Abstract
Acinetobacter sp. strain ADP1 is a naturally transformable gram-negative bacterium with simple culture requirements, a prototrophic metabolism and a compact genome of 3.7 Mb which has recently been sequenced. Wild-type ADP1 can be genetically manipulated by the direct addition of linear DNA constructs to log-phase cultures. This makes it an ideal organism for the automation of complex strain construction. Here, we demonstrate the flexibility and versatility of ADP1 as a genetic model through the construction of a broad variety of mutants. These include marked and unmarked insertions and deletions, complementary replacements, chromosomal expression tags and complex combinations thereof. In the process of these constructions, we demonstrate that ADP1 can effectively express a wide variety of foreign genes including antibiotic resistance cassettes, essential metabolic genes, negatively selectable catabolic genes and even intact operons from highly divergent bacteria. All of the described mutations were achieved by the same process of splicing PCR, direct transformation of growing cultures and plating on selective media. The simplicity of these tools make genetic analysis and engineering with Acinetobacter ADP1 accessible to laboratories with minimal microbial genetics expertise and very little equipment. They are also compatible with complete automation of genetic analysis and engineering protocols.
INTRODUCTION
Manipulative genetics is a primary method in both descriptive and constructive molecular biological investigations. Whole-gene function is often identified through phenotypic analysis of gene deletions and complementary replacements. The replacement of wild-type genes with alternate alleles is also used in vivo to identify the function of specific features of genes and proteins. Biochemical analysis of naturally expressed protein requires the addition of sequences coding for binding tags to chromosomal genes. The development of new biochemical pathways for biomedical and biotechnological industries requires highly reiterative genetic manipulation, including insertion and deletion of many genes in the same strain, and often alteration of those genes in the process. These uses of manipulative genetics are essential to the current progress of biological research, and often determine the cost and efficiency of the experimental process.
Many fields of biology have either chosen or happened upon primary model organisms for which there are straightforward, user-friendly methods for genetic manipulation. Caenorhabditis elegans and Drosophila are relatively challenging, but the complexity of animal development and metabolism makes increased difficulties in these organisms inevitable. The Agrobacterium/Arabidopsis system provides a reasonably simple way to test genetic hypotheses in plants. Saccharomyces cerevisiae offers the same to mycologists, and serves as the model organism for all eukaryotes. Among bacteria, the primary gram-positive model Bacillus subtilus offers a relatively easy target for genetic manipulation. However, the primary gram-negative model organism, the archetypal model organism for all genetics, Escherichia coli, is relatively resistant to genetic manipulation.
E.coli has been the primary genetic model since the first functional description of a mapped genetic locus, the lac operon (1). Since then, researchers have struggled to overcome the genetic obstacles presented by this model, obstacles created by two specific traits of this bacterium. Due to a lack of natural competence, E.coli must be manipulated to allow transformation. The second obstacle is a lack of natural recombination capabilities. This must be overcome by the addition of recombination functions from other organisms and the simultaneous deletion or inhibition of native nuclease activities that prevent recombination through direct destruction of the introduced DNA construct (2,3). The manipulations needed to achieve recombination are deleterious and have considerable epistatic effects, necessitating their reversal after the desired genetic changes have been achieved (4). All of these steps take considerable time and are subject to unpredictable failure in the hands of less-experienced researchers. Even transduction, the simplest method for transfer of alleles from one strain of E.coli to another, requires the maintenance of phage stocks, and the results are genetically inexact (the recombining region is unpredictable), requiring post-transfer screening of multiple markers to assure accuracy (5).
Here we present evidence that Acinetobacter ADP1 is a superior model system for studies requiring genetic manipulation of gram-negative bacteria. We show that the unique natural characteristics of ADP1 avoid the problems associated with E.coli, allowing all common types of chromosomal genetic manipulations to be achieved with a very simple, automatable protocol. Furthermore, we show that the close relationship between E.coli and ADP1, combined with the newly available whole-genome sequence of ADP1, allows the tremendous amount of existing knowledge related to gene function and metabolism of E.coli to be applied directly to ADP1.
Acinetobacter ADP1 shares most of the features that make E.coli a desirable laboratory organism. It is a common constituent in aquatic and soil environments. It is fully prototrophic, aerobic, and is able to grow overnight on both rich and minimal salts media. It can grow slowly at room temperature, but grows optimally between 30 and 37°C. It has a small genome of 3.7 Mb, which has recently been completely sequenced (6). Well-studied lab strains like ADP1 are apparently harmless to humans and carry no obvious pathogenicity or virulence factors. Most of the common antibiotics used with E.coli are effective against Acinetobacter ADP1, and many of the antibiotic resistance cassettes used in E.coli also transfer functionally (7). There are two relevant differences between the two species, and together they make Acinetobacter ADP1 a much more effective model organism for research and engineering requiring manipulative genetics in bacteria.
The first advantage of Acinetobacter ADP1 is the property of natural competence (8), which extends to both plasmid DNA and linear fragments. Several genes necessary for this process have been identified. One example is comP, which encodes a pilus-like protein (9), suggesting that ADP1 has an active DNA uptake system. The second advantage is a strong natural tendency towards homology-directed recombination (10). The only requirement for expression of natural competence and recombination in ADP1 is that transformation must occur during exponential growth (11). In this state, in our hands, ADP1 is approximately 10–100 times as competent as calcium chloride treated E.coli (cell−1 μg−1). Combined, these properties allow genetic manipulation by simple addition of linear PCR products to small volumes of growing cell culture, followed by a few hours of incubation and plating on appropriate selective media.
In this work, splicing PCR (12) was used to create recombinant constructs for transformation of Acinetobacter ADP1 (13), and these constructs were used to generate a wide variety of mutants, including marked and unmarked gene deletions, chromosomal protein tags for purification, replacements of wild-type alleles with mutant alleles, interspecific gene complementations, recursive serial deletions, whole operon insertions and nonpolar deletions. These manipulations required only a PCR machine, shaking and stationary incubators, and oligonucleotide synthesis. The efficiency and broad applicability of this simple technique suggest that ADP1 is an ideal organism for genetic analysis and engineering. The tools needed to generate specific mutant strains of ADP1 are available to virtually any researcher, student or industrial worker in need of such a system. All manipulations in this work were performed in very small culture volumes with raw PCR products, straightforward selections and simple post-manipulation analytical methods, in order to demonstrate the possibility of automating these processes with existing robotic technologies.
