Abstract
Functionally related homologues of known genes can be difficult to identify in divergent species. In this paper, we show how multi-character analysis can be used to elucidate the relationships among divergent members of gene superfamilies. We used probabilistic modelling in conjunction with protein structural predictions and gene-structure analyses on a whole-genome scale to find gene homologies that are missed by conventional similarity-search strategies and identified a variant gene superfamily in six species of malaria (Plasmodium interspersed repeats, pir). The superfamily includes rif in P.falciparum, vir in P.vivax, a novel family kir in P.knowlesi and the cir/bir/yir family in three rodent malarias. Our data indicate that this is the major multi-gene family in malaria parasites. Protein localization of products from pir members to the infected erythrocyte membrane in the rodent malaria parasite P.chabaudi, demonstrates phenotypic similarity to the products of pir in other malaria species. The results give critical insight into the evolutionary adaptation of malaria parasites to their host and provide important data for comparative immunology between malaria parasites obtained from laboratory models and their human counterparts.
INTRODUCTION
The study of host–pathogen interactions very often relies on laboratory model systems. This holds especially true for many parasitic diseases, for which the pathogen must be matched to suitable laboratory animals for experimental immunology or other studies that require experimental infections and manipulation of both host and parasite. Very often, the parasites suitable for such laboratory study are very closely related to the human disease-causing parasites, but are derived from a different isolate, strain or even species. The identification of the human parasite's functional homologues of genes, relevant to pathogenesis or other aspects of interest, in the laboratory model parasite is essential for the meaningful interpretation of experimental results in terms of human disease. Clues to the relationships among genes can be found by looking at common links of sequence motif modules, gene and protein structure, and patterns of conservation within gene or protein sequences (1). These characteristics of genes and their products can give an insight into evolutionary relationships and perhaps functional relationships. Although functions may have changed for genes derived from a common ancestor, they may still point to the presence of catalytic pathways or their components. A sizeable proportion of genes of related organisms are presumed to be derived from genetic starting material of common ancestry. Sorting out the relationships among genes from closely and distantly related genomes will help towards elucidating enzyme pathways and structural components of cells.
In this paper, we describe the use of multi-character analyses to examine the relationships among multi-gene families in the parasites of the genus Plasmodium that are the causative agents of malaria (2). Using whole-genome analyses, we systematically examined sequence motif modules, gene and protein structure, and patterns of conservation within gene or protein sequences to build up a picture of relationships among some of the major multi-gene families found in the genomes of human, monkey and rodent malarias. The discovery of several multi-gene families, implicated or potentially implicated in host immune system interactions in various species of human, monkey and rodent malaria parasites, including var and rif/stevor in P.falciparum (3,4), the most important malaria parasite of humans, vir in P.vivax (5), sicavar in P.knowlesi (6) and the cir/bir/yir family in rodent malarias (7) has raised important questions about the evolutionary origin of multi-gene families in the genus Plasmodium. The human malarias will only infect man and a small number of monkeys and apes, which limits the in vivo investigations of these parasites and necessitates the use of animal models. Any relationship, or lack thereof, in the structures, functions and evolution of these families would have important implications for experimental and comparative immunological studies. Our understanding of the rate of generation of diversity of variant genes would be greatly enhanced by the ability to assess the rate of evolution of the gene families with respect to speciation within the genus, and the related phenomenon of host-switching. It is also important to assess the impact of the potential differences in immune response of the different host species on the evolution of variant multi-gene families.
The analyses that we present not only provide important data for comparative immunology between laboratory model malarias and their human counterparts, but, also allow us to describe a scheme of analyses that will prove useful to other researchers in the identification of functional homologues of other genes in other organisms.
