Abstract
RUNX transcription factors are key regulators of lineage-specific gene expression and might be involved in autoimmune diseases. Runx3 plays a role during the development of sensory neurons and T cells and regulates transforming growth factor β (TGF-β) signaling in dendritic cells. Here, we report that at 4 weeks of age, Runx3 knockout (KO) mice spontaneously develop inflammatory bowel disease (IBD) characterized by leukocyte infiltration, mucosal hyperplasia, formation of lymphoid clusters, and increased production of IgA. Additionally, at a considerably older age (8 months), the KO mice also develop progressive hyperplasia of the gastric mucosa associated with disturbed epithelial differentiation and cellular hyaline degeneration. Analysis of cytokines in the colonic mucosa of Runx3 KO mice revealed a mixed T helper 1/T helper 2 response. By using immunohistochemistry and RNA in situ hybridization, Runx3 expression in the gastrointestinal tract is detected in lymphoid and myeloid populations but not in the epithelium. The data indicate that loss of leukocytic cell-autonomous function of Runx3 results in IBD and gastric lesion in the KO mice. IBD in humans is viewed as a complex genetic disorder. Several susceptibility loci were identified on different human chromosomes including the chromosomal region 1p36 where RUNX3 resides. It is thus tempting to speculate that mutations in RUNX3 may constitute an IBD risk factor in humans.
RUNX3 belongs to the runt domain family of transcription factors, which are key regulators of lineage-specific gene expression and more recently were linked to human autoimmunity (1). The RUNX genes arose early in evolution, and the three mammalian members maintained extensive structural similarities (2–5). RUNX3 is the smallest of the three mammalian genes and could be considered the evolutionary founder of the mammalian RUNX family (3, 4, 6). In the developing mouse embryo, Runx3 displays a distinct tissue-specific expression pattern. It is expressed in hematopoietic organs, developing bones, peripheral nervous system, and skin appendages (7). Comparative analysis showed that when expressed during development, Runx3 is confined to mesenchymal components in contrast to Runx1, which is also expressed in epithelial components (7).
Studies in knockout (KO) mice have delineated several cell-autonomous functions of Runx3. In neurogenesis, Runx3 is required for the development and survival of the dorsal root ganglia (DRG) TrkC neurons (8, 9). Loss of Runx3 expression in TrkC neurons results in severe limb ataxia (8, 9). In thymopoiesis, Runx3 is expressed in immature double-negative and in single-positive CD8+ or CD4+ thymocytes (10–12). In the absence of Runx3, the silencing of CD4 during T cell lineage decisions is impaired, as is the function of mature peripheral CD8 T cells (10–12). Runx3 also is highly expressed in dendritic cells (DC), where it functions as a component of the transforming growth factor β (TGF-β) signaling cascade (13). Runx3 KO DC do not respond to TGF-β; their maturation is accelerated and accompanied by an increased efficacy to stimulate T cells. The abnormalities in Runx3 KO DC function constitute a primary immune system defect associated with spontaneous development of lung inflammation in the KO mice (13). At birth, Runx3 KO mice were also reported to exhibit hyperplasia of the gastric epithelium attributed to a lower sensitivity of the Runx3-deficient epithelial cells to TGF-β signals (14, 15).
The human idiopathic inflammatory bowel diseases (IBD) are relatively common chronic relapsing inflammations of the gastrointestinal tract (GIT). The absence of simple Mendelian inheritance suggests that inappropriate function of multiple gene products contributes to the risk of developing IBD (16–18). Several susceptibility loci for IBD were identified on different human chromosomes, including the chromosomal region 1p36 where RUNX3 resides (16, 19, 20). Spontaneous colitis development in various strains of KO and transgenic mice not only further indicates that multiple gene loci are involved but also highlights the role that dysregulated immune response plays in the etiology of the disease (18, 21–23).
Here, we report that, starting at 4 weeks of age, Runx3 KO mice spontaneously develop IBD characterized by leukocyte infiltration, epithelial hyperplasia, formation of mucosal B cell clusters, and increased production of IgA. Interestingly, in addition to the early-onset colitis, the KO mice develop at a much older age (>8 months) a progressive hyperplasia of the glandular mucosa of the stomach. This lesion probably develops secondary to the colitis. Because Runx3 is normally not expressed in the epithelium of the GIT but is highly expressed in the resident leukocytic populations, we conclude that loss of leukocytic cell-autonomous function of Runx3 causes IBD.
Materials and Methods
Pathology, Histopathology, Immunohistochemistry, and RNA in Situ Hybridization. Analyses of KO (n = 70) and heterozygous plus WT mice (n = 60) were carried out as described (8). Animal experiments followed the Institutional Animal Care and Use Committee guidelines of the Weizmann Institute. GIT was examined in situ and after removal fixed in toto in 10% neutral-buffered formalin or in Bouin's solution. Representative samples of the GIT were embedded in paraffin, trimmed at 4 μm, and stained with hematoxylin/eosin (H&E). Where appropriate, sections were stained with Masson's trichrome, periodic acid/Schiff, and Alcian blue. Immunohistochemistry (IHC) was performed on paraffin sections by using the various antibodies listed in Supporting Text, which is published as supporting information on the PNAS web site. Radioactive in situ hybridization on paraffin sections by using riboprobes labeled with 5′-[α-[35S]thio]UTP was performed as described (24) by using the murine Runx1 region spanning the 650-bp region between 1114 and 1765 in GenBank accession no. D13802 and a Runx3 probe spanning the 769-bp region between 1035 and 1804 in GenBank accession no. AF155880.