MATERIALS AND METHODS
Culture of Acinetobacter ADP1
Acinetobacter ADP1 was routinely grown in standard E.coli media and conditions (either Luria–Bertani (LB) medium or MS/0.2% glucose (MSglc) (14), liquid or agar plates, in 30 or 37°C incubators or on benchtop at 25°C, 250–300 r.p.m. shaking when appropriate). Because liquid cultures grown in rolling incubators tend to granulate (cells clump), shaking is recommended for aeration. Media were supplemented with antibiotics or negative selection agents as appropriate (Table 1). All auxotrophy tests were done in MSglc supplemented with 300 μM of the appropriate amino acids. One important difference in culturing was noted: ADP1 does not survive well at 4°C. Strains were reisolated with the appropriate selective plate twice after manipulation and initial selection, then frozen without antibiotic in LB liquid with 10% dimethyl sulfoxide (DMSO) at −80°C.
Table 1. Selective agents and resistance cassettes.
| Antibiotic | Selection cassette/source | Concentration(s) |
|---|---|---|
| Kanamycin | KanR/pUC4K | 15–50 μg/ml |
| Spectinomycin | SpecR/pOmegaSpec | 200–400 μg/ml |
| Tetracycline | TetR/pBR322 | 10–20 μg/ml |
| Trimethoprim | TrimR/pR67D3 | 200–250 μg/ml |
| AZT | PvalStdkHi/pKLC4 | 200–250 μg/ml |
| Sucrose | PKanR(/pUC4K)sacBBs(/pIB279) | 6% (wt/vol) in LB w/o NaCl |
Chromosomal replacement (marked deletion)
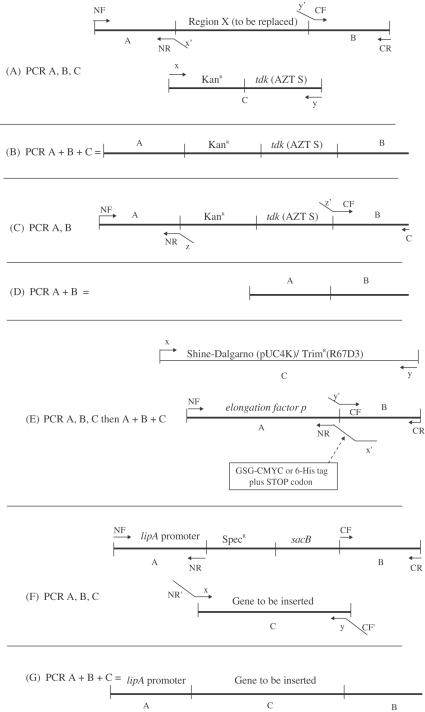
The primary method used to manipulate the Acinetobacter ADP1 chromosome was treatment of exponentially growing cultures with spliced PCR products (see ‘PCR, Splicing PCR and Transformation of Acinetobacter ADP1’, below). The technique used for splicing was developed by Murphy and coworkers for use in recombination-proficient (ΔrecBCD::red), electroporation-competent E.coli (12). Identical protocols were followed in this work, except as described. The basic strategy for marked deletion is shown in Figure 1A and B. Briefly, a cassette carrying both positive and negative selection markers, such as the KanR/tdk and KanR/sacB cassettes described below, was amplified by PCR (Figure 1A). Simultaneously, 1 kb regions were amplified from the upstream (N) and downstream (C) flanks of the region targeted for replacement, using primers that included 20 bp 5′ extensions complementary to the primers used for cassette amplification (Figure 1A). These first-stage PCRs included an excess of external primers, and these became the primers for the second (splicing) stage. In the second stage, the three first-stage PCRs were combined and reamplified together to generate a spliced product consisting of the N- and C-terminal flank regions surrounding the cassette (Figure 1B). The final spliced PCR product was then used to directly transform the target Acinetobacter strain (see below). Selection on antibiotic plates was used to isolate marked deletions. Correct insertion was verified by PCR using primers NF and CR. Products were run on agarose gels and analyzed for length against wild-type (parental) controls. Because the cassettes contained two genes and the targets were usually single genes, this test was usually definitive due to product size.
Figure 1.
(A–G) Strategies for genetic manipulation in Acinetobacter ADP1 using splicing PCR. NF, NR, CF, CR, x, y and z represent primer sequences. The associated arrows represent the primers themselves in the drawings. Designations followed by a prime (′) represent reverse complements. A and B when otherwise undesignated, represent ∼1 kb regions of sequence flanking the target sequence in the ADP1 genome.
The initial cassettes were used to generate marked and unmarked deletions of the ilvC locus of Acinetobacter ADP1, using the methods described below (chromosomal replacement) and the primers shown below. Upper case primer sequences are initial primer sequences for simple PCR amplification, lower case portions act as overlap primers for subsequent splicing PCR; CF and CR primers amplify C-terminal flank of the ilvC gene, while NF and NR amplify the N-terminal flank. These primers exemplify the design elements used throughout this work.
Mutants were checked by both locus-specific PCR and phenotype screening on MSglc plates with and without supplemental isoleucine, valine and leucine. The two marked deletion strains (ΔilvC::KanR/tdk and ΔilvC::KanR/sacB) were then used to expand the set of selection cassettes in vivo, by using the technique described above to replace the Kanamycin resistance cassette with other antibiotic resistance cassettes. Both the promoter and the open reading frame (ORF) were replaced in the case of the AbR/tdk cassettes, whereas only the ORF was replaced in the case of the AbR/sacB cassettes. These mutants were tested for both gain of the new marker and loss of the old one. Resistance cassettes tested included Spectinomycin resistance from pOmegaSpec (15), Trimethoprim resistance from R67D3 (16) and Tetracycline resistance from pBR322 (17). Selective agents were used at the concentrations listed in Table 1. Ampicillin was not used for chromosomal manipulations because it exhibited weak and noisy selection when tested using plasmids, and the Acinetobacter ADP1 genome bears several β-lactam resistance factors. Selection with other agents was free of background growth in most cases, the noted exception being AZT (see ‘Unmarked deletion’, below).
PCR, splicing PCR and transformation of Acinetobacter ADP1
Target genes and flanking sequences were identified by either (i) BLASTing homologous genes from other organisms against the unannotated sequence contigs archived at Genoscope (www.genoscope.cns.fr) prior to publication of the genome or (ii) searching the ERGO database (18). Primers were chosen following the strategy of Murphy et al. (12) to produce amplicons of approximately 1 kb regions of the genome flanking the region targeted for replacement.
With the following exceptions, the techniques used to generate spliced PCR products were identical to those described previously (12). The strategies behind the various applications of splicing PCR used here are shown in Figure 1A–G. Pfu Turbo polymerase (Stratagene) was used, along with its standard buffer, to maximize fidelity. All primers were chosen to be as close to 20 bp and 50% GC content as possible to maintain consistent annealing temperatures between constructions (58°C for fragment amplifications, 55°C for multi-fragment splicing amplifications). Efforts were made to avoid repetitive regions, GC runs, hairpins and heterodimers. Primers were adjusted so that no more than 4 nt of the 3′ end of any primer formed a perfect reverse-complement match to any internal sequences on other primers, in order to avoid primer-templated extension. In cases where initial attempts to generate spliced PCR products failed, primer redesign was usually the most efficient solution. In general, primers were chosen such that a few codons of the target gene remained on either side of the cassette, in an effort to prevent disruption of neighboring gene sequences or other functional elements. During deletion of the cassette (see ‘Unmarked deletion’, below), these codons were brought back together to form a short ORF, otherwise retaining the functional structure of the operon. Restriction site linkers were not added to the overlap primers, as our scheme did not require cloning at any step. Successfully amplified products were used to directly transform growing cultures of Acinetobacter ADP1, which were then selected via plating on appropriate media.