MATERIALS AND METHODS
Overview
We carried out comparative genomic studies on multi-gene families in six species of Plasmodium: P.falciparum (human host), P.vivax (human host), P.knowlesi (monkey host), P.berghei (rodent host), P.yoelii (rodent host), and P.chabaudi (rodent host). Whole-genome analyses were carried out in all instances, except with P.vivax, for which only partial information was available in the form of EST, a YAC clone, and GST. Sequence data from the P.yoelii genome (strain 17X NL, clone 1.1) was obtained from The Institute for Genomic Research website (www.tigr.org). Sequences of the P.vivax, IVD10 YAC clone, P.berghei of clone 15cy1 of ANKA strain, P.chabaudi AS strain and P.knowlesi H strain were produced by the Pathogen Sequencing Group at the Sanger Centre and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/. The P.falciparum genome data and P.vivax GST were obtained from PlasmoDB (8). The accession numbers for genome sequences are AL844501–AL844509, AE001362.2, AE014185–AE014187 and AE014188 for the P.falciparum genome, AABL00000000 (project accession number) for P.yoelii. The remaining rodent malaria genomes are currently submitted for publication; sequence data are available from ftp://ftp.sanger.ac.uk/pub/pathogens/ and annotated genes can be browsed at GeneDB http://www.genedb.org/. Databases of all genomes were created locally, and all analyses were carried out on local computers using whole-genome data. All probability models and motifs were checked against the Non-redundant database of protein sequences (Non-redundant GenBank, DDBJ and EMBL CDS translations + PDB + SwissProt + PIR), referred to as NR from here on, available from the National Center for Biotechnology Information (NCBI) (ftp://ftp.ncbi.nlm.nih.gov/blast/db/). NR was downloaded and used as a local database.
Probabilistic models and gene annotations
The gene families that we examined were rif/stevor (P.falciparum) (4), var (P.falciparum) (9), vir (P.vivax) (5), sicavar (P.knowlesi) (6) kir (novel finding) (P.knowlesi), cir (P.chabaudi) (7), bir (P.berghei) (7) and yir (P.yoelii) (7). rif and stevor were treated as one family since, from a probabilistic model view, STEVOR and RIFINS are identical. This is reflected in the Pfam model of RIFIN/STEVOR, submitted by Bateman and Lawson (Accession number PF02009), which does not discriminate between the two protein families. All references to rif and its products from here on in refer to the rif/stevor family or respective protein products. All gene families were annotated on the local genome databases using the same methodology to ensure consistency. Annotation was carried out using sequence similarity search (FASTA) (10) in the first instance, followed by construction of hidden Markov models (HMMs) and HMM searches (11), and annotation using the genewisedb algorithm of the Wise 2 package (12) with gene-family specific HMMs. In detail, conceptual translations of exons from gene-family sequences were aligned by the Dialign program (13). The alignment was used for generating a HMM using the HMMER 2.2 software package (11), building both global and fragment HMMs. All HMMs are available on request from c.janssen@bio.gla.ac.uk. A composite CIR/YIR HMM was constructed from an alignment of translations of second and third exons from 40 yir and 33 cir sequences. Two RIFIN HMMs were built; one from 9 EST sequences and one from 24 sequences taken from the 3D7 P.falciparum genome. In addition, the RIFIN/STEVOR HMM available from Pfam (14), was also used. The VIR HMM was made from 29 randomly chosen divergent sequences. Both PfEMP1 (erythrocyte membrane protein 1 of P.falciparum) (var products) and SICAVAR HMMs were built from 50 sequences each, taken from the 3D7 and P.knowlesi H strain genomes, respectively. These HMMs were not only used in the annotation of the genomes from which they were derived, but also used for gene finding in all other genomes with the HMMER 2.2 software package.
Motif finding
Conserved amino acid motifs were identified in the putative protein sequences of each gene family using the MEME and MAST programs (15–17). Motifs were found using the ‘zero or one expected occurrence of each motif per sequence’ model of MEME, with a maximum set width ranging from 10 to 25 amino acids, and a maximum of seven motifs per sequence. Only var gene translations were also searched with the ‘two-component mixture’ model. When relationships between gene families were evident, based on the sequence and structure similarities, sequences from different families were combined and used as a training set for finding motifs. The malaria genomes were searched with the motifs derived from all gene families using the MAST program. In brief, the MAST program performs a position specific probability matrix (PSPM) search of the target database with the query motifs. Sets of motifs identified within a gene family were treated as modules, which are defined as sets of motifs with distinct connectivity relationships. Sensitivity and robustness of the motif modules as unique identifiers of each gene family's predicted protein products were tested by searching all open reading frames (ORFs) (stop to stop codon, minimum size 50 amino acids) that were derived from all the available local malaria genome databases as well as all protein sequences from the NR protein database while controlling for compositional bias (built in option of MAST). To further test the robustness of the PSPM motifs, the most statistically significant PSPMs were translated to HMMs and used to search the NR protein database with HMMER 2.2.