Cytokine Expression. RNA was isolated from the colons of WT or KO mice by using an EZ-RNA isolation kit (Biological Industries, Kibbutz Beit-Haemmek, Israel). cDNA was generated by using SuperScript II RNaseH– reverse transcriptase (Invitrogen), and PCR products were derived by using the sets of oligonucleotides and PCR conditions detailed in Supporting Text.
Results
Runx3 KO Mice Develop Chronic Colitis and Hyperplasia of the Gastric Glandular Mucosa. Runx3 KO mice were maintained under specific pathogen-free (SPF) conditions. When crossed onto a C57BL/6 or BALB/c background, homozygotes did not survive postnatally. Homozygous Runx3 KO mice on outbred ICR or MF1 background had congenital sensory ataxia, reduced growth, and a high rate of mortality during the first 2 weeks of life (8). Surviving KO mice lived to old age (24 months). Between 1 and 8 weeks of age, KO mice developed spontaneous eosinophilic airway inflammation (13). From postnatal day 3 onward, KO mice were invariably smaller than controls, often weighing ≈50% of WT littermates.
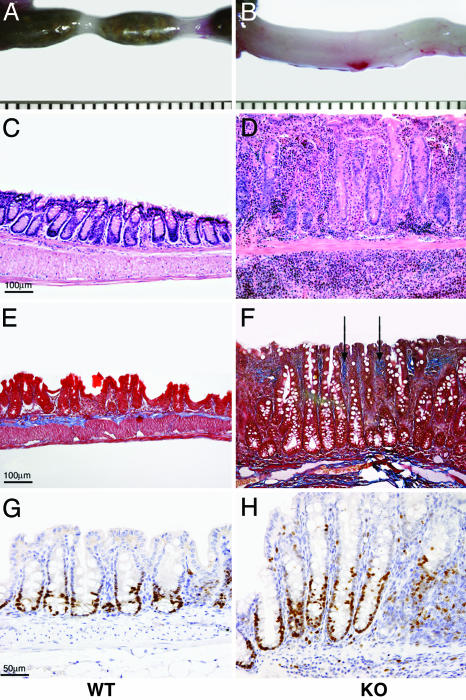
Postmortem examination showed that KO mice had minimal fat stores and that their GIT contained little ingesta. The cecal wall was frequently thickened, rigid, and opaque, and the colon exhibited variable tubular thickening (Fig. 1 A and B). Neither perineal soiling by diarrhea or blood nor rectal prolapse was seen in any of the mice. Mesenteric lymph nodes in the KO mice often were enlarged (2- to 3-fold) and contained many more cells, compared with WT (data not shown). Microscopically, starting at ≈4 weeks of age, all KO mice examined (48/48) and none of the WT or heterozygotes had colitis, characterized by multifocal and coalescing mixed mucosal and submucosal infiltration of plasma cells, lymphocytes, histiocytes, and eosinophils (Fig. 1 C and D). Frequently, formation of lymphocytic clusters was observed. Inflammatory cellular infiltration was associated with mucosal hyperplasia (2- to 5-fold), crypt loss (Fig. 1 C and D), and increased mitotic figures (Ki-67) (Fig. 1 G and H). In some cases, fibrosis of the lamina propria also was evident (Fig. 1 E and F). Affected segments were present in the cecum and ascending and descending colon from the earliest stage. Maintenance of the KO mice in a separate facility under conventional conditions did not exacerbate the colitis. Inflammation of the small intestine was seen in ≈20% of the KO mice. Intestinal neoplasia was not found in any of the KO mice.
Fig. 1.
Pathologic findings in the large intestines of Runx3 KO mice in comparison with WT mice. (A and B) Gross specimens of the descending colon and rectum show marked thickening of the intestinal wall and absence of fecal pellets in the KO. (C and D) H&E-stained sections of the colon show severe mucosal and submucosal thickening in KO due to mixed inflammatory cellular infiltration with crypt hyperplasia and loss. (E and F) Masson's trichrome stain demonstrates abnormal deposition of collagen (arrows in F), stained blue, in the KO lamina propria. (G and H) Immunostaining with anti-Ki-67, which directly monitors cell divisions, shows an increase in proliferating cells (stained brown) in crypts of the KO cecum. (Magnification: C–F, ×10; G and H, ×20.)
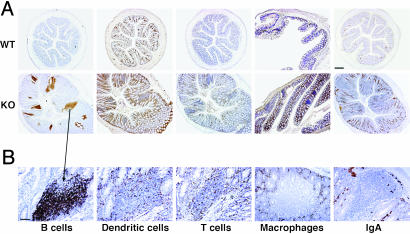
The leukocytic infiltrate in the KO large intestine was characterized by immunostaining with anti-CD45R and Pax5 (B lymphocytes), anti-GFP (DC, by using Runx3–/–/CX3CR1+/GFP mice (13), anti-CD3 (T lymphocytes), and anti-F4/80 (macrophages) (Fig. 2). An increase in T lymphocytes, macrophages, and DC was seen throughout the inflamed KO mucosa (Fig. 2 A). In the KO, a marked increase was observed in lymphocytic clusters that contained predominantly B cells (Fig. 2). They also contained scattered T cells and DC but were free of F4/80-positive macrophages (Fig. 2B). The clusters' B cells did not express CD5 (data not shown) and thus were identified as the B-2 type. The majority of these clusters, which are scattered throughout the large intestine, are unorganized tertiary lymphoid follicles formed de novo (25, 26). The number of organized clusters corresponding to isolated lymphoid follicles (26, 27) was also increased in the KO. Outside the clusters, a substantial increase in plasma cells and in production of IgA was observed in the KO (Fig. 2). A substantial increase (2.5-fold) in IgA level was also detected in the serum and fecal pellets of the KO mice (data not shown).