Transformation was done following the conditions suggested by the experiments of Palmen et al. in their investigations of plasmid transformation in Acinetobacter (19). The initial strain was grown overnight on an LB plate at 37°C. Multiple colonies were inoculated into 2 ml of LB and grown for 12–16 h at 30°C in a shaking incubator. Stationary phase had to be reached at this step for transformation to be efficient, as suggested by previous studies, but further incubation (36 h) also decreased efficiency dramatically. An aliquot of 20 μl of the stationary culture was diluted into 300 μl of fresh LB in a glass test tube. This dilution was incubated with shaking at 30°C for 2.5 h; 100 μl of raw PCR product was added to the tube, which was then returned to shaking incubation at 30°C for 2 h. The entire 400 μl culture was then spread on an appropriate selective plate, allowed to fully absorb, then placed in a 30°C incubator until colonies appeared or the plate dried up, whichever came first.
Construction of dual positive/negative selection cassettes
KanR/tdk cassette
The 1339 bp Acinetobacter ADP1 replication origin (20) was cloned into the AatII site of pUC18 (21) to generate the Ac/Ec shuttle vector pKT40. The PvalSHi/tdkHi selection cassette (22) was cloned into pKT40 using SalI and XhoI to generate pKLC4.
pKLC4 was purified from E.coli (Qiagen) and linearized upstream of the PvalSHi/tdkHi construct with EcoRI (4860 bp). The Kanamycin resistance (KanR) cassette (1282 bp) was removed from pUC4K with EcoRI. Both were agarose gel-purified (Qiagen) and ligated (Boehringer). CaCl2-competent E.coli TG1 were transformed with the ligation product, selected on LB + Ampicillin (Amp, 300 μg/ml) + Kanamycin (Kan, 30 μg/ml), and reisolated twice on the same plates. Clones were tested for the correct relative orientation of the KanR cassette and the tdk cassette (divergent polarity with both transcribed outward from the middle) using EcoRI and SmaI.
The tdk gene can be counter-selected based on the ability of the Tdk protein to activate the toxic base analog 3′-Azido-3′-deoxythymidine (AZT) (23). Clones were tested for AZT sensitivity by the halo test method (24). In brief, 1/500 dilutions of the bacteria to be tested were poured on MSglc + Amp plates and then poured off to remove excess. After drying, a 50 μl well was made in the center and filled with AZT (6.8 mg/ml) dissolved in water. The plates were allowed to grow overnight and then observed for the presence of an inhibition halo around the well. The most sensitive clone was retained as pDLM02.1. The cassette, as amplified by primers KanTdkF2 (5′-caagtcagcgtaatgctctgcc) and KanTdkR (5′-ggtcgacttaaatttgaccgcac), is 1878 bp long.
KanR/sacB cassette
The KanR/sacB selection cassette was constructed using PCR, and was designed to contain the B.subtilis saccharase B (sacB) ORF from pIB279 (25) inserted between the promoter and the KanR resistance gene of pUC4K (Pharmacia) to generate an operon with co-transcribed positive and negative selection cassettes. The sacB ORF was amplified from pIB279 using the primers SacBF (5′-aaaggagacatgaacgatgaac) and SacBR (5′-ttatttgttaactgttaattgtcc). The primers pUC4KlinF (5′-atacaaggggtgttatgagcc) and pUC4KlinR (5′-tactgtttatgtaagcagacag) were used to amplify linear pUC4K divergently from a site 15 bp upstream of the KanR start codon. The sacB PCR product was treated with DpnI and phosphorylated with T4 PNK, then both PCR products were gel purified with the Qiaquick Gel Extraction kit (Qiagen). DNA fragments were ligated (Boehringer) and used to transform CaCl2-competent E.coli TG1. Clones were selected on LB + Kan (50 μg/ml), reisolated twice, then screened for sucrose sensitivity (SacS) by cross-streaking on LB + Sucrose plates (25) at 30°C. KanR/SacS clones were tested for the correct cassette orientation using EcoRI, and the most sensitive was retained as pDLM03. The final cassette, as amplified by primers SacB/KanF (5′-ggaaagccacgttgtgtctc) and SacB/KanR (5′-gctctgccagtgttacaacc) is 2401 bp long.
Unmarked deletion
A strategy very similar to that used for gene replacement with cassettes was used to evict the cassettes. In this case, the overlap regions of primers NR and CF were designed to complement each other such that the second PCR stage yielded a product in which the N- and C-terminal flank regions were spliced together with no intervening insert. This was accomplished by adding the reverse complement of primer NR to the 5′ end of CF. In some cases, a unique 21 bp coding sequence was inserted to provide a strain identifier by using complementary 21 bp overlap regions as done in (12). Recombinants were isolated on plates that selected for the loss of the negative marker. Correct eviction was verified by loss of antibiotic resistance, and tested by length-specific PCR with primers NF and CR.
Tdk expression results in the toxic incorporation of AZT, a chain-terminating base analogue, but it is a very noisy and subtle selection. This noise may be due to the low efficiency of the tdk construct used, as opposed to any natural resistance, or may be due to a reduction in the toxicity of chain termination in the presence of the Acinetobacter ADP1 recombination machinery. On AZT, tdk+ and tdk− cultures would both inevitably yield a lawn of cells. Random streaks were taken from these lawns and restreaked on AZT to isolate single colonies, which were themselves further reisolated. Throughout this process, control and experimental lines yielded visually identical results. However, PCR screening of the resulting clones generally showed that the majority of experimental lines were tdk− mutants (unmarked deletions from which the cassette had successfully been removed), while the control lines were inevitably wild type.
Sucrose counter-selection of the sacB gene (25) yielded isolated colonies of strongly resistant cells in most cases, but occasionally failed completely, either because of epistasis between sacB and the specific chromosomal mutations associated with the strain being manipulated, or because of local regulatory effects decreasing SacB expression. However, while sacB counter-selection never yielded background growth, approximately 70% of the colonies from sucrose counter-selection plates could be identified as spontaneous loss-of-function mutants, as opposed to the desired targeted replacements, and hence this method of counter-selection required considerably more post-selection PCR- and phenotype-based screening to identify the correct clones than did AZT counter-selection. This type of screening is generally required for negative selection in other bacteria, and in our case it was often quite straightforward, as spontaneous loss-of-function mutants could be readily identified on the basis of retained antibiotic resistance from the positive selection markers that were also present in our cassettes. Such phenotypes could easily be screened in a robotic format by replica plating or spot testing on selective media.
