Abstract
Biogenesis of eukaryotic ribosomal subunits proceeds via a series of precursor ribonucleoprotein particles that correspond to different stages in the maturation pathway. The different pre-ribosomal particles each contain a distinct complement of non-ribosomal, trans-acting factors that are crucial for correct and efficient progress of the maturation process. Although in recent years we have gained considerable insight into the composition of the pre-ribosomal particles, our knowledge how the ordered association with and their dissociation from the pre-ribosome of these trans-acting factors is controlled is still quite limited. Here, we have studied the mutual dependence between three of these factors, Rrp5p, U3 snoRNP and Rok1p, all essential for the early stages of pre-rRNA processing/assembly, for association with the 35S pre-rRNA in Saccharomyces cerevisiae. Using co-immunoprecipitation assays, we show that Rrp5p and U3 snoRNP associate independently of each other and that the two factors do not detectably interact prior to incorporation into the pre-ribosome. In contrast, association of the putative RNA helicase Rok1p, which is known to genetically interact with Rrp5p, is absolutely dependent on the presence of the latter protein but does not require U3.
INTRODUCTION
Eukaryotic ribosomes are largely produced in a specialized region of the nucleus, the nucleolus. Here, the pre-rRNA is transcribed and assembled into pre-ribosomes, which then mature on their journey to the cytoplasm by modification and processing of the pre-rRNA and stepwise assembly of further ribosomal proteins (r-proteins). Studies using Saccharomyces cerevisiae as a model organism have made it clear that the biogenesis of ribosomes in eukaryotic cells is considerably more complex than in prokaryotes. As shown by the pioneering work of Nomura and Held (1) and Nierhaus and Dohme (2), the assembly of prokaryotic ribosomes can be accomplished in vitro using only purified rRNA and r-proteins, although recent data indicate that in vivo some non-ribosomal proteins, in particular a protein chaperone and RNA helicases, facilitate this process (3–6). In contrast, attempts to reconstitute eukaryotic ribosomes in vitro have remained unsuccessful, and a large body of evidence demonstrates that formation of mature ribosomes in S.cerevisiae critically depends upon a set of more than 150 trans-acting factors, proteins as well as ribonucleoprotein particles (RNPs), that are not retained in the finished product (7,8).
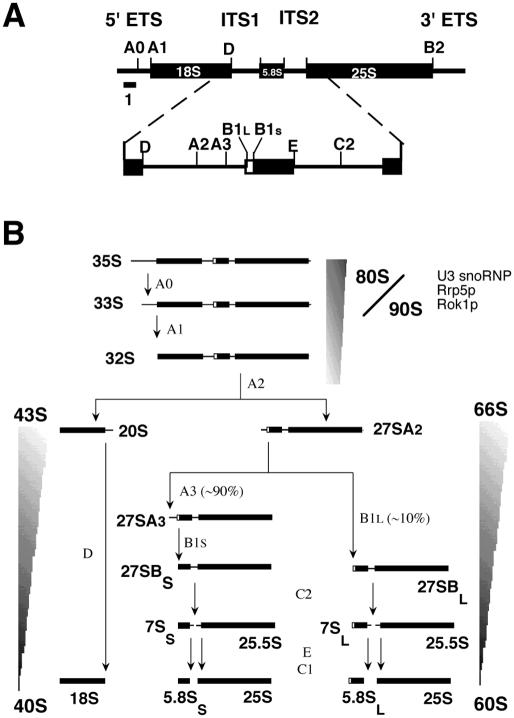
Three of the four mature yeast rRNA molecules (18S, 5.8S and 25S rRNA) are produced from a single 35S precursor that is cotranscriptionally modified and assembled into a large pre-ribosomal particle (∼90S), which contains a substantial number of trans-acting factors as well as a subset of r-proteins (Figure 1). This pre-ribosome probably goes through several stages characterized by the association and dissociation of particular (sets of) trans-acting factors and cleavage of the pre-rRNA at sites A0 and A1 and is then split into a 43S and a 66S pre-ribosome, containing 20S and 27SA2 pre-rRNA, respectively, by pre-rRNA processing at site A2. The 43S particle goes through one or more additional stages in the nucleoplasm before being exported to the cytoplasm for final maturation, which in yeast, but not in higher eukaryotes, includes the processing of 20S pre-rRNA into 18S rRNA (9–11). The 66S pre-ribosome first incorporates the 5S rRNA-containing RNP, followed by processing of the 27SA2 to 27SB pre-rRNA, which is converted into 7S and 25.5S pre-rRNA by cleavage at site C2. Exonucleolytic removal of the remaining spacer sequences from these precursors then results in mature 5.8S and 25S rRNA, respectively (12–14). Analysis of the protein content of immunopurified 66S particles using different TAP-tagged trans-acting factors has revealed at least four distinct intermediate pre-ribosomal particles in this pathway, which ends in an export-competent pre-60S particle that undergoes some final conformational adjustments in the cytoplasm (15–17).