Position-specific variability in amino acid sequence and the overall pattern of conservation within whole protein sequences among members of the gene families was analysed using the Plotcon program in the EMBOSS package (18). Overall (average) variation among members of the gene family was measured using the BLOSUM62 matrix (19) as implemented by the Infoalign program from the EMBOSS package. Consensus secondary structures of members of the protein families of each species were generated by the jnet (20) and DSC (21) algorithms.
Similarities among the intron sequences found in all members of the rif, vir, cir, yir and kir families were examined by motif analysis using the MEME and MAST system, as well as phylogenetic analysis of alignments of intron sequences generated with the Dialign program. Trees were generated using the maximum likelihood method implemented by the dnaml algorithm (22) and the DNA maximum parsimony method implemented by the dnapars algorithm of the Phylip package (23).
Phylogenetic analyses
A total of 157 amino acid sequences of members from rif, vir, kir, yir and cir were aligned using clustalW (24) and Dialign. The best global alignment (clustalW) was chosen for phylogenetic analysis. Poorly aligned positions and divergent regions of the alignment were eliminated and only conserved regions were chosen (185 amino acids) using the program Gblocks (25). Thus, the variant domains of the proteins were eliminated from the analyses. Phylogenetic analysis was performed on the dataset using the maximum likelihood method applied to pairwise sequence distances calculated using quartet puzzling, which automatically assigns estimations of support to each internal branch (26,27). Trees were also generated using the maximum likelihood method implemented by the proml algorithm and the maximum parsimony method implemented by the protpars algorithm of the Phylip package (23). Trees were drawn using the ATV (28) and TreeView (29) programs.
Protein detection and localization
Parasite extracts and erythrocyte ghosts were obtained by saponin lysis (2 volumes of 0.15% saponin/phosphate-buffered saline (PBS) of late trophozoite infected blood of P. chabaudi and subsequent centrifugation. Late trophozoites were extracted on ice for 30 min with PBS containing 1% Triton X-100 and protease inhibitor cocktail (Sigma). After the extraction, the sample was centrifuged at 7500 g for 10 min, and the supernatant discarded. The pellet was re-suspended in 2% SDS/PBS and protease inhibitors and incubated for 45 min at room temperature with frequent vortexing. The Triton X-100 insoluble/SDS soluble fraction obtained from the parasite/ ghost pellet was electrophoresed on a 5–20% gradient SDS–PAGE gel and blotted onto nitrocellulose. The blot was probed with anti-CIR peptide antibodies (described in Supplementary Material) and pre-immune control serum using standardized methods. Briefly, the membrane was blocked for 1 h at room temperature in 1× TBS, 0.1% Tween-20, and 5% w/v non-fat dry milk. After washing three times in TBS, the membrane was incubated with primary antibody in 1× TBS, 0.1% Tween-20 and 5% BSA at 4°C overnight. Subsequent to the three washes in PBS, the membrane was incubated with secondary antibody that was conjugated to Horseradish peroxidase (HRP) (Cell Signalling Tech.) for 1 h according to the manufacturer's instructions. The membrane was washed three times in TBS and bound antibodies were detected using the Phototope_HRP detection system (Cell Signalling Tech.) according to the manufacturer's instructions.
CIR protein was localized in the infected erythrocytes of P.chabaudi using indirect fluorescent antibody techniques (IFAT), as described in the Supplementary Material.
RESULTS
Overview
Until now, little data has been available on the overall family characteristics of cir and its homologues in other rodent malarias, kir, sicavar and vir. The annotation of these genes in the genome databases and the subsequent generation of probabilistic models to statistically describe the families generated the first whole-genome overview of their characteristics. Copy number estimates of genes per genome are as follows: about 160–200 genes for cir and bir, ∼800 yir, 35 kir (plus at least 42 pseudogenes or gene fragments), 200–300 vir, and ∼300 sicavar. Copy numbers are estimates since none of the genomes have been taken to completion, but currently each give about five times the coverage. Rif and var have been well described previously, with 175 and ∼60 copies per genome, respectively. All gene families can be predominantly found in sub-telomeric regions, as indicated by the proximity of sub-telomeric repeats (30,31) to those genes identified on larger contigs. When taken as individual families, the rodent malaria genes, rif, kir and vir all have ∼30% sequence identity at the amino acid level as revealed by the alignment of the conceptual translations of all genes available from each family. However, the range of amino acid-level sequence identity within each family is about 6–90%.