Fig. 2.
Characterization of leukocyte infiltrates in the inflamed Runx3 KO colon. IHC on WT and KO colon sections (×4) (A) and on a KO B cell cluster (×10) (B) with anti-CD45R for detection of B cells, anti-IgA for detection of IgA, anti-GFP for detection of DC (13), anti-CD3 for detection of T cells, and anti-F4/80 for detection of macrophages. (Scale bars: A, 250 μm; B, 50 μm.)
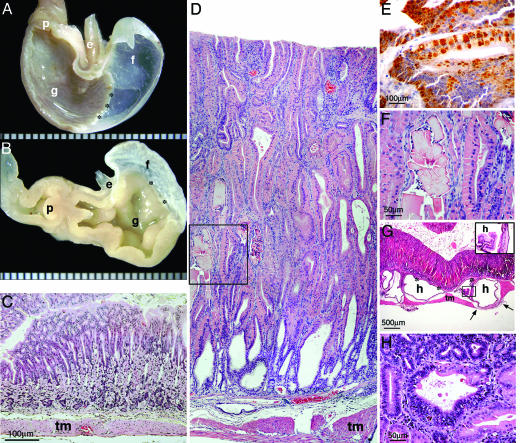
In addition to colitis, most (12/15) Runx3 KO mice older than 8 months developed progressive hyperplasia of the gastric glandular mucosa, which was not seen in any of the WT mice (Fig. 3). The extent and region affected correlated with the age of the mouse. The lesion began in the pyloric region and by 1 year of age involved the wall of the entire glandular stomach (Fig. 3 A and B). Microscopically, hyperplastic mucosa exhibited disturbed epithelial differentiation, pronounced elongation of the gastric pits and glands (up to 2 mm high) (Fig. 3 C and D), and, in advanced cases, was completely devoid of parietal and chief cells. Mitotic figures were numerous, and a marked increase in Ki-67-expressing cells was noted (data not shown). There was multifocal mucosal and submucosal inflammatory cellular infiltration comprising lymphocytes, plasma cells, and eosinophils, but contrary to the colitis, it was usually mild.
Fig. 3.
Pathologic findings in the stomach of the Runx3 KO. (A and B) Gross specimens of hemisected stomachs show contraction and marked thickening of the entire glandular portion in the KO (B), compared with WT (A). e, Esophagus; f, forestomach; g, glandular stomach; and p, pylorus. (C and D) H&E-stained sections of the gastric wall in the fundic region. There is prominent mucosal hyperplasia and hyalinosis in the KO. tm, Tunica muscularis. (E) IHC with anti-YM1/2 protein demonstrates positive globular material within the cytoplasm of tall columnar epithelial cells. (F) High magnification of the area indicated in D by a square shows accumulation of eosinophilic rectangular crystalline material within the lumen of hyperplastic glands. (G) Cystic dilation and collapse of abnormal mucosa into subjacent layers, or “mucosal herniation.” Asterisks denote the muscularis mucosa below, in which five dilated glands are seen, two of which are indicated by h. The four glands on the left are above the tunica muscularis (tm). One gland reaches but does not invade the serosa (in the area between the two arrows). Inset shows the epithelial lining of the cysts (area of Inset indicated by a square in the main photo). (H) A focus of mucosal dysplasia. In the gland at the center of the field, there is cellular atypia in the form of loss of orderly stratification, nuclear crowding, and increased nuclear/cytoplasmic ratio. Relatively well differentiated glands are seen at the bottom right and left-hand side of the field. (Magnification: C and D, ×10; E, ×40; F, ×20; G, ×2; Inset, ×20; and H, ×20.)
Within the hyperplastic mucosa, there was prominent hyaline degeneration, whereby the cytoplasm of epithelial cells expanded and became smooth and hypereosinophilic (28) (Fig. 3F). The hypereosinophilic material, which filled the entire cytoplasm or formed discrete globular accumulations, was identified as Ym2 protein by IHC (Fig. 3E). Ym2 is a member of the chitinase family and is believed to act as stomach-specific eosinophil chemotactic cytokine (29). In advanced cases, the hyperplastic mucosa protruded into subjacent layers up to, but not through, the serosa (Fig. 3G).
Dysplastic foci were occasionally recognized (Fig. 3H). Nevertheless, apart from isolated cases with mucosal polyps, gastric neoplasms were not observed in any of the KO mice including those who lived to old age (>2 years). Taken together, the gross and histological features of Runx3 KO stomach lesion are consistent with adenomatous hyperplasia of the glandular mucosa with mucous metaplasia, a well recognized lesion of murine stomach, also referred to as proliferative gastritis (23, 30–32).
Expression of both T Helper (Th)1 and Th2 Cytokines Is Elevated in the Colon of Runx3 KO Mice. Th lymphocytes differentiate into two distinct subgroups, Th1 and Th2. The functional differences between the two subgroups are mediated by the activities of the cytokines that they secrete. IBD is characterized by perturbations of the normal balance between inflammatory and regulatory cytokines (18, 33).