Serial deletions
Three techniques were employed to generate strains with multiple genetic knockouts. One involved simple repetition of the procedure used to generate single unmarked deletions. The second method relied on transformation of one mutant with a normal PCR product generated by amplification of a manipulated allele from a second mutant using primers NF and CR. A third strategy was to transform Acinetobacter ADP1 directly with purified genomic DNA. Genomic DNA was prepared from the donor strain (26). Adding a microliter of unquantified, purified chromosomal DNA to a growing ADP1 culture was sufficient to generate hundreds of transformants with the desired genotype.
Nonpolar deletion
Nonpolar marked deletions were accomplished in the same way as regular marked deletions, with the following exceptions. The cassette used was always KanR/sacB, with the two genes transcribed as an operon from the PKan promoter of pUC4K. The cassette was always inserted such that the marker operon was immediately upstream of the next co-operonic gene downstream of the target. The cassette was amplified using primers that included only a few base pairs beyond the end of the KanR gene, and the insertion was designed such that the end of the KanR gene was closely followed by the Shine–Dalgarno sequence preceding the downstream gene. Correct replacement was verified by length-specific PCR with primers NF and CR.
Chromosomal gene tagging
PCR was used to generate the gene targeted for tagging, and, separately, the first 1000 bp immediately following the gene. Overlap primers were designed to replace the stop codon of the target gene with sequences encoding either the c-myc antigen (with a glycine–serine–glycine linker) or a 6-His tag, followed by a stop codon. These products were generated with regions of homology to allow splicing of an extremely short Trimethoprim resistance ORF from R67D3 (16) with a preceding sequence encoding the KanR Shine–Dalgarno region from pUC4K. It was expected that the TrimR allele would be expressed as part of an operon with the tagged gene.
RESULTS
The operations required in most genome engineering applications are gene disruption, gene deletion, ectopic expression of heterologous genes, tagging of chromosomally encoded genes and various combinations of these operations. The purpose of this work was to demonstrate that the unique properties of Acinetobacter ADP1 allow all of these operations to be accomplished using simple, rapid and automatable PCR-based techniques.
Gene disruption and deletion
Cassettes containing both positive and negative selection markers were constructed as described in Materials and Methods. Initial cassettes were made using traditional in vitro cloning methods in E.coli, with later cassettes being constructed by direct transformation of Acinetobacter ADP1 with spliced PCR products. The positive selection markers used were TetR, KanR, SpecR and TrimR. The negative selection markers were sacB, which confers sensitivity to sucrose, and tdk, which confers sensitivity to the base analogue AZT. Both polar and nonpolar cassettes were constructed (see Materials and Methods).
Positive selection markers were used to select for the insertion of PCR-amplified cassettes into the chromosome by homologous recombination (see Figure 1A and B). Negative selection markers were used to select for the subsequent elimination of the cassette and the generation of unmarked gene deletions (see Figure 1C and D). A list of the insertion and deletion mutants generated in this study is given in Table 2.
Table 2. Mutants constructed with PCR-mediated gene replacement strategies in Acinetobacter ADP1.
| Strain | Genotype | Tested phenotype |
|---|---|---|
| PS6290 | ΔlipA::KanR/tdk | No halo on egg-yolk plates |
| PS6293 | ΔlipA | No halo on egg-yolk plates |
| PS6298 | ΔilvC::KanR/tdk | Requires Ile, Val, Leu |
| PS6301 | ΔthyA::KanR/tdk | Requires thymidine and tdk |
| PS6304 | ΔilvC::TetR/tdk | Requires Ile, Val, Leu |
| PS6308 | ΔilvC::KanR/sacB | Requires Ile, Val, Leu |
| PS6312 | ΔilvC | Requires Ile, Val, Leu |
| PS6315 | ΔilvC::SpecR/tdk | Requires Ile, Val, Leu |
| PS6322 | ΔthyA::TetR/tdk | Requires thymidine and tdk |
| PS6324 | ΔilvC::SpecR/sacB | Requires Ile, Val, Leu |
| PS6331 | ΔhptA::KanR/sacB | None |
| PS6334 | ΔykvK::KanR/sacB | tRNA does not contain queuosine |
| PS6337 | ΔykvM::KanR/sacB | tRNA does not contain queuosine |
| PS6345 | ΔilvC::TetR/sacB | Requires Ile, Val, Leu |
| PS6347 | ΔlysX::KanR/sacB | Extremely slow growth |
| PS6352 | ΔlipA::ileSEc/KanR, ΔileS::SpecR/tdk | No halo on egg-yolk plates |
| PS6353 | ΔgluX::KanR/sacB | None |
| PS6355 | ΔlipA::ileSEc/KanR ΔileS | No halo on egg-yolk plates |
| PS6359 | ΔyjeK::KanR/sacB | Extremely slow growth |
| PS6363 | ΔilvC::TrimR/sacB | Requires Ile, Val, Leu |
| PS6369 | ΔthrB::KanR/tdk | None |
| PS6372 | ΔtrpGDC::KanR/tdk | Requires Trp |
| PS6375 | Δtgt::KanR/tdk | tRNA does not contain queuosine |
| PS6378 | ΔykvL::KanR/tdk | tRNA does not contain queuosine |
| PS6382 | ΔykvJ::KanR/tdk | tRNA does not contain queuosine |
| PS6387 | ΔlysX::KanR/tdk | Extremely slow growth |
| PS6390 | ΔthrB | |
| PS6393 | ΔtrpGDC | Requires Trp |
| PS6396 | Δtgt | tRNA does not contain queuosine |
| PS6399 | ΔlysX | Extremely slow growth |
| PS6403 | Δtgt, ΔykvJ::KanR/tdk | tRNA does not contain queuosine |
| PS6406 | Δtgt, ΔykvL::KanR/tdk | tRNA does not contain queuosine |
| PS6409 | Δtgt, ΔykvM::KanR/tdk | tRNA does not contain queuosine |
| PS6412 | Δtgt, ΔykvK::KanR/tdk | tRNA does not contain queuosine |
| PS6421 | efp/C-MYC/TrimR | |
| PS6423 | efp/6-His/TrimR | EFP purifies on Cobalt-NTA column |
| PS6427 | Δefp::KanR/sacB (nonpolar) | Extremely slow growth |
| PS6434 | ΔlipA::SpecR/sacB (using lipA promoter) | No halo on egg-yolk plates |
| PS6437 | ΔlipA::ykvJKLMBs (using lipA promoter) | |
| PS6439 | ΔlysX::KanR/sacB, efp/6-His/TrimR | EFP purifies on Cobalt-NTA column |
| PS6443 | ΔyjeK::KanR/sacB, efp/6-His/TrimR | EFP purifies on Cobalt-NTA column |
