) Structure 7, 891–902. [DOI] [PubMed] [Google Scholar]
- 20.Mancia, F., Keep, N. H., Nakagawa, A., Leadlay, P. F., McSweeney, S., Rasmussen, B., Bosecke, P., Diat, O. & Evans, P. R. (1996) Structure 4, 339–350. [DOI] [PubMed] [Google Scholar]
- 21.Shibata, N., Masuda, J., Tobimatsu, T., Toraya, T., Suto, K., Morimoto, Y. & Yasuoka, N. (1999) Structure 7, 997–1008. [DOI] [PubMed] [Google Scholar]
- 22.Berkovtich, F., Nicolet, Y., Wan, J. T., Jarrett, J. T. & Drennan, C. L. (2004) Science 303, 76–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Budisa, N., Steipe, B., Demange, P., Eckerskorn, C., Kellermann, J. & Huber, R. (1995) Eur. J. Biochem. 230, 788–796. [DOI] [PubMed] [Google Scholar]
- 24.Chang, C. H. & Frey, P. A. (2000) J. Biol. Chem. 275, 106–114. [DOI] [PubMed] [Google Scholar]
- 25.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 26.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McRee, D. E. (1999) J. Struct. Biol. 123, 156–165. [DOI] [PubMed] [Google Scholar]
- 28.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 55, 905–921. [DOI] [PubMed] [Google Scholar]
- 29.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 30.Baker, J. J., van der Drift, C. & Stadtman, T. C. (1973) Biochemistry 12, 1054–1063. [DOI] [PubMed] [Google Scholar]
- 31.Drennan, C. L., Huang, S., Drummond, J. T., Matthews, R. G. & Ludwig, M. L. (1994) Science 266, 1669–1674. [DOI] [PubMed] [Google Scholar]
- 32.Evans, J. C., Huddler, D. P., Hilgers, M. T., Romanchuk, G., Matthews, R. G. & Ludwig, M. L. (2004) Proc. Natl. Acad. Sci. USA 101, 3729–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider, G., Käck, H. & Lindqvist, Y. (2000) Structure 8, R1–R6. [DOI] [PubMed] [Google Scholar]
- 34.Tang, K. H., Harms, A. & Frey, P. A. (2002) Biochemistry 41, 8767–8776. [DOI] [PubMed] [Google Scholar]
- 35.Champloy, F., Gruber, K., Jogl, G. & Kratky, G. (2000) J. Synchrotron Radiat. 7, 267–273. [DOI] [PubMed] [Google Scholar]
- 36.Gruber, K., Reitzer, R. & Kratky, G. (2001) Angew. Chem. Int. Ed. Engl. 40, 3377–3380. [DOI] [PubMed] [Google Scholar]
- 37.Mancia, F. & Evans, P. R. (1998) Structure 6, 711–720. [DOI] [PubMed] [Google Scholar]
- 38.Tang, K. H., Casarez, A. D., Wu, W. & Frey, P. A. (2003) Arch. Biochem. Biophys. 418, 49–54. [DOI] [PubMed] [Google Scholar]
- 39.Bandarian, V., Ludwig, M. L. & Matthews, R. G. (2003) Proc. Natl. Acad. Sci. USA 100, 8156–8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vlasie, M. D. & Banerjee, R. (2003) J. Am. Chem. Soc. 125, 5431–5435. [DOI] [PubMed] [Google Scholar]
- 41.Tang, K. H., Chang, C. H. & Frey, P. A. (2001) Biochemistry 40, 5190–5199. [DOI] [PubMed] [Google Scholar]
- 42.DeLano, W. L. (2002) The PyMOL Molecular Graphics System (DeLano Scientific, San Carlos, CA).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.