Figure 1.
Pre-ribosomal RNA processing in S.cerevisiae. (A) Structure of the rDNA transcription unit. Thick bars represent the mature 18S, 5.8S and 25S rRNA sequences and thin bars represent the spacer sequences. Processing sites and the location of probe 1 used for the primer extension assay are depicted. Note that this primer is located upstream from site A0 and, thus, will only detect unprocessed (35S) pre-rRNA. (B) The pre-rRNA processing pathway. The pre-ribosomal particles containing the different processing intermediates are indicated.
In a large majority of cases, genetic depletion or mutation of an individual trans-acting factor inhibits the formation of either the small or the large ribosomal subunit. So far, only two factors, Rrp5p and Rrp12p, are known to be required for the production of both subunits (18,19). The RRP5 gene was identified in a synthetic lethality screen with a deletion of the gene encoding the snR10 snoRNA. Genetic depletion of Rrp5p inhibits the early processing cleavages at sites A0–A2, which are essential for the production of 18S rRNA, as well as the RNase MRP-dependent cleavage at site A3, involved in 5.8SS rRNA synthesis (18). Structural analysis showed Rrp5p to possess two distinct regions, an N-terminal one containing 12 S1 RNA-binding motifs and a C-terminal one consisting of seven tetratricopeptide (TPR) motifs thought to be involved in protein–protein interactions (20,21). Mutational analysis demonstrated that each region represents a specific functional domain: deletions in the S1-containing region cause inhibition of processing at either site A3 or A2 (22,23), whereas mutant Rrp5p proteins lacking parts of the TPR region are unable to support cleavage at A0–A2 (20,21). Mutations in several of the TPR motifs cause a temperature-sensitive phenotype that can be suppressed by overexpression of the putative ATP-dependent RNA helicase Rok1p (21). Strikingly, Rok1p is itself essential for the A0–A2 processing steps and ROK1 genetically interacts with SNR10, establishing a further link with Rrp5p (24).
Proteomic studies indicate that Rrp5p associates with the pre-rRNA at a very early stage of ribosome biogenesis. Using sequential immunoprecipitation of two epitope-tagged components of the U3 snoRNP, which is essential for the early cleavages at A0–A2, Dragon et al. (25) isolated an 80S particle containing 35S pre-rRNA, most of the U3 snoRNP core proteins, Rrp5p and 17 novel proteins (Utp1-17), all of which were proved to be required for 18S rRNA synthesis. Formation of this 80S particle, dubbed the small subunit (SSU) processome, depends absolutely upon association of U3 with the 35S pre-rRNA, which requires specific base-pairing between U3 snoRNA and the precursor (26,27) as well as the assistance of Pwp2p, which corresponds to the Utp1p protein (28). Rrp5p as well as Rok1p have also been detected in 90S particles that were immunopurified using various other tagged trans-acting factors (11,16,29). The precise relationship between the two types of particle is not quite clear, but comparison of their protein content suggests that they are subsequent stages in the ribosome biogenesis pathway.
The simultaneous occurrence of U3 snoRNP, Rrp5p and Rok1p in early pre-ribosomes, as well as the functional and genetic links between these three trans-acting factors prompted us to investigate as to what extent their association with the 35S pre-rRNA is mutually dependent. Using co-immunoprecipitation, we show that U3 and Rrp5p associate independently. Incorporation of Rok1p into the pre-ribosome requires the prior association of Rrp5p but not U3.
MATERIALS AND METHODS
Strains and plasmids
Escherichia coli strain MH1 was used for cloning and propagation of plasmids. Yeast strains used in this study are listed in Table 1. The plasmids pHIS3-ProtA::rrp9 and pTRP1-ProtA::rok1 have been described previously (24,30). Yeast transformation was performed according to Gietz et al. (31). Integration of ProtA::rrp5 into JH84 (32) was performed as described previously (23), except that we used the HIS3 marker from YDP-H (33). YJV182 and YJV184 are segregants of the diploid RS453 ROK1/Δrok1 strain, in which the deletion was made by replacing the NcoI–BamHI fragment of ROK1 by the HIS3 marker of YDP-H, transformed with pTRP1-ProtA::Rok1.