Gene family annotations and descriptions
Genewise analysis of the rodent malaria genomes using the CIR/YIR HMM confirmed the general conservation of the intron splice junction sequences that we have previously determined experimentally (7). Second introns of average length of 99 nt, were found to contain several conserved sequence motifs (Supplementary Table 1), all or some of which can be found in each family of cir/yir/bir genes. Average sequence identity of second introns is 72%. First introns, of average length of 135 nt, were also shown to have some sequence conservation, average sequence identity being 67%. All sequences contained some or all of the conserved sequence motifs shown in Supplementary Table 1. Overall, more diversity was found in sequences from the first intron compared with second introns.
Sequence analysis revealed that the rif single intron sequence is most probably derived from a sequence sharing ancestry with the rodent malaria cir/bir/yir second intron sequence. Four sequence motifs conserved between most rodent malaria cir-homologue and rif introns could be identified using neural network analysis. Most rif introns share at least some of the motifs with the rodent malaria sequences, whereas at least 40 contain all four. Motif consensus sequences, derived from a best-fit annotation of the training set with the PSPM, are as follows: CATATATATGCGACC, TTTTATATTAGTAT, ACATCGTACGCTTCT and AAAAATTATTTTCAT. Tree-reconstruction analysis of an alignment of 503 rif and cir/yir second introns shows 35 rif introns as more similar to the rodent malaria introns than those from other rifs (see Supplementary Material). No significant similarity could be found between rodent malaria cir-homologue first introns, or kir and vir second introns, and rif introns.
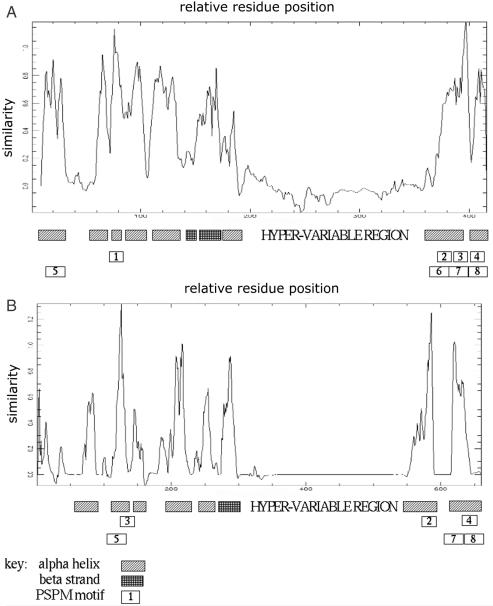
All rodent malaria CIR/BIR/YIR sequences are predicted to have a trans-membrane domain, spanning ∼29 amino acids, coded at the end of the second exon. Secondary structure prediction indicated that CIR/BIR/YIR proteins consist of a string of fairly evenly distributed alpha helices (numbering 7–8), averaging 12–20 amino acids in length, separated by coiled–coil regions (Figure 1). The general sequence conservation and variation pattern in the protein family alignment is shown in Figure 1: a highly variant region is flanked by two conserved regions.
Figure 1.
Plots of sequence conservation in alignments of (A) 276 rif amino acid sequences and (B) 1016 cir, yir and bir amino acid sequences, measured with the PAM250 scoring matrix, using a window size of four amino acids. The hypervariable regions are artificially long due to extensive gapping in the alignments. The predicted conserved secondary structure is given beneath each conservation plot (see key). Sequence motifs conserved among rifins and rodent malaria cir protein homologues, identified using neural network analysis (see Table 1), are shown in open boxes containing motif numbers.
Both vir and kir share the characteristics detailed for the rodent malaria cir/bir/yir families above, with a few notable exceptions. Conservation of intron sequence is less evident in both vir and kir sequences. Vir and kir introns are more variant, containing variable stretches of low complexity AT repeat sequences, and do not have the same intron motif patterns as the other gene families. The predicted secondary structure for vir also varies slightly from the other families, being more beta-strand rich: Some of the structured regions upstream of the hypervariable region are predicted to be beta-stranded in VIR.
RIFIN family members share the conserved predicted secondary structure of CIR/BIR/YIR sequences. Further, predicted secondary structure correlates well with the intra-family sequence conservation pattern (see Figure 1).