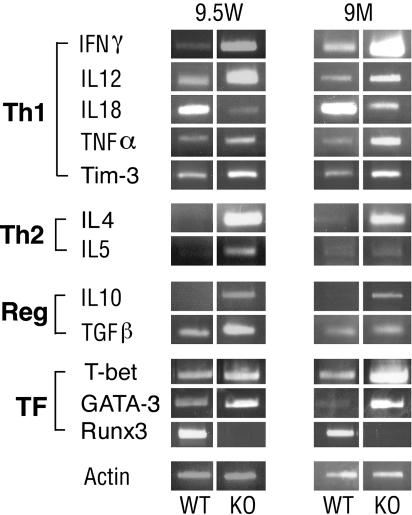
Employing RT-PCR on colon mRNA, we compared the changes in cytokine expression in young and old KO mice with those of WT littermates. Expression of IFN-γ, the signature cytokine of Th1 cells, was markedly increased in the colon of young and old KO mice (Fig. 4 and Fig. 6, which is published as supporting information on the PNAS web site). The level of two other proinflammatory cytokines of the Th1 domain, TNF-α and IL-12, was also increased in the KO, as was the expression of Tim-3, a surface marker of differentiated Th1 cells (34). Surprisingly, however, the level of IL-18, another Th1-response cytokine, decreased (Figs. 4 and 6).
Fig. 4.
Expression of Th1 and Th2 cytokines in inflamed Runx3 KO colon. RT-PCR analysis of cytokines and transcription factors (TF), in colon RNA of young (9.5 weeks) and adult (9 months) WT and KO mice. Reg, regulatory cytokines. A detailed account of primers used for PCR is available in Supporting Text. Depicted are representative examples of several independent repetitions.
As for the Th2 cytokines, an increase of the Th2 signature cytokine IL-4 was noted in the colon of young and old KO mice (Figs. 4 and 6), but there was only a small change in the levels of IL-5. Expression of the regulatory cytokines TGF-β and IL-10 was slightly increased in KO colon, compared with WT (Figs. 4 and 6). Together, the data indicate that the Runx3 KO colitis was associated with an increase in Th1 and to a lesser degree in Th2 responses in the colon of young and old KO mice, without substantial changes in regulatory cytokines. This conclusion is supported by the increase in the KO of two transcription factors, T-bet and GATA-3 (Figs. 4 and 6), known to be required for induction of Th1 and Th2 differentiation, respectively (35–37). It is tempting to speculate that Runx3 negatively regulates transcription of these two key regulators of the Th1/Th2 response.
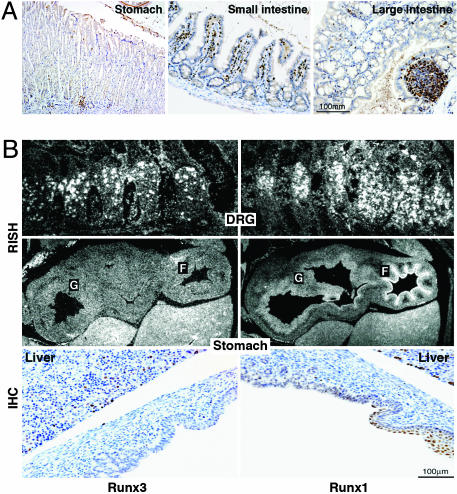
Runx3 Is Highly Expressed in the GIT Lymphoid and Myeloid Cells. In search for the cause of the colitis and gastric hyperplasia in the Runx3 KO mice, we set out to identify the cells within the GIT that normally express Runx3. Expression of Runx3 is readily detected by RT-PCR on colon RNA (Fig. 4). IHC analysis of Runx3 during embryonic and postnatal development of GIT in WT mice is summarized in Fig. 5. Strong nuclear staining of Runx3 is detected in the GIT resident lymphoid and myeloid cells, either single or clustered in lymphoid follicles (Fig. 5A). In contrast, Runx3 is neither expressed in the epithelium of the large and small intestine nor detected in the gastric epithelium (Fig. 5 A and B). Cytoplasmic staining of Runx3 was not observed.
Fig. 5.
Expression of Runx3 and Runx1 in GIT and DRG of WT mice. (A) Expression of Runx3 in adult GIT (stomach, small intestine, and large intestine). (B) Expression of Runx3 and Runx1 in embryonic day 16.5 embryos. Shown are RISH analysis of DRG (Top) and stomach (Middle) and IHC analysis of stomach (Bottom). F, forestomach; G, glandular stomach.
We next assessed Runx3 expression in the GIT by using RNA in situ hybridization (RISH), compared with its expression in DRG. RISH signals are readily detected in DRG but not in gastric epithelium (Fig. 5B). IHC of the stomach and the adjacent liver shows Runx3 staining only in lymphocytes (Fig. 5B). Interestingly, expression of Runx1, another member of the Runx family, is detected in the DRG as well as in gastric epithelium by both RISH and IHC. Runx1 signals are high in the forestomach and lower in the glandular region (Fig. 5B). These Runx1 and Runx3 expression data are in good agreement with the previously reported IHC and β-galactosidase staining (7, 8) and with the nonradioactive RISH data presented in the digital atlas of gene expression patterns (see www.genepaint.org).
Discussion
IBD is a chronic relapsing inflammation of the GIT. The fundamental cause of human IBD is unknown, but the current view is that it involves genetic predisposition, which causes a dysregulated GIT immune response to environmental agents (18). Here, we show that at an early age, 100% of Runx3 KO mice spontaneously develop IBD characterized by leukocytic infiltration, epithelial hyperplasia, and increased production of IgA. In several genetically modified mouse models of IBD, the host genetic background affects the susceptibility to GIT inflammation (18). Runx3 deficiency, on the other hand, results in 100% incidence of early-onset colitis when KO mice are bred onto two outbred strains (ICR and MFI). However, unlike the situation in several other mutant IBD mice (38–40), an increased incidence of intestinal neoplasia was not observed in Runx3 KO mice.