Table 1. Yeast strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| YJV153 | MATα ade2 his3 leu2 trp1 ura3 URA3-Gal::rrp5 | (18) |
| YJV154 | MATa ade2 his3 leu2 trp1 ura3 URA3-Gal::rrp5 | (18) |
| YJV182 | MATa ade2 his3 leu2 trp1 ura3 rok1::HIS3 + pTRP1-ProtA::rok1 | This paper |
| YJV184 | MATα ade2 his3 leu2 trp1 ura3 rok1::HIS3 + pTRP1-ProtA::rok1 | This paper |
| JH84 | MATα ade2-1 his3-Δ leu2-3,12 ura3-52 can1-100 URA3-Gal::U3A U3B::LEU2 | (32) |
| YRB501 | MATα ade2-1 his3-Δ leu2-3,12 ura3-52 can1-100 URA3-Gal::U3A U3B::LEU2 + pHIS3-ProtA::rrp9 | This paper |
| YHV159 | MATa ade2 his3 leu2 trp1 ura3 URA3-Gal::rrp5 + pHIS3-ProtA::rrp9 | This paper |
| YHV200 | Mating type unknown ade2 his3 leu2 trp1 ura3 URA3-Gal::rrp5 rok1::HIS3 + pTRP1-ProtA::rok1 | This paper |
| YHV300 | MATα ade2-1 his3-Δ leu2-3,12 ura3-52 can1-100 URA3-Gal::U3A U3B::LEU2 HIS3::ProtA::rrp5 | This paper |
| YHV400 | Mating type unknown his3 leu2 ura3 Gal::U3A::URA3 U3B::LEU2 rok1::HIS3 + pTRP1-ProtA::rok1 | This paper |
Growth of cells
All yeast strains were grown at 30°C on appropriately supplemented, galactose-based medium to mid-exponential phase and then shifted to glucose-based medium to deplete the pertinent factor. Strain YHV504 was grown in YPD at 30°C and shifted to 37°C for 6 h.
Immunoprecipitation and isolation of RNA and proteins
Approximately 100 OD600 units of cells were harvested by centrifugation and washed with ice-cold water. The pellet was resuspended in IP buffer [50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.1% (v/v) Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 mM DTT, 0.2% protease inhibitor cocktail (Sigma)] at a concentration of 10 μl/OD600 unit of cells. The cells were lysed by vortexing for 5 min using glass beads (diameter 0.5 mm). Lysates were cleared by centrifugation at 15 000 g at 4°C and used immediately. Immunoprecipitation was performed by the addition of 50 μl immunoglobulin G (IgG)–Sepharose beads (Amersham). After extensive washing with IP buffer, bound material was eluted with 500 μl guanidinium thiocyanate (GTC) solution (4 M GTC, 50 mM Tris–HCl, pH 8.3, 10 mM EDTA, 20% sarkosyl, 1% β-mercapto-ethanol, diluted with an equal volume of IP buffer). Phenol extractions were performed using phenol–chloroform–isoamylalcohol (Fluka), and RNA was precipitated overnight with 2.5 vol of ethanol and 1/10 (v/v) 3 M NaAc, pH 5.3 at −20°C using 2 μg of glycogen and 2 μg of E.coli tRNA (Roche) as carrier. RNA was dissolved in 30 μl of DEPC-treated water. Proteins were precipitated from the phenol phase with 3 vol of ethanol. The pellet was washed extensively with ethanol to remove the phenol and dissolved in loading buffer for denaturing SDS–PAGE.
Analysis of RNA and protein
Northern hybridization and primer extension analysis were carried out using oligonucleotides 1 [5′-GATCACCTAGCGACTCTCTCCACC-3′; complementary to a region of the external transcribed spacer (5′-ETS) of 35S pre-rRNA directly upstream from A0; cf. Figure 1A] and SD14 (5′-AACCGCTAAGGATTGCGGAC-3′; complementary to the U3 snoRNA) as described previously (23). Western analysis was performed using PAP antibodies (Sigma) recognizing the ProtA tag.
RESULTS
The 80S SSU processome has been reported to represent a very early, if not the first, stage in the ribosome biogenesis pathway in S.cerevisiae. Formation of this particle, which appears to correspond to the ‘terminal knobs’ seen in electron micrographs of actively transcribed rDNA units, depends absolutely upon U3 (25), a box C/D snoRNP essential for the early pre-rRNA cleavages at sites at A0–A2. In addition to the known core proteins of the U3 snoRNP, the 80S particle contained 17 novel proteins (Utp1-17) as well as Rrp5p, suggesting that association of the latter might be U3-dependent. On the other hand, Rrp5p was also detected in a large complex with RNA polymerase I (34), indicating that the protein associates with the pre-rRNA transcript prior to U3. Thus, like Pwp2p (28), it might be required for subsequent association of U3 with 35S pre-rRNA.
Incorporation of U3 snoRNP and Rrp5p into the pre-ribosome is mutually independent
To investigate whether Rrp5p and U3 snoRNP are mutually dependent for incorporation into the pre-ribosome, we decided to use a co-immunoprecipitation approach employing yeast strains in which production of one or the other factor can be conditionally inhibited.