Comparative genomics
Our report on characterization of sicavar limits itself to the comparative genomics using probabilistic models and motif matrices, since other details are to be published elsewhere by the Sanger Institute Pathogen Sequencing Unit when reporting on the P.knowlesi genome. Comparative genomic sequence searching relied on two main methods: HMM searches and searches with PSPMs of motifs identified by MEME. The search for related sequences in other genomes using the above probabilistic models of sicavar genes and conceptual translations did not result in the identification of similar genes in any of the other Plasmodium genomes. In all seven SICAVAR motifs were used in the search, with statistical value ranging from llr = 1636 and E-value = 1.9e−478 (motif 1, best) to llr = 1226 and E-value = 1.4e−283 (motif 7, weakest). llr is the log likelihood ratio, which is the logarithm of the ratio of the probability of the occurrences of the motif given the motif model (likelihood given the motif) versus the probability given the background model (training-set of 43 sequences). HMM searches using both a fragment and a global hmm also failed to identify SICAVAR in any other Plasmodium genome.
Comparative genomics of the two most-studied and best characterized gene families in our dataset, var and rif did yield very different results. HMMs of the conserved second exon of var, global var HMMs, and var fragment HMMs, both from DNA and from conceptual translations, all failed to detect any significantly similar sequences in any of the other Plasmodium genomes. Further, none of the 10 PSPM motifs generated from conserved domains of var and PfEMP1, including the duffy-binding like domains and the conserved second exon, detected similar motif-modules of three or more motifs connected within any ORFs in any of the other genomes. Rif, on the other hand, was identified by the CIR/YIR global HMM when searched against the P.falciparum genome, although at statistically marginal significance only (e = 5 × 10−3). However, key conserved amino acid motifs were common to the CIR/YIR/BIR family and predicted products of rif. A distribution of motifs common to CIR and RIF predicted proteins can be seen in Figure 1. These motifs are best described as PSPMs, available in the Supplementary Material. Probability values for the motifs common to both CIRs and RIFINs are as follows: (motif 2) width = 12, sites = 192, llr = 2999, E-value = 5.6e−561, (motif 3) width = 11, sites = 177, llr = 3668, E-value = 7.1e−922, (motif 4) width = 12, sites = 135, llr = 2737, E-value = 1.3e−616, (motif 5) width = 10, sites = 136, llr = 2197, E-value = 5.2e−407, (motif 7) width = 10, sites = 143, llr = 2447, E-value = 1.7e−497, (motif 8) width = 10, sites = 198, llr = 3592, E-value = 3.7e−856. llr is the log likelihood ratio, which is the logarithm of the ratio of the probability of the occurrences of the motif given the motif model (likelihood given the motif) versus the probability given the background model. The training set consisted of 205 sequences; equal numbers of RIFIN and CIR, all balanced for equal intra-family variability. These motifs are unique to cir- and rif- homologous gene products, and do not occur in any other ORFs found in Plasmodium genomes, nor in other protein sequences of the public NR. MAST searches of NR, taking into account any compositional bias of sequences, with two PSPM sets of three motifs taken as modules gave following results: (a) module 1 (motifs 2, 3 and 4 above), E-value range from 4.4e−23 (rif) to 0.001 (yir): 369 hits in E-value range, consisting of rif, yir, bir, cir only, (b) module 2 (motifs 5, 7 and 8 above), E-value range from 1.2e−18 (yir) to 0.001 (yir): 513 hits in E-value range, consisting of rif, stevor, cir, yir only. The transformation of the two strongest cir/rif motifs (motifs 3 and 2 above) from PSPMs to HMMs, as well the construction of a composite fragment HMM from module 1 above, maintained robustness when searched against NR (using HMMER 2.2), but lost sensitivity. Nonetheless, the fragment HMM of module 1 hit 513 sequences within the E-value cut-off 10, all of which are stevor, rif, yir, bir or cir. The individual motif HMMs from motifs 3 and 2 hit 24 and 89 sequences respectively, all stevor, rif, yir, bir or cir. Further motifs are supplied in the Supplementary Material, including motifs that are conserved in VIR and KIR.