Although severe histological lesions were common in the IBD of Runx3 KO mice, ulceration did not occur, and apart from a probable contribution to poor growth, the clinical course of the disease was indolent. Runx3 KO mice develop IBD even when kept in clean facilities free of known murine viral and bacterial pathogens. On the other hand, the disease was not aggravated when KO mice were kept in a conventional facility. Interestingly, in addition to the early-onset colitis, the KO mice develop at an older age (>8 months) progressive hyperplasia of the glandular mucosa of the stomach.
Microscopically, the Runx3 KO IBD is characterized by mononuclear and eosinophilic infiltration, cecal and colonic mucosal hyperplasia, formation of B cell clusters, and increased production of IgA. De novo formed B cell clusters were previously seen in cases of IBD and are common in autoimmune diseases (25, 41, 42). B cells clusters are also found in IBD of several transgenic and KO mouse models (39, 43–46). Blocking TGF-β signaling in T cells by transgenic expression of dominant-negative form of TGF-β receptor II (CD4-dnTGF-β-RII transgenics) results in an autoimmune condition characterized by colitis, lung inflammation, Th1 and Th2 responses, and presence of autoimmune antibodies (47). The similarities between the phenotypic features of these CD4-dnTGF-β-RII transgenics and those of Runx3 KO mice are particularly intriguing given that Runx3 functions as a component in the TGF-β pathway (13, 14). Specifically, Runx3 mediates TGF-β signaling in DC, and when absent, KO DC display accelerated maturation, overresponsiveness to innocuous antigens, and increased potency to stimulate T cells (13). Moreover, the KO mice develop spontaneous lung inflammation, as do the CD4-dnTGF-β-RII mice. Because abrogation of TGF-β signaling leads to spontaneous activation of autoimmune response and an exaggerated mucosal immune response plays a role in IBD development, it is possible that Runx3 KO DC are involved in the etiology of the colitis.
Mice that are deficient for the T cell receptor α (TCR-α) develop spontaneous colitis with features resembling those of Runx3 KO mice, including increased amounts of B cells and IgA and enhanced expression of both Th1 and Th2 signature cytokines IFN-γ and IL-4, respectively (48). Of potential relevance to these findings are the observations that Runx proteins are involved in regulation of TCR-α and TCR-β transcription (49, 50) and that Runx3 KO mice display abnormalities in T cell development and function (10–12). Specifically, Runx3 is highly expressed in CD8+ T cells comprising the majority of intraepithelial lymphocytes, known to play a protective role against intestinal inflammation (51).
In Runx3 KO, a marked increase occurs in the production of colon and serum IgA. This in vivo increase of IgA in mice lacking Runx3 was previously observed in the alveolar lavage of Runx3 KO mice (13). On the other hand, it was previously found that in vitro Runx3 participates in TGF-β mediated Ig class switching to IgA (13, 52). It thus appears that, in vivo, other cytokines/pathways besides TGF-β play a role in regulating the switch to IgA. Consistent with this occurrence, mice lacking the TGF-β signal transducer Smad3 have a normal number of IgA-producing plasma cells (53).
Runx3 KO mice older than ≈8 months develop progressive hyperplasia of the gastric glandular mucosa. The gastric and intestinal lesions differ in the following aspects: (i) the colitis is early-onset, whereas the gastric hyperplasia is late-onset; (ii) the severity of the colitis is individual (severe cases were seen at early ages, and moderate cases were seen in older mice), whereas the gastric mucosal hyperplasia is progressive and its severity is age-related; (iii) the incidence of the colitis is 100% vs. 80% of the gastric lesion; and (iv) in the large intestine, the degree of inflammatory cellular infiltration is commensurate with the extent of mucosal hyperplasia, whereas in the stomach, inflammatory infiltration is usually modest and florid hyperplasia often coexists with mild gastritis.
Given the absence of Runx3 expression in WT gastric mucosa, we postulate that the late-onset gastric mucosal hyperplasia seen in Runx3 KO mice is secondary to the colitis or represents an independent autoimmune process (54). Compatible with this thesis, gastric mucosal hyperplasia in association with colitis was previously observed in other mutant mice (23, 55).
The gastric lesion in adult (8-month-old) Runx3 KO MF1 or ICR mice described here completely differs from the stomach lesion observed in newborn Runx3 KO C57BL/6 mice by Li et al. (15). According to the report by Li et al. (15), gastric mucosa of newborn Runx3 KO mice displays hyperplasia, attributed to lack of Runx3 expression in the KO epithelium. Newborn Runx3 KO MF1 or ICR mice do not display gastric hyperplasia (56). Moreover, no Runx3 expression was detected in epithelia of various mouse strains (ICR, MF1, C57BL/6) by either IHC or by β-galactosidase (LacZ) staining in Runx3+/LacZ embryos (7, 8, 56). In an attempt to further investigate the cause for this discrepancy, we performed, in addition to the IHC, a RISH analysis on stomach sections by using side-by-side Runx3 and Runx1 probes. The results showed that, in contrast with Runx1, Runx3 was not readily detected in gastric mucosa or in the epithelium of the large and small intestine. On the other hand, Runx3 is highly expressed in the GIT lymphoid and myeloid cells. We therefore conclude that Runx3 KO colitis and gastric lesion result from the loss of a leukocyte cell-autonomous function of Runx3. The high expression of Runx3 in GIT leukocytes should be considered when expression levels in tissue samples are assessed by using RT-PCR or Western blotting, particularly when expression in gastric tumors is compared with the adjacent control stomach tissue, which is frequently inflamed.