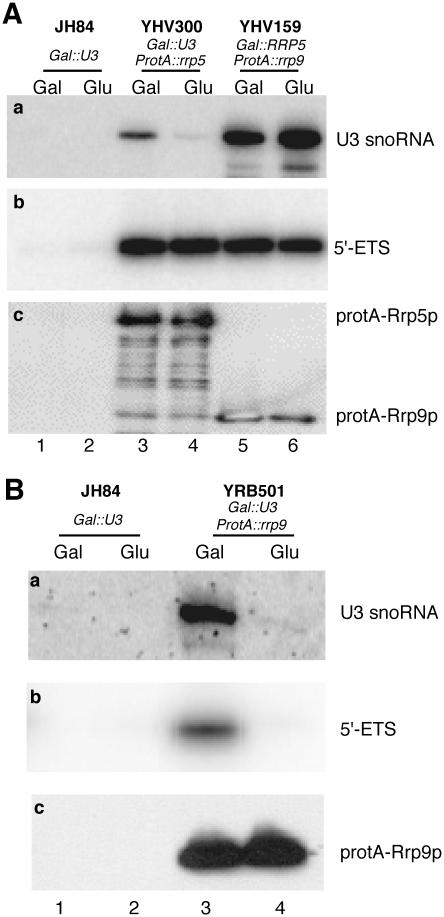
Strain YHV300 contains a single U3 snoRNA gene under control of the GAL promoter as well as a genomic ProtA-rrp5 allele (Table 1) allowing production of U3 to be shut down by shifting the cells from a galactose- to a glucose-based medium. Extracts were prepared from equal amounts of YHV300 cells harvested immediately before and 24 h after the shift, and treated with IgG–agarose beads. Bound material was eluted from the beads with GTC solution, after which protein and RNA were separated by phenol extraction and analyzed by western and northern blotting, respectively. Control extracts prepared from galactose- and glucose-grown JH84 (GAL-U3, RRP5) cells were analyzed in the same manner.
As shown in Figure 2A, similar amounts of tagged Rrp5p were recovered from both the galactose- and glucose-grown YHV300 cells (panel c, lanes 3 and 4), whereas no IgG-bound Rrp5p could be recovered from the control cells (lanes 1 and 2). Northern analysis showed that, in accordance with previous data (25), U3 snoRNA co-precipitated with Rrp5p in extracts from YHV300 cells harvested before the shift (panel a, lane 3). Virtually no U3 snoRNA was recovered by precipitation of ProtA-Rrp5p from extracts of cells prepared after the shift demonstrating the effective shutdown of U3 synthesis (panel a, lane 4).
Figure 2.
Co-immunoprecipitation analysis of extracts prepared from (A) JH84 (Gal::U3), YHV300 (GAL::U3 ProtA::rrp5) and YHV159 (GAL::rrp5 pHIS3-ProtA::rrp9) and (B) JH84 (Gal::U3) and YRB501 (GAL::U3 pHIS3-ProtA::rrp9) cells immediately before (Gal) and 24 h after (Glu) a shift from galactose- to glucose-based medium. (a) Northern analysis using a probe complementary to the U3 snoRNA. (b) Primer extension analysis using probe 1, complementary to the 5′-ETS. (c) Western blot analysis.
To assay for the presence of pre-rRNA in the co-precipitate, we performed reverse transcription assays using a primer complementary to a sequence located directly upstream from site A0 in the 5′-ETS of the precursor transcript (Figure 1A). Consequently, this primer will only detect 35S pre-rRNA that has not yet undergone any of the initial processing steps removing the 5′-ETS (cf. Figure 1B). Signals of identical strength are clearly visible in both lane 3 and lane 4 of Figure 2A (panel b), demonstrating that depletion of U3 does not affect the ability of Rrp5p to co-precipitate 35S pre-rRNA. Thus, incorporation of Rrp5p into the pre-ribosome does not appear to depend upon the presence of U3.
It has recently been reported (28) that the association of U3 snoRNP with the pre-rRNA, in addition to base-pairing interactions, also requires Pwp2p, one of the novel proteins detected in the 80S SSU processome (Utp1p). In order to see whether Rrp5p might play a similar role in U3 association, we introduced a plasmid encoding ProtA-Rrp9p, a known core component of the U3 snoRNP (30), into strain YHV159, which contains a genomic GAL-rrp5 gene (Table 1). Extracts prepared from galactose- and glucose-grown cells were treated with IgG–agarose beads after which the bound material was analyzed. As shown by a comparison of lanes 5 and 6 in Figure 2A, the depletion of Rrp5p resulting from this shift has no effect on the ability of Rrp9p to co-precipitate 35S pre-rRNA as measured by reverse transcription. We, therefore, conclude that incorporation of Rrp5p and U3 snoRNP into the pre-ribosome can occur fully independently.