| PS6445 | ΔlipA::ykvJKLMBs, ΔykvJ::KanR/tdk | tRNA has restored queuosine |
| PS6448 | ΔlipA::ykvJKLMBs, ΔykvL::KanR/tdk | tRNA has restored queuosine |
| PS6456 | ΔyceG::KanR/sacB (nonpolar) | None |
| PS6459 | metGΔtrbp::KanR/tdk | Slow growth |
| PS8045 | ΔtrpGDC, ΔlipA::SpecR/sacB | Requires Trp, no halo on egg-yolk plates |
| PS8054 | ΔtrpGDC, ΔlipA | Requires Trp, no halo on egg-yolk plates |
| PS8082 | ΔproC::KanR/tdk | Requires Pro |
| PS8089 | ΔproC | Requires Pro |
| PS8092 | ΔtrpGDC, ΔlipA, ΔproC::KanR/tdk | Requires Trp and Pro, no halo on egg-yolk plates |
| PS8101 | ΔtrpGDC, ΔlipA, ΔproC | Requires Trp and Pro, no halo on egg-yolk plates |
| PS8105 | ΔilvC, ΔlipA, ΔproC | Requires Ile, Leu, Val, Pro, no halo on egg-yolk plates |
| PS8108 | ΔtrpGDC, ΔilvC, ΔproC | Requires Trp, Ile, Leu, Val, Pro |
| PS8119 | ΔtrpGDC, ΔlipA, ΔilvC | Requires Trp, Ile, Leu, Val, no halo on egg-yolk plates |
Examples of PCR results comparing the same locus from wild-type and mutant strains are shown in Figure 2. PCR products amplified with the same primers (IlvCNF and IlvCCR, the two outside flank primers surrounding the affected locus, ilvC, an isoleucine/valine biosynthetic gene) are shown for ADP1 (wild-type), PS6301 (ΔilvC::KanR/tdk), PS6304 (ΔilvC::TetR/tdk) and PS6308 (ΔilvC::KanR/sacB), along with the strains resulting from the subsequent cassette evictions (unmarked deletions with 21 bp in-frame insertions) using each one of the depicted marked deletion strains. These were all of the expected length. In most constructions, approximately 70% of the selected clones yielded locus-specific PCR products of the correct length.
Figure 2.
PCR of the IlvC region of the Acinetobacter ADP1 genome from wild-type (ADP1), three marked deletion mutants with different replacement cassettes (PS6301, PS6304, PS6308), and three independently generated unmarked deletions constructed from the three marked deletions (PS6312, PS6313, PS6314). All PCRs were done with the same primers, IlvCNF and IlvCCR, which amplify the ilvC ORF and ∼1 kb of flanking DNA from either side.
Splicing PCR was very efficient, with initial primer choices yielding functional quantities of spliced PCR products in standardized conditions approximately 90% of the time. Transformation of ADP1 with spliced PCR products resulted in very efficient recombination and chromosomal manipulation. Colonies were selected and reisolated on appropriate selective plates (Table 1). Reisolated mutants were tested by PCR using primers NF and CR (see Figure 1 legend), with the parental strain as a control, to test for the length changes that would be expected, given replacement of the targeted region with the cassette. This test was only ambiguous when the targeted region was similar in size to the replacement cassette. In these cases, amplification by the pairs {x and CR} and {NF and y} was used as a test for replacement. In cases where a phenotypic change such as antibiotic resistance, substrate sensitivity or auxotrophy was expected to accompany the replacement, the phenotype was also tested by streaking on appropriate plates. Selection was generally free of background growth. That is, no colonies grew on selective plates if the parent cultures were not transformed with appropriate constructs. However, approximately 3 in 10 colonies from experimental transformations appeared to result from nonspecific integration of the marker sequences. These were readily identified and removed by either PCR testing or testing of secondary phenotypes.
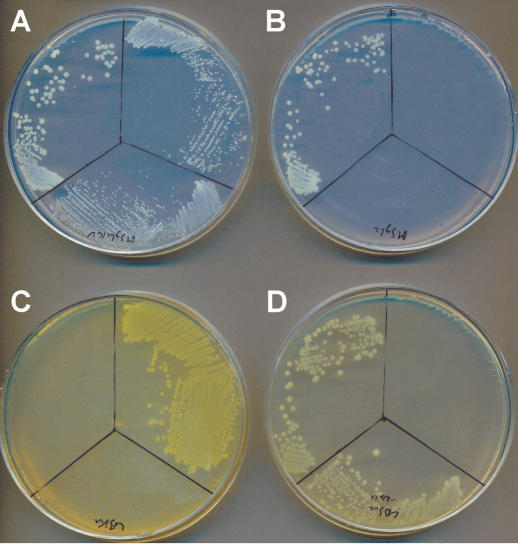
Examples of phenotypic screen results are shown in Figure 3. The growth of ADP1 (upper left sector—wild-type), PS6308 (upper right sector—ΔilvC::KanR/sacB) and PS6312 (lower sector—ΔilvC) are compared on several specific plates. IlvC is an essential gene coding for a key enzyme in the isoleucine and valine biosynthetic pathways. All three strains can grow on MSglc with isoleucine, valine and leucine (Figure 3A). On MSglc (Figure 3B), only ADP1 can grow, while the ilvC knockouts are shown to be isoleucine/valine auxotrophs. A comparison of the same strains is shown on LB + Kan (Figure 3C), where only the insertional deletion mutants can grow because they carry the resistance allele. On LB + Suc (Figure 3D) PS6308 is poisoned because the sacB allele in the cassette converts sucrose to toxic levans, allowing counter-selection.
Figure 3.
Demonstration of cassette insertion and function using phenotypic screens. The same three clones were streaked on (A) MSglc supplemented with isoleucine, valine and leucine, (B) MSglc, (C) LB + Kan, and (D) LB + Suc. Top left sectors are streaked with ADP1 (wild-type), top right sectors with PS6308 (ΔilvC::KanR/sacB), and lower sectors with PS6312 (ΔilvC).
The number of mutants constructed (Table 2) allows us to provide some approximate statistics regarding the robustness of the described strategy. For all manipulations, 90% of the primers chosen resulted in effectively amplified and spliced PCR products (see Figure 4 for an example), and 70% of the positively selectable marked chromosomal replacements described were achieved on the first attempt. The remainder worked either on the second attempt or, in one case, on a third attempt, using newly chosen primers. The only failure involved an attempted deletion of a gene, purH, that is likely to be essential in Acinetobacter ADP1. The antibiotic cassettes described gave positively selectable phenotypes at all tested loci, though a few particularly deleterious mutations such as ΔthyA and ΔlysX could not be isolated on any but the lowest listed concentrations of antibiotics (e.g. 15 μg/ml Kan). These concentrations still yielded background-free selection. Twenty-eight independent marked chromosomal replacements were generated in this study, including several that resulted in the construction of new selection cassettes (the ΔilvC mutants constructed during the in vivo cassette library expansion).