Development of IBD was found to be attenuated in some mutant mice when kept in a SPF environment. Runx3 KO IBD develops spontaneously in mice kept under SPF conditions. Antigens produced by the normal GIT microflora have been implicated in induction of the inflammatory process (57). Because these antigens are present in the body from birth and could thus be considered as self-antigens, it was argued that spontaneously developed IBD could be viewed as an autoimmune disease (18). Intriguingly, an autoimmune disease-associated regulatory single nucleotide polymorphism (SNP) affecting the RUNX binding site was recently found in three different autoimmune diseases (58–60). Thus, the potential involvement of RUNX family members in autoimmunity is an intriguing possibility (1). Further analysis of the etiological similarities of Runx3 KO mice IBD and stomach lesion to autoimmune disorders, including the presence of autoimmune antibodies, may prove revealing.
In humans, similarities between the genesis of IBD and asthma have been noted (61). The current understanding is that both diseases are multifactorial and involve defects in multiple genes, which could interact with one another in a complex manner (62, 63). Several susceptibility loci for IBD and asthma were identified on different human chromosomes, including the chromosomal region 1p36 where RUNX3 resides (19, 20, 64, 65). However, a specific gene at 1p36 that predisposes humans to asthma and/or IBD has not been identified. It is tempting to speculate that RUNX3 deficiency, which in the mouse causes phenotypes with hallmarks of human IBD (present report) and asthma (13), constitutes a susceptibility factor for both asthma and IBD in humans.
Supplementary Material
Acknowledgments
We thank Judith Chermesh and Rafi Saka for help in animal husbandry; Dorit Nathan, Tamara Berkuzki, and Calanit Raanan for technical assistance; Dr. Alon Harmelin, Dr. Yehuda Chovers, Dr. Joseph Lotem, and Francisco Quintana for helpful discussions; and Dr. J. M. Ward (National Institutes of Health) for IHC of Ym2. This work was supported by grants from the Commission of the European Union, the Israel Science Foundation, Minerva Foundation Germany, and the Shapell Family Biomedical Research Foundation at the Weizmann Institute.
Author contributions: O.B., D.L., V.N., O.G., O.F., and E.W. performed research; O.B., D.L., V.N., and Y.G. analyzed data; and Y.G. wrote the paper.
Abbreviations: DC, dendritic cells; DRG, dorsal root ganglia; GIT, gastrointestinal tract; IBD, inflammatory bowel disease; IHC, immunohistochemistry; KO, knockout; RISH, RNA in situ hybridization; SPF, specific pathogen-free; TGF, transforming growth factor; Th, T helper.
References
- 1.Alarcon-Riquelme, M. E. (2004) Arthritis Res. Ther. 6, 169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coffman, J. A. (2003) Cell Biol. Int. 27, 315–324. [DOI] [PubMed] [Google Scholar]
- 3.Levanon, D., Glusman, G., Bettoun, D., Ben-Asher, E., Negreanu, V., Bernstein, Y., Harris-Cerrruti, C., Brenner, O., Eilam, R., Lotem, J., et al. (2003) Blood Cells Mol. Dis. 30, 161–163. [DOI] [PubMed] [Google Scholar]
- 4.Levanon, D. & Groner, Y. (2004) Oncogene 23, 4211–4219. [DOI] [PubMed] [Google Scholar]
- 5.Rennert, J., Coffman, J. A., Mushegian, A. R. & Robertson, A. J. (2003) BMC Evol. Biol. 3, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bangsow, C., Rubins, N., Glusman, G., Bernstein, Y., Negreanu, V., Goldenberg, D., Lotem, J., Ben-Asher, E., Lancet, D., Levanon, D. & Groner, Y. (2001) Gene 279, 221–232. [DOI] [PubMed] [Google Scholar]
- 7.Levanon, D., Brenner, O., Negreanu, V., Bettoun, D., Woolf, E., Eilam, R., Lotem, J., Gat, U., Otto, F., Speck, N. & Groner, Y. (2001) Mech. Dev. 109, 413–417. [DOI] [PubMed] [Google Scholar]
- 8.Levanon, D., Bettoun, D., Harris-Cerruti, C., Woolf, E., Negreanu, V., Eilam, R., Bernstein, Y., Goldenberg, D., Xiao, C., Fliegauf, M., et al. (2002) EMBO J. 21, 3454–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue, K., Ozaki, S., Shiga, T., Ito, K., Masuda, T., Okado, N., Iseda, T., Kawaguchi, S., Ogawa, M., Bae, S. C., et al. (2002) Nat. Neurosci. 5, 946–954. [DOI] [PubMed] [Google Scholar]