To validate this conclusion, we performed a co-precipitation experiment with strain YRB501 (GAL-U3) transformed with the ProtA-Rrp9p plasmid. The results, depicted in Figure 2B, show that in these cells depletion of U3 snoRNA, by shifting the cells to a glucose-based medium, does result in a complete loss of the 35S pre-rRNA signal (panel b, cf. lane 4 with lane 3). Thus, co-precipitation of 35S pre-rRNA by Rrp9p only occurs when the protein is part of the snoRNP particle and, therefore, is a reliable indicator of U3/35S association.
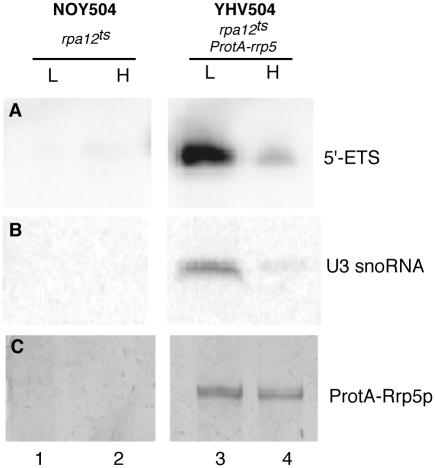
Although the results described above show that incorporation of U3 snoRNP and Rrp5p into the pre-ribosome are mutually independent, they do not exclude a direct interaction between the two factors. To further investigate this aspect, we constructed strain YHV504, in which the wild-type RRP5 gene of strain NOY504 was replaced by a ProtA::rrp5 allele. NOY504 lacks the RPA12 gene, which encodes a subunit of RNA polymerase I that is dispensable at 30°C but becomes essential at 37°C (35). Consequently, in YHV504 cells, transcription of 35S pre-rRNA can be conditionally inhibited by a temperature shift, allowing us to determine whether co-precipitation of U3 by ProtA-Rrp5p depends on the presence of the pre-rRNA.
Although, as shown in Figure 3, very similar amounts of ProtA-Rrp5p were recovered from extracts prepared before and after the shift, inhibiting polymerase I transcription strongly reduced the amount of co-precipitated U3 snoRNA, in concert with the reduction in 35S pre-rRNA detected by reverse transcription (cf. lane 4 with lane 3). Thus, co-precipitation of U3 by ProtA-Rrp5p depends upon the presence of the pre-rRNA and there is no evidence for a direct interaction between the two factors.
Figure 3.
Co-immunoprecipitation analysis of extracts prepared from NOY504 (rpa12::LEU2) and YHV504 (rpa12::LEU2 ProtA::rrp5) cells immediately before (L) and 6 h after (H) a shift from 30 to 37°C. (A) Primer extension analysis using probe 1. (B) Northern analysis using a probe complementary to the U3 snoRNA. (C) Silver-stained SDS polyacrylamide gel.
Rrp5p but not U3 snoRNP is essential for incorporation of Rok1p into the pre-ribosome
The putative ATP-dependent RNA helicase Rok1p has multiple genetic links with Rrp5p. First, like RRP5, a mutation in ROK1 is synthetically lethal with deletion of the gene encoding the RNA component of the snR10 snoRNP (24). Second, the overexpression of Rok1p suppresses several temperature-sensitive mutations in the TPR domain of Rrp5p that affect processing at sites A0–A2 (20,21), while Rok1p itself is essential for these cleavages (24). The protein has not been detected in the 80S SSU processome (25) but was found in 90S pre-ribosomes (11), though no components of the latter particle were immunopurified using a TAP-tagged version of Rok1p (36). Nevertheless, preliminary experiments in our laboratory showed co-precipitation of U3 snoRNA with ProtA-Rok1p (cf. Figure 4) which encouraged us to investigate whether incorporation of this protein into the pre-ribosome depends upon U3 and/or Rrp5p.
Figure 4.
Co-immunoprecipitation analysis of extracts prepared from JH84 (Gal::U3), YHV400 (GAL::U3 rok1::HIS3 + pTRP1-ProtA::rok1) and YHV200 (GAL::rrp5 rok1::HIS3 + pTRP1-ProtA::rok1) cells immediately before (Gal) and 24 h after (Glu) a shift from galactose- to glucose-based medium. (A) Northern analysis using a probe complementary to the U3 snoRNA. (B) Primer extension analysis using probe 1, complementary to the 5′-ETS. (C) Western blot analysis.
To that end, we constructed two additional strains (Table 1). YHV400 contains a single GAL::U3 allele as well as a deletion of ROK1, but remains viable due to a plasmid-encoded ProtA::rok1 gene. YHV200 is the result of a cross between YJV184 (ProtA::rok1) and YJV153 (GAL::rrp5).