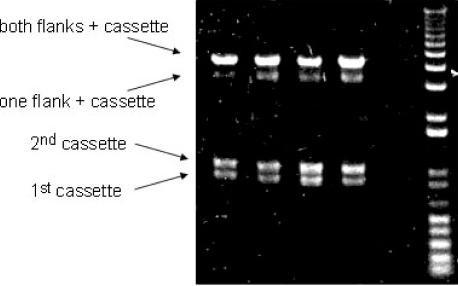
Figure 4.
An example of an unpurified splicing PCR product (top band) with three components, including two flanks and an insert cassette. The lower two bands are inevitably present and represent flank PCR products that remain unincorporated. The band just below the complete product is assumed to be partially spliced fragments including one flank and the cassette.
Negative selection as a tool for cassette eviction (to generate unmarked deletions) was considerably less efficient, probably due to consistency problems involving expression of the negatively selectable markers (see Materials and Methods). In most cases, however, unmarked deletion was achieved in one or two attempts, and in cases where selection against one of the negative markers failed multiple times, the other marker worked well. Seven unique unmarked deletions were generated during this study.
Gene deletion using a nonpolar cassette
This strategy was used in the construction of the ΔyceG deletion mutant (PS6456). YceG is just upstream of the co-transcribed essential gene tmk (thymidylate kinase), and a polar insertion here would be expected to be lethal. Using a nonpolar version of the KanR/sacB cassette, the yceG deletion was easily achieved and yielded strains with no noticeable phenotypic defect.
Multiple deletions
The ability to efficiently and quickly generate unmarked gene deletions, a process that took approximately 10 days, allows for successive iterations of the deletion process and the reuse of a limited set of markers. Furthermore, independent mutations existing in different strains can be readily combined, so long as they do not share markers and at least one of them has a positively selectable marker. Any locus bearing a positively selectable marker can be transferred to another strain by transforming the recipient strain with either a PCR amplification of the marked locus along with 1 kb homologous flanking sequences or a chromosomal DNA preparation of the strain carrying the marked locus. Two examples of these strategies are described.
Four strains, each one independently deleted for one of four genes in the queuosine biosynthesis pathway, were generated as part of a published study and tested for loss of queuosine in tRNA (26). Multiple deletion mutants were subsequently generated by pairing these deletions with a deletion of tgt, the protein that transfers the queuosine base to tRNA. The four double deletion mutants (PS6403, PS6406, PS6409 and PS6412) generated for this study were produced by repetitive marked deletion with KanR-containing cassettes (as follows), and demonstrate the effectiveness of the tdk negative-selection cassette. Strain PS6375 (Δtgt::KanR/tdk) was constructed as above (‘Gene disruption and deletion’), as were the individual queuosine-biosynthesis deletions (PS6334, PS6347, PS6378, PS6382). The deleted alleles from the four biosynthesis mutants were then paired with the tgt deletion as follows: The methods discussed in the section on umarked deletion were used to evict the KanR/tdk cassette from PS6375, to give strain PS6396 (Δtgt). The four individual marked biosynthesis-pathway deletions were then independently moved into PS6396 by simply amplifying the allele and 1000 bp on either side with the two external primers used in their original construction, then transforming the resulting PCR product directly into PS6396 and selecting on LB + Kan plates.
An alternate approach was also investigated. In order to generate strains deficient in multiple metabolic processes, chromosomal DNA preparations from marked and clean deletions were alternately used to transform strains in order to introduce cassette insertions, and then to generate the unmarked isogenic strains. This process was used to generate all triple-mutant combinations of ΔlipA, ΔproC, ΔtrpGDC and ΔilvC. As one example, here we describe the process to generate PS8101 (ΔtrpGDC, ΔlipA, ΔproC). Strain PS6393 (ΔtrpGDC) was transformed with genomic DNA of strain PS6434 (ΔlipA::SpecR/sacB) and selected on LB + Spec plates, resulting in the double mutant, strain PS8045 (ΔtrpGDC, ΔlipA::SpecR/sacB). The cassette was evicted from the lipA locus by transformation with genomic DNA from strain PS6293 (ΔlipA) followed by selection on LB + Suc plates, generating strain PS8054 (ΔtrpGDC, ΔlipA). This double mutant was further transformed with genomic DNA from strain PS8082 (ΔproC::KanR/tdk) and selected on LB + Kan plates, generating strain PS8092 (ΔtrpGDC, ΔlipA, ΔproC::KanR/tdk). PS8092 was transformed with genomic DNA from strain PS8089 (ΔproC) and selected on LB AZT plates to generate the triple mutant, strain PS8101, (ΔtrpGDC, ΔlipA, ΔproC). A similar strategy yielded strains PS8105 (ΔproC, ΔlipA, ΔilvC), PS8108 ((ΔtrpGDC, ΔilvC, ΔproC) and PS8119 (ΔtrpGDC, ΔlipA, ΔilvC). Transformation efficiencies using chromosomal DNA are vastly improved relative to the use of PCR products. Phenotypic testing of strains was always as expected.
Phenotypic testing of multiple-knockout strains can be a powerful tool in assessing interactions between genes. This technique has been underutilized in the past, primarily because the difficulties and inefficiencies associated with chromosomal mutation in E.coli increase exponentially with the multiplicity of deletion. The rapid turnover, high success rate and homogeneous methodology of the process demonstrated here should allow expansion and automation of combinatorial mutation assessments to the whole-genome level.
Chromosomal gene tagging
Chromosomal gene tagging has many applications. These include purification of protein for in vitro functional analysis, the study of post-translational modifications that are not clearly defined by the sequence of the associated gene, and identification of protein complexes by co-purification on affinity columns.
The insertions of 6-His and c-myc tags at the 3′ end of the efp gene were successful. Following the introduction of the tag and the co-operonic TrimR marker (Figure 1E), it was found that EFP could be purified on a Cobalt-NTA column. The tagged chromosomal gene was expressed at low levels and appeared as both full-length and apparently N-terminal-proteolyzed products. Neither the full-length nor the proteolyzed band appeared in controls using wild-type Acinetobacter ADP1 (Figure 5). This mutant was later successfully combined with two deletion mutations (ΔlysX::KanR/sacB and ΔyjeK::KanR/sacB) by the strategies described above for multiple deletion.
Figure 5.