- 10.Taniuchi, I., Osato, M., Egawa, T., Sunshine, M. J., Bae, S. C., Komori, T., Ito, Y. & Littman, D. R. (2002) Cell 111, 621–633. [DOI] [PubMed] [Google Scholar]
- 11.Woolf, E., Xiao, C., Fainaru, O., Lotem, J., Rosen, D., Negreanu, V., Bernstein, Y., Goldenberg, D., Brenner, O., Berke, G., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 7731–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehlers, M., Laule-Kilian, K., Petter, M., Aldrian, C. J., Grueter, B., Wurch, A., Yoshida, N., Watanabe, T., Satake, M. & Steimle, V. (2003) J. Immunol. 171, 3594–3604. [DOI] [PubMed] [Google Scholar]
- 13.Fainaru, O., Woolf, E., Lotem, J., Yarmus, M., Brenner, O., Goldenberg, D., Negreanu, V., Bernstein, Y., Levanon, D., Jung, S. & Groner, Y. (2004) EMBO J. 23, 969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito, Y. & Miyazono, K. (2003) Curr. Opin. Genet. Dev. 13, 43–47. [DOI] [PubMed] [Google Scholar]
- 15.Li, Q. L., Ito, K., Sakakura, C., Fukamachi, H., Inoue, K., Chi, X. Z., Lee, K. Y., Nomura, S., Lee, C. W., Han, S. B., et al. (2002) Cell 109, 113–124. [DOI] [PubMed] [Google Scholar]
- 16.Parkes, M. & Jewell, D. (2001) Expert Rev. Mol. Med. 2001, 1–18. [DOI] [PubMed] [Google Scholar]
- 17.Podolsky, D. K. (2002) N. Engl. J. Med. 347, 417–429. [DOI] [PubMed] [Google Scholar]
- 18.Bouma, G. & Strober, W. (2003) Nat. Rev. Immunol. 3, 521–533. [DOI] [PubMed] [Google Scholar]
- 19.Cho, J. H., Nicolae, D. L., Gold, L. H., Fields, C. T., LaBuda, M. C., Rohal, P. M., Pickles, M. R., Qin, L., Fu, Y., Mann, J. S., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 7502–7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho, J. H., Nicolae, D. L., Ramos, R., Fields, C. T., Rabenau, K., Corradino, S., Brant, S. R., Espinosa, R., LeBeau, M., Hanauer, S. B., et al. (2000) Hum. Mol. Genet. 9, 1425–1432. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann, J. C., Pawlowski, N. N., Kuhl, A. A., Hohne, W. & Zeitz, M. (2002) Pathobiology 70, 121–130. [DOI] [PubMed] [Google Scholar]
- 22.Hibi, T., Ogata, H. & Sakuraba, A. (2002) J. Gastroenterol. 37, 409–417. [DOI] [PubMed] [Google Scholar]
- 23.Fox, J. G., Dangler, C. A. & Schauer, D. B. (2000) in Pathology of Genetically Engineered Mice, eds. Ward, J. M., Mahler, J. F. & Maronpot, R. R. (Iowa State Univ. Press, Ames), pp. 299–315.
- 24.Yamashiro, T., Åberg, T., Levanon, D., Groner, Y. & Thesleff, I. (2002) Gene Expr. Patterns 2, 109–112. [DOI] [PubMed] [Google Scholar]
- 25.Hjelmstrom, P. (2001) J. Leukocyte Biol. 69, 331–339. [PubMed] [Google Scholar]
- 26.Lorenz, R. G., Chaplin, D. D., McDonald, K. G., McDonough, J. S. & Newberry, R. D. (2003) J. Immunol. 170, 5475–5482. [DOI] [PubMed] [Google Scholar]
- 27.Hamada, H., Hiroi, T., Nishiyama, Y., Takahashi, H., Masunaga, Y., Hachimura, S., Kaminogawa, S., Takahashi-Iwanaga, H., Iwanaga, T., Kiyono, H., et al. (2002) J. Immunol. 168, 57–64. [DOI] [PubMed] [Google Scholar]
- 28.Leninger, J. R., Jokinen, M. P., Dangler, C. A. & Whiteley, L. O. (1999) in Pathology of the Mouse, eds. Maronpot, R. R., Boorman, G. A. & Gaul, B. W. (Cache River Press, Vienna, IL), pp. 36–37.
- 29.Ward, J. M., Yoon, M., Anver, M. R., Haines, D. C., Kudo, G., Gonzalez, F. J. & Kimura, S. (2001) Am. J. Pathol. 158, 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Betton, G. R., Whiteley, L. O., Anver, M. R., Brown, R., Deschl, U., Elwell, M., Germann, P. G., Hartig, F., Kuettler, K., Mori, T., et al. (2001) in International Classification of Rodent Tumors: The Mouse, ed. Mohr, U. (Springer, Berlin), p. 38.
- 31.Greaves, P. & Boiziau, J. L. (1984) Vet. Pathol. 21, 224–228. [DOI] [PubMed] [Google Scholar]
- 32.Maehler, M., Rozell, B., Mahler, J. F., Merlino, G., Devor-Henneman, D. & Ward, J. M. (2000) in Pathology of Genetically Engineered Mice, eds. Ward, J. M., Mahler, J. F., Maronport, R. R. & Sundberg, J. P. (Iowa State Univ. Press, Ames), pp. 269–297.