YHV400 and YHV200 cultures were subjected to a 24 h shift from galactose- to glucose-based medium to deplete U3 or Rrp5p, respectively, and immunoprecipitation was performed as described above. As shown in Figure 4, ProtA-Rok1p is able to co-precipitate U3, as well as 35S pre-rRNA, from extracts of galactose-grown YHV400 and YHV200 cells (panels A and B, lanes 3 and 5). After shifting YHV400 (GAL::U3) cells to glucose, the U3 signal is lost, demonstrating effective inhibition of U3 snoRNA synthesis, but the primer extension signal remains unaffected (lane 4). Thus, lack of U3 does not prevent the association of Rok1p with the pre-rRNA. In contrast, ProtA-Rok1p is unable to co-precipitate either U3 or pre-rRNA from extracts prepared from glucose-grown YHV200 (GAL::rrp5) cells. We conclude that under these conditions Rok1p no longer associates with the pre-ribosome and, therefore, that incorporation of Rok1p into the pre-ribosome depends upon the presence of Rrp5p in the particle.
DISCUSSION
Although the first reports on eukaryotic pre-ribosomal particles date from the 1970s (37,38), techniques required for purification of these low-abundant particles and analysis of their protein composition were developed only recently (39). Nevertheless, already a sizeable number of such proteomic studies have made it clear that these particles represent consecutive stages in the ribosomal maturation pathway that arise by the ordered association and dissociation of the large number of trans-acting factors long known to be essential for ribosome biogenesis in eukaryotic cells (7,8,17,40). At present, our picture of the dynamics of the ribosomal processing/assembly machinery, however, is still at relatively low resolution. Much work remains to be performed to establish the precise order in which the individual factors associate and dissociate as well as their interrelationships governing this process.
In this paper, we have used an immunological approach to study the possible mutual dependence between three of these trans-acting factors of yeast: U3 snoRNP, Rrp5p and Rok1p, which were known to be present at an early stage of ribosome biogenesis (11,25), to be required for the initial processing events at sites A0–A2 (18,21,26) and, as far as the last two are concerned, to be genetically linked (18,21,24). Although previous experiments failed to detect co-precipitation of pre-ribosomal components with TAP-tagged versions of either Rok1p or Rrp5p (36,41), we identified both U3 and 35S pre-rRNA in the IgG-bound fraction prepared from cell extracts that express ProtA-Rrp5p or ProtA-Rok1p (Figures 2 and 4).
Using yeast strains that conditionally express either U3 snoRNA or Rrp5p, we show that association of the latter protein with the 35S pre-rRNA is not affected by genetic depletion of the snoRNA (Figure 2A), even though Rrp5p is found in the 80S SSU processome, assembly of which depends absolutely upon the U3 snoRNP (25). Conversely, association of the U3 snoRNP with the 35S precursor is not diminished when cells are depleted of Rrp5p (Figure 2A), indicating that, in contrast to many, if not all, of the other components of the 80S processome (25,28,42), Rrp5p is not essential for assembly of this particle. Co-precipitation of U3 with Rrp5p becomes impossible when RNA polymerase I transcription in blocked (Figure 3), which demonstrates that the two trans-acting factors do not stably interact prior to their association with the pre-RNA. This does not exclude the possibility, however, that U3 and Rrp5p are in contact after their incorporation into the pre-ribosome.
Rok1p was not identified in the 80S processome (25) but was found to be present in the 90S pre-ribosome (11), indicating that it is incorporated somewhat later than U3 and Rrp5p. As shown in Figure 4, co-precipitation of 35S pre-rRNA with ProtA-Rok1p is unaffected in cells depleted of U3 snoRNA, indicating that association of Rok1p with the pre-rRNA can occur independently of U3. Furthermore, the absence of Rok1p from the 80S processome (25) indicates that association of U3 with 35S pre-rRNA does not require Rok1p. This conclusion is further supported by our finding that U3 can associate in the absence of Rrp5p and, thus, according to the data presented in this paper, failure of Rok1p to be incorporated into the pre-ribosome. Depletion of Rrp5p, however, did completely abolish co-precipitation of both U3 and the pre-rRNA by tagged Rok1p. We conclude that pre-ribosomes lacking Rrp5p are unable to acquire Rok1p, whereas failure to assemble the 80S processome due to lack of U3 does not affect the association of Rok1p with an Rrp5p/35S pre-rRNA complex.