Detection of a chromosomal introduction of a 6-His tag to elongation factor P. Western blots with 6-his tag-specific monoclonal antibody against naturally expressed EFP protein in lysates of strains carrying a 3′ (C-terminal) His-tag and no tag, as indicated.
Ectopic gene insertion and expression
Another useful application of the techniques described here is the ectopic expression of foreign genes or mutant alleles of native genes. Ectopic expression can allow testing of specific point mutations, insertion of foreign genes in order to build new pathways and functionalities, and testing of heterologous genes for complementation of natural or deletion-based phenotypic deficiencies.
In this work, two strains were constructed that allow the integration and expression of foreign genes in the Acinetobacter ADP1 chromosome. The first construct was derived from plasmid pZR80 (7) by subcloning of the E.coli isoleucyl tRNA synthetase gene (ileS) from pVDC433 (27). In this construction, the ileS gene was transcribed from the PTet promoter of pBR322 and followed by the KanR marker from pUC4K, and both ileS and KanR were located between the two ADP1 lipA gene homology regions of pZR80. This region was then transferred to the ADP1 genome by homologous recombination, yielding strain PS6226 (ΔlipA::ileSEc/KanR). The native and essential ADP1 ileS (22) was then replaced with a SpecRtdk cassette, which was subsequently evicted, generating strain PS6355 (ΔlipA::ileSEc/KanR, ΔileS).
The second construction was designed to be more universal, allowing for the unmarked insertion and ectopic expression of any ORF. A promoterless SpecR/sacB cassette was used to replace most of the native Acinetobacter ADP1 lipA gene, such that the cassette was transcribed from the lipA promoter, yielding a ‘gene acceptor’ strain, PS6434 (ΔlipA::SpecR/sacB). The Bacillis subtilus queuosine biosynthesis operon was then used to replace the cassette (using the basic splicing PCR strategy used to introduce cassettes), and mutants were selected on LB + Suc plates, yielding the complementation strain PS6437 (ΔlipA::ykvJKLMBs ). Two of the queuosine-biosynthesis gene deletions (from PS6445 and PS6448) were then transferred to PS6437 by amplification of the marked deletion alleles with NF and CR primers, and subsequent transformation into PS6437 and selection on LB + Kan plates. These strains, PS6445 (ΔlipA::ykvJKLMBs, ΔykvJ::KanR/tdk) and PS6448 (ΔlipA::ykvJKLMBs, ΔykvL::KanR/tdk) were then analyzed by HPLC and shown to have restored queuosine biosynthesis (27). The general usefulness of this technique is supported elsewhere, by independent experiments in which sacB was also used as a negative selection for unmarked gene insertion in ADP1 (13).
One further example of heterologous gene expression is provided by PS6322 (ΔthyA::TetR/tdk), in which the expression of the tdk gene, together with supplemental thymidine, is essential in the absence of thyA activity as Acinetobacter ADP1 does not contain an endogenous tdk allele.
DISCUSSION
Acinetobacter ADP1 provides a remarkably simple, inexpensive and robust model system for genetic manipulation. Most of the existing antibiotics and antibiotic resistance cassettes tested here were functional in ADP1. These markers were used to build positive/negative selection cassettes, which were in turn used to efficiently construct a wide variety of mutations including gene disruptions and deletions, expressed chromosomal insertions, tagged chromosomal genes and various combinations of these types of mutations. Moving mutations from one strain to another was as straightforward as amplifying the mutation from the donor strain and inoculating a growing culture of the recipient strain with the raw PCR product, or even simply transforming with purified genomic DNA from the donor strain. All constructions shown here utilized approximately 1 kb flanking regions to specifically integrate constructs into the ADP1 genome. Attempts to use shorter flanks were generally unsuccessful. Splicing PCRs with shorter flanks resulted in high yields of product, but no transformants were recovered in selection. This limitation may be due to the minimal volume of our transformations.
The techniques used in this paper were reiterative and highly similar. All manipulations aside from the initial cassette constructions were performed using splicing PCR and selective plating only. Primers were chosen using very simple rules based on melting temperature, GC content, potential inter-primer misextension and position with regard to the affected ORF. All PCRs were performed in identical conditions. Furthermore, all direct manipulations of ADP1 cells were performed in minimal volumes similar to those found in standard 96-well plate formats. The high rate of success under these conditions suggests that this system could be readily adapted to an automated platform, allowing for all steps to be achieved robotically. Similarly, the simplicity of the ADP1 genetic engineering protocols developed here should allow this system to be adopted by both training institutions and laboratories that have a need for an inexpensive and user-friendly method for generating genetically manipulated strains. The Acinetobacter constructions described here required only a PCR machine, incubators and access to oligonucleotide synthesis. It is notable that the majority of all constructions, including the cassette constructions, antibiotic tests, resistance allele tests and associated design efforts, were achieved by one researcher (D.Metzgar), with very little previous genetic manipulation experience in the course of one year. Attempts by other researchers to use the same system were generally equally successful, but it was noted that success was dependent on careful and consistent choice of primer sequences (see Materials and Methods).
Together, the paired traits of natural competence and recombination allow for rapid production of genetically engineered strains. Replacement of existing genetic models with Acinetobacter ADP1 should be straightforward, as ADP1 presented no particular challenges with respect to culturing conditions, bioinformatic prediction of metabolic pathways, or mutational stability in culture, even in conditions that were optimized for E.coli rather than Acinetobacter. In the short time during which this model has been under development in our laboratory, it has allowed us to test a number of biological questions (27) in a much more efficient manner than would have been possible with previously utilized model organisms.
| IlvCCR | 5′-GTGAAAAAGCCAGGCCAGAACC |
| IlvC:KanTdkCF | 5′-gtgcggtcaaatttaagtcgaccGATGCCTTGGATTCAAGCCAAC |
| IlvC:KanSacBCF | 5′-ggttgtaacactggcagagcGATGCCTTGGATTCAAGCCAAC |
| IlvC:KanSacBNR | 5′-gagacacaacgtggctttccCGTGACCTTGTGAACCGTAAC |
| IlvC:KanTdkNR | 5′-ggcagagcattacgctgacttgCGTGACCTTGTGAACCGTAAC |
| IlvCNF | 5′-CCAACAAGTCGTCTGTATCAC |
| IlvCCF/21BPIF | 5′-gtacttggagttctaggccttGATGCCTTGGATTCAAGCCAAC |
| IlvCNR/21BPIF | 5′-aaggcctagaactccaagtacCGTGACCTTGTGAACCGTAAC |
Acknowledgments
ACKNOWLEDGEMENTS
We thank L. Nicholas Ornston for critical reading of this work, and Integrated Genomics for access to ERGO. This work was supported by National Institute of Health Grant GM23562, National Science Foundation Grant MCB-0128901 and a fellowship from the National Foundation for Cancer Research.