- 33.Neurath, M. F., Finotto, S. & Glimcher, L. H. (2002) Nat. Med. 8, 567–573. [DOI] [PubMed] [Google Scholar]
- 34.Monney, L., Sabatos, C. A., Gaglia, J. L., Ryu, A., Waldner, H., Chernova, T., Manning, S., Greenfield, E. A., Coyle, A. J., Sobel, R. A., et al. (2002) Nature 415, 536–541. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, D. H., Cohn, L., Ray, P., Bottomly, K. & Ray, A. (1997) J. Biol. Chem. 272, 21597–21603. [DOI] [PubMed] [Google Scholar]
- 36.Zheng, W. & Flavell, R. A. (1997) Cell 89, 587–596. [DOI] [PubMed] [Google Scholar]
- 37.Szabo, S. J., Kim, S. T., Costa, G. L., Zhang, X., Fathman, C. G. & Glimcher, L. H. (2000) Cell 100, 655–669. [DOI] [PubMed] [Google Scholar]
- 38.Berg, D. J., Davidson, N., Kuhn, R., Muller, W., Menon, S., Holland, G., Thompson-Snipes, L., Leach, M. W. & Rennick, D. (1996) J. Clin. Invest. 98, 1010–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hermiston, M. L. & Gordon, J. I. (1995) Science 270, 1203–1207. [DOI] [PubMed] [Google Scholar]
- 40.Rudolph, U., Finegold, M. J., Rich, S. S., Harriman, G. R., Srinivasan, Y., Brabet, P., Boulay, G., Bradley, A. & Birnbaumer, L. (1995) Nat. Genet. 10, 143–150. [DOI] [PubMed] [Google Scholar]
- 41.Kaiserling, E. (2001) Lymphology 34, 22–29. [PubMed] [Google Scholar]
- 42.Carlsen, H. S., Baekkevold, E. S., Johansen, F. E., Haraldsen, G. & Brandtzaeg, P. (2002) Gut 51, 364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sadlack, B., Merz, H., Schorle, H., Schimpl, A., Feller, A. C. & Horak, I. (1993) Cell 75, 253–261. [DOI] [PubMed] [Google Scholar]
- 44.Alonzi, T., Newton, I. P., Bryce, P. J., Di Carlo, E., Lattanzio, G., Tripodi, M., Musiani, P. & Poli, V. (2004) Cytokine 26, 45–56. [DOI] [PubMed] [Google Scholar]
- 45.Kawamura, T., Kanai, T., Dohi, T., Uraushihara, K., Totsuka, T., Iiyama, R., Taneda, C., Yamazaki, M., Nakamura, T., Higuchi, T., et al. (2004) J. Immunol. 172, 6388–6397. [DOI] [PubMed] [Google Scholar]
- 46.Wang, J., Anders, R. A., Wu, Q., Peng, D., Cho, J. H., Sun, Y., Karaliukas, R., Kang, H. S., Turner, J. R. & Fu, Y. X. (2004) J. Clin. Invest. 113, 826–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorelik, L. & Flavell, R. A. (2000) Immunity 12, 171–181. [DOI] [PubMed] [Google Scholar]
- 48.Mizoguchi, A., Mizoguchi, E., Chiba, C., Spiekermann, G. M., Tonegawa, S., Nagler-Anderson, C. & Bhan, A. K. (1996) J. Exp. Med. 183, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giese, K., Kingsley, C., Kirshner, J. R. & Grosschedl, R. (1995) Genes Dev. 9, 995–1008. [DOI] [PubMed] [Google Scholar]
- 50.Levanon, D., Goldstein, R. E., Bernstein, Y., Tang, H., Goldenberg, D., Stifani, S., Paroush, Z. & Groner, Y. (1998) Proc. Natl. Acad. Sci. USA 95, 11590–11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das, G., Augustine, M. M., Das, J., Bottomly, K., Ray, P. & Ray, A. (2003) Proc. Natl. Acad. Sci. USA 100, 5324–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi, M. J. & Stavnezer, J. (1998) J. Immunol. 161, 6751–6760. [PubMed] [Google Scholar]
- 53.Yang, X., Letterio, J. J., Lechleider, R. J., Chen, L., Hayman, R., Gu, H., Roberts, A. B. & Deng, C. (1999) EMBO J. 18, 1280–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Judd, L. M., Gleeson, P. A., Toh, B. H. & van Driel, I. R. (1999) Am. J. Physiol. 277, G209–G218. [DOI] [PubMed] [Google Scholar]
- 55.Fernandez-Salguero, P. M., Ward, J. M., Sundberg, J. P. & Gonzalez, F. J. (1997) Vet. Pathol. 34, 605–614. [DOI] [PubMed] [Google Scholar]
- 56.Levanon, D., Brenner, O., Otto, F. & Groner, Y. (2003) EMBO Rep. 4, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sartor, R. B. (1997) Res. Immunol. 148, 567–576. [DOI] [PubMed] [Google Scholar]
- 58.Prokunina, L., Castillejo-Lopez, C., Oberg, F., Gunnarsson, I., Berg, L., Magnusson, V., Brookes, A. J., Tentler, D., Kristjansdottir, H., Grondal, G., et al. (2002) Nat. Genet. 32, 666–669. [DOI] [PubMed] [Google Scholar]
- 59.Tokuhiro, S., Yamada, R., Chang, X., Suzuki, A., Kochi, Y., Sawada, T., Suzuki, M., Nagasaki, M., Ohtsuki, M., Ono, M., et al. (2003) Nat. Genet. 35, 341–348. [DOI] [PubMed] [Google Scholar]
- 60.Helms, C., Cao, L., Krueger, J. G., Wijsman, E. M., Chamian, F., Gordon, D., Heffernan, M., Daw, J. A., Robarge, J., Ott, J., et al. (2003) Nat. Genet. 35, 349–356. [DOI] [PubMed] [Google Scholar]
- 61.Eliakim, R. & Rachmilewitz, D. (1996) Inflamm. Bowel Dis. 2, 122–132. [PubMed] [Google Scholar]
- 62.Barnes, K. C. (2000) J. Allergy Clin. Immunol. 106, S192–200. [DOI] [PubMed] [Google Scholar]
- 63.Bonen, D. K. & Cho, J. H. (2003) Gastroenterology 124, 521–536. [DOI] [PubMed] [Google Scholar]
- 64.Haagerup, A., Bjerke, T., Schiotz, P. O., Binderup, H. G., Dahl, R. & Kruse, T. A. (2002) Allergy 57, 680–686. [DOI] [PubMed] [Google Scholar]
- 65.Levanon, D., Negreanu, V., Bernstein, Y., Bar-Am, I., Avivi, L. & Groner, Y. (1994) Genomics 23, 425–432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.