The simplest explanation for this finding would be a direct interaction between Rok1p and Rrp5p involving the TPR motifs of the latter, which would also explain why overexpression of Rok1p can suppress the temperature-sensitive phenotype of mutations in the TPR domain. However, we have been unable to detect direct interaction between the two proteins using two-hybrid analysis with either intact Rrp5p or its TPR domain (H. R. Vos, unpublished data), and in fact no biochemical interactions of Rrp5p with Rok1p, apart from their simultaneous presence in 90S pre-ribosomes, are reported in the literature (SGD database: http://www.yeastgenome.org/). These observations also make it unlikely that Rrp5p and Rok1p associate with the pre-rRNA as a heterodimer. In fact, literature data clearly indicate that Rrp5p can recognize the precursor in the absence of Rok1p, because depletion of the latter protein blocks cleavage at site A0, A1 and A2, but not at site A3 (24), a cleavage for which Rrp5p is essential (18,21). Therefore, it seems that association of Rrp5p with the 35S precursor is required to unmask the binding site for Rok1p, either on the protein or on the pre-rRNA. Furthermore, our data suggest that the requirement of Rrp5p for pre-rRNA processing at site A0–A2 may be due to its essential role in the association of Rok1p and, thus, be indirect.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Jaap Venema for construction of yeast strains YJV182 and YJV184 and Drs Nomura and Samarsky for providing strains NOY504 and JH84, respectively.
REFERENCES
- 1.Nomura M. and Held,W.A. (1974) In Nomura,M., Tissières,A. and Lengyel,P. (eds), Ribosomes. Cold Spring Harbor Laboratories, Cold Spring Harbor NY, pp. 193–224. [Google Scholar]
- 2.Nierhaus K.H. and Dohme,F. (1974) Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc. Natl Acad. Sci. USA, 71, 4713–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maki J.A., Southworth,D.R. and Culver,G.M. (2003) Demonstration of the role of the DnaK chaperone system in assembly of 30S ribosomal subunits using a purified in vitro system. RNA, 9, 1418–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charollais J., Dreyfus,M. and Iost,I. (2004) CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res., 32, 2751–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charollais J., Pflieger,D., Vinh,J., Dreyfus,M. and Iost,I. (2003) The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol., 48, 1253–1265. [DOI] [PubMed] [Google Scholar]
- 6.Hage A.E. and Alix,J.H. (2004) Authentic precursors to ribosomal subunits accumulate in Escherichia coli in the absence of functional DnaK chaperone. Mol. Microbiol., 51, 189–201. [DOI] [PubMed] [Google Scholar]
- 7.Fromont-Racine M., Senger,B., Saveanu,C. and Fasiolo,F. (2004) Ribosome assembly in eukaryotes. Gene, 313, 17–42. [DOI] [PubMed] [Google Scholar]
- 8.Raué H.A. (2004) In Olson,M.O. (ed.), The Nucleolus. Kluwer Academic/Plenum Publishers, NY, pp. 199–222. [Google Scholar]
- 9.Moy T.I. and Silver,P.A. (1999) Nuclear export of the small ribosomal subunit requires the Ran-GTPase cycle and certain nucleoporins. Genes Dev., 13, 2118–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanrobays E., Gleizes,P.-E., Bousquet-Antonelli,C., Noaillac-Depeyre,J., Caizergues-Ferrer,M. and Gélugne,J.-P. (2001) Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J., 20, 4204–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schäfer T., Strauss,D., Petfalski,E., Tollervey,D. and Hurt,E. (2003) The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J., 22, 1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell P., Petfalski,E. and Tollervey,D. (1996) The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev., 10, 502–513. [DOI] [PubMed] [Google Scholar]
- 13.Geerlings T., Vos,J.C. and Raué,H.A. (2000) The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′→3′ exonucleases. RNA, 6, 1698–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faber A.W., van Dijk,M., Raué,H.A. and Vos,J.C. (2002) Ngl2p is a Ccr4p-like RNA nuclease essential for the final step in 3′-end processing of 5.8S rRNA in Saccharomyces cerevisiae. RNA, 8, 1095–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Senger B., Lafontaine,D.L.J., Graindorge,J.-S., Gadal,O., Camasses,A., Sanni,A., Garnier,J.-M., Breitenbach,M., Hurt,E. and Fasiolo,F. (2001) The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell, 8, 1363–1373. [DOI] [PubMed] [Google Scholar]
- 16.Nissan T.A., Bassler,J., Petfalski,E., Tollervey,D. and Hurt,E. (2002) 60S Pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J., 21, 5539–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fatica A. and Tollervey,D. (2002) Making ribosomes. Curr. Opin. Cell Biol., 14, 313–318. [DOI] [PubMed] [Google Scholar]
- 18.Venema J. and Tollervey,D. (1996) RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J., 15, 5701–5714. [PMC free article] [PubMed] [Google Scholar]