REFERENCES
- 1.Beckwith J.R. (1967) Regulation of the lac operon. Recent studies on the regulation of lactose metabolism in Escherichia coli support the operon model. Science, 156, 597–604. [DOI] [PubMed] [Google Scholar]
- 2.Murphy K.C. (1998) Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol., 180, 2063–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y., Buchholz,F., Muyrers,J.P. and Stewart,A.F. (1998) A new logic for DNA engineering using recombination in Escherichia coli. Nature Genet., 20, 123–128. [DOI] [PubMed] [Google Scholar]
- 4.Court D.L., Sawitzke,J.A. and Thomason,L.C. (2002) Genetic engineering using homologous recombination. Annu. Rev. Genet., 36, 361–388. [DOI] [PubMed] [Google Scholar]
- 5.Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 6.Barbe V., Vallenet,D., Fonknechten,N., Kreimeyer,A., Oztas,S., Labarre,L., Cruveiller,S., Robert,C., Duprat,S., Wincker,P. et al. (2004) Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res., 32, 5766–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kok R.G., van Thor,J.J., Nugteren-Roodzant,I.M., Vosman,B. and Hellingwerf,K.J. (1995) Characterization of lipase-deficient mutants of Acinetobacter calcoaceticus BD413: identification of a periplasmic lipase chaperone essential for the production of extracellular lipase. J. Bacteriol., 177, 3295–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmen R. and Hellingwerf,K.J. (1997) Uptake and processing of DNA by Acinetobacter calcoaceticus—a review. Gene, 192, 179–190. [DOI] [PubMed] [Google Scholar]
- 9.Porstendorfer D., Gohl,O., Mayer,F. and Averhoff,B. (2000) ComP, a pilin-like protein essential for natural competence in Acinetobacter sp. Strain BD413: regulation, modification, and cellular localization. J. Bacteriol., 182, 3673–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Vries J. and Wackernagel,W. (2002) Integration of foreign DNA during natural transformation of Acinetobacter sp. by homology-facilitated illegitimate recombination. Proc. Natl Acad. Sci. USA, 99, 2094–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamb D.C., Kelly,D.E., Masaphy,S., Jones,G.L. and Kelly,S.L. (2000) Engineering of heterologous cytochrome P450 in Acinetobacter sp.: application for pollutant degradation. Biochem. Biophys. Res. Commun., 276, 797–802. [DOI] [PubMed] [Google Scholar]
- 12.Murphy K.C., Campellone,K.G. and Poteete,A.R. (2000) PCR-mediated gene replacement in Escherichia coli. Gene, 246, 321–330. [DOI] [PubMed] [Google Scholar]
- 13.Jones R.M. and Williams,P.A. (2003) Mutational analysis of the critical bases involved in activation of the AreR-regulated sigma54-dependent promoter in Acinetobacter sp. strain ADP1. Appl. Environ. Microbiol., 69, 5627–5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richaud C., Mengin-Lecreulx,D., Pochet,S., Johnson,E.J., Cohen,G.N. and Marlière,P. (1993) Directed evolution of biosynthetic pathways. Recruitment of cysteine thioethers for constructing the cell wall of Escherichia coli. J. Biol. Chem., 268, 26827–26835. [PubMed] [Google Scholar]
- 15.Blondelet-Rouault M.H., Weiser,J., Lebrihi,A., Branny,P. and Pernodet,J.L. (1997) Antibiotic resistance gene cassettes derived from the omega interposon for use in E. coli and Streptomyces. Gene, 190, 315–317. [DOI] [PubMed] [Google Scholar]
- 16.Martinez M.A., Pezo,V., Marlière,P. and Wain-Hobson,S. (1996) Exploring the functional robustness of an enzyme by in vitro evolution. EMBO J., 15, 1203–1210. [PMC free article] [PubMed] [Google Scholar]
- 17.Bolivar F., Rodriguez,R.L., Greene,P.J., Betlach,M.C., Heyneker,H.L. and Boyer,H.W. (1977) Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene, 2, 95–113. [PubMed] [Google Scholar]
- 18.Overbeek R., Larsen,N., Walunas,T., D'Souza,M., Pusch,G., Selkov,E., Jr., Liolios,K., Joukov,V., Kaznadzey,D., Anderson,I. et al. (2003) The ERGO genome analysis and discovery system. Nucleic Acids Res., 31, 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmen R., Buijsman,P. and Hellingwerf,K.J. (1994) Physiological regulation of competence induction for natural transformation in Acinetobacter calcoaceticus. Arch. Microbiol., 162, 344–351. [DOI] [PubMed] [Google Scholar]
- 20.Hunger M., Schmucker,R., Kishan,V. and Hillen,W. (1990) Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene, 87, 45–51. [DOI] [PubMed] [Google Scholar]
- 21.Vieira J. and Messing,J. (1982) The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene, 19, 259–268. [DOI] [PubMed] [Google Scholar]
- 22.Pezo V., Metzgar,D., Hendrickson,T.L., Waas,W.F., Hazebrouck,S., Döring,V., Marlière,P., Schimmel,P. and De Crécy-Lagard,V. (2004) Artificially ambiguous genetic code confers growth yield advantage. Proc. Natl Acad. Sci. USA, 101, 8593–8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balzarini J., Herdewijn,P. and De Clercq,E. (1989) Differential patterns of intracellular metabolism of 2′,3′-didehydro-2′,3′-dideoxythymidine and 3′-azido-2′,3′-dideoxythymidine, two potent anti-human immunodeficiency virus compounds. J. Biol. Chem., 264, 6127–6133. [PubMed] [Google Scholar]
- 24.Nangle L.A., De Crécy-Lagard,V., Döring,V. and Schimmel,P. (2002) Genetic code ambiguity. Cell viability related to the severity of editing defects in mutant tRNA synthetases. J. Biol. Chem., 277, 45729–45733. [DOI] [PubMed] [Google Scholar]
- 25.Blomfield I.C., Vaughn,V., Rest,R.F. and Eisenstein,B.I. (1991) Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol., 5, 1447–1457. [DOI] [PubMed] [Google Scholar]
- 26.Ausubel F.M. (1997) Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology, 3rd edn. Wiley, New York. [Google Scholar]
- 27.Reader J.S., Metzgar,D., Schimmel,P. and de Crécy-Lagard,V. (2004) Identification of four genes necessary for biosynthesis of the modified nucleoside queuosine. J. Biol. Chem., 279, 6280–6285. [DOI] [PubMed] [Google Scholar]
- 28.Hendrickson T.L., Nomanbhoy,T.K., de Crécy-Lagard,V., Fukai,S., Nureki,O., Yokoyama,S. and Schimmel,P. (2002) Mutational separation of two pathways for editing by a class I tRNA synthetase. Mol, Cell, 9, 353–362. [DOI] [PubMed] [Google Scholar]