- 19.Oeffinger M., Dlakic,M. and Tollervey,D. (2004) A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev., 18, 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torchet C., Jacq,C. and Hermann-le Denmat,S. (1998) Two mutant forms of the S1/TPR-containing protein Rrp5p affect the 18S rRNA synthesis in Saccharomyces cerevisiae. RNA, 4, 1636–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eppens E.A., Rensen,S., Granneman,S., Raué,H.A. and Venema,J. (1999) The roles of Rrp5p in the synthesis of yeast 18S and 5.8S rRNA can be functionally and physically separated. RNA, 5, 779–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eppens E.A., Faber,A.W., Rondaij,M., Jahangir,R.S., van Hemert,S., Vos,J.C., Venema,J. and Raué,H.A. (2002) Deletions in the S1 domain of Rrp5p cause processing at a novel site in ITS1 of yeast pre-rRNA that depends on Rex4p. Nucleic Acids Res., 30, 4222–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vos H.R., Faber,A.W., De Gier,M.D., Vos,J.C. and Raué,H.A. (2004) Deletion of the three distal S1 motifs of yeast Rrp5p abolishes pre-rRNA processing at site A2 without reducing the production of functional 40S subunits. Eukaryotic Cell, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venema J., Bousquet-Antonelli,C., Gelugne,J.-P., Caizergues-Ferrer,M. and Tollervey,D. (1997) Rok1p is a putative RNA helicase required for rRNA processing. Mol. Cell. Biol., 17, 3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dragon F., Gallagher,J.E.G., Compagnone-Post,P.A., Mitchell,B.A., Porwancher,K.A., Wehner,K.A., Wormsley,S., Settlage,R.E., Shabanowitz,J., Osheim,Y. et al. (2002) A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature, 417, 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beltrame M. and Tollervey,D. (1995) Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J., 14, 4350–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma K. and Tollervey,D. (1999) Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell. Biol., 19, 6012–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dosil M. and Bustelo,X.R. (2004) Functional characterization of Pwp2, A WD family protein essential for the assembly of the 90S pre-ribosomal particle. J. Biol. Chem., 279, 37385–37397. [DOI] [PubMed] [Google Scholar]
- 29.Grandi P., Rybin,V., Baßler,J., Petfalski,E., Strauss,D., Marzioch,M., Schäfer,T., Kuster,B., Tschochner,H., Tollervey,D. et al. (2002) 90S Pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell, 10, 105–115. [DOI] [PubMed] [Google Scholar]
- 30.Venema J., Vos,H., Faber,A.W., van Venrooij,W.J. and Raué,H.A. (2000) Yeast Rrp9p is an evolutionarily conserved U3 snoRNP protein essential for the early pre-rRNA processing cleavages and requires box C for its association. RNA, 6, 1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gietz D., St Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Improved methods for high efficiency transformation of intact yeast cells. Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samarsky D.A. and Fournier,M.J. (1998) Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 3431–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berben G., Dumont,J., Gilliquet,V., Bolle,P.A. and Hilger,F. (1991) The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast, 7, 475–477. [DOI] [PubMed] [Google Scholar]
- 34.Fath S., Milkereit,P., Podtelejnikov,A.V., Bischler,N., Schultz,P., Bier,M., Mann,M. and Tschochner,H. (2000) Association of yeast RNA polymerase I with a nucleolar substructure active in rRNA synthesis and processing. J. Cell Biol., 149, 575–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nogi Y., Yano,R., Dodd,J., Carles,C. and Nomura,M. (1993) Gene RRN4 in Saccharomyces cerevisiae encodes the A12.2 subunit of RNA Polymerase I and is essential only at high temperatures. Mol. Cell. Biol., 13, 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gavin A.-C., Bösche,M., Krause,R., Grandi,P., Marzioch,M., Bauer,A., Schultz,J., Rick,J.M., Michon,A.-M., Cruciat,C.-M. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 414, 141–147. [DOI] [PubMed] [Google Scholar]
- 37.Kumar A. and Warner,J.R. (1972) Characterization of ribosomal precursor particles from HeLa cell nucleoli. J. Mol. Biol., 63, 233–246. [DOI] [PubMed] [Google Scholar]
- 38.Trapman J., Retèl,J. and Planta,R.J. (1975) Ribosomal precursor particles from yeast. Exp. Cell Res., 90, 95–104. [DOI] [PubMed] [Google Scholar]
- 39.Rigaut G., Shevchenko,A., Rutz,B., Wilm,M., Mann,M. and Séraphin,B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol., 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- 40.Warner J.R. (2001) Nascent ribosomes. Cell, 107, 133–136. [DOI] [PubMed] [Google Scholar]
- 41.Krogan N.J., Peng,W.-T., Cagney,G., Robinson,M.D., Haw,R., Zhong,G., Guo,X., Zhang,X., Canadien,V., Richards,D.P. et al. (2004) High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell, 13, 225–239. [DOI] [PubMed] [Google Scholar]
- 42.Wehner K.A., Gallagher,J.E.G. and Baserga,S.J. (2002) Components of an interdependent unit within the SSU processome regulate and mediate its activity. Mol. Cell. Biol., 22, 7258–7267. [DOI] [PMC free article] [PubMed] [Google Scholar]