Phylogenetic analyses
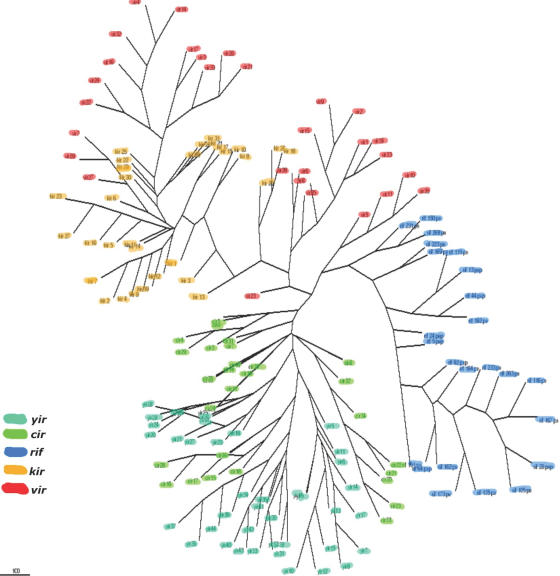
Tree reconstruction from the conserved regions (185 amino acids) of the alignment of 157 amino acid sequences of members from RIFIN, VIR, KIR, YIR and CIR sequences shows distinct clustering of sequences into three main similarity groups. The rodent malaria CIR-related sequences cluster together closely, as do KIR and VIR sequences, forming a separate group, whereas RIFINS, which form their own group, appear more divergent (see Figure 2). The same groupings were observed whether trees were built using the maximum likelihood methods or maximum parsimony (see the Supplementary Material for maximum likelihood trees).
Figure 2.
A total of 157 amino acid sequences of superfamily members from P.falciparum, P.vivax, P.knowlesi, P.yoelii and P.chabaudi were aligned using ClustalW. Poorly aligned positions and divergent regions of the alignment were eliminated (see text), and a tree generated using the maximum parsimony method implemented by the protpars algorithm of the Phylip package (23).
Protein detection and localization
The membrane fraction of late trophozoite P.chabaudi parasites and erythrocyte ghosts was probed for the CIR proteins using Western blotting techniques. Probing the blot of the Triton X-100 insoluble/SDS soluble protein fraction with anti-CIR peptide antibodies led to the detection of a single band predicted to be about 32 kDa (Supplementary Figure 4). Control sera failed to label any proteins on the blot.
Localization of CIR protein in the parasite and infected erythrocyte using anti-CIR peptide antibodies in immunofluorescence assays revealed that CIR proteins are exported from the parasite into the erythrocyte and appear to localize with the outer cell membrane of the erythrocyte (see Supplementary Material).
DISCUSSION
Although animal malarias have been used as experimental models for the human disease for a long time, few links regarding molecular mechanisms responsible for pathogenesis or immune system interactions have been made between Plasmodium species that exclusively infect animals or humans, until now. One of the most important factors in malaria parasite–host interactions, the phenomenon of antigenic variation (32), was first described in animal malarias. Although the first indication that asexual blood stage malaria parasites could undergo antigenic variation was the work of Cox (33) with P.berghei, the formative work of Brown and Brown (34) with the simian malaria P.knowlesi showed that repeated antigenic variation could occur during the course of a chronic infection. Subsequently antigenic variation was shown to occur in two further monkey malarias, P.cynomolgi bastianelli (35) and P.fragile (36), in the murine malaria P.chabaudi AS strain (37) and in P.falciparum in the squirrel monkey (38) and in vitro (39). The variant antigen in P.falciparum is thought to be the erythrocyte membrane protein 1 (PfEMP1) (40) and the encoding genes belong to a diverse family called var genes (3). The surface variant antigen of P.knowlesi has been found to be encoded by another multigene family, the sicavar gene family (6). However, recently several other multi-gene families linked to antigenic variation were described in the human malarias P.vivax (the vir gene family) (5), and P.falciparum (rif and stevor) (4), and the vir-related gene families in the rodent malarias P.chabaudi, P.berghei, and P.yoelii (7). The discoveries of these gene families have raised important questions about the relationships of gene families coding for proteins that are putatively involved in antigenic variation, found in different species of Plasmodium. Any relationships, or lack thereof, in the structures, functions and evolution of these families would have important implications in comparative immunological studies.
The initial hypothesis that we set out to test was that, unless previously shown otherwise, the gene families under study do not share common ancestry and have evolved independently. Further, we wanted to test whether evidence of genes, either present or past (pseudo-genes) that are homologous to gene families found in each species, could be found in the other species. Our whole-genome analysis of cir, its homologues in other rodent malarias, kir, sicavar, var, rif/stevor and vir has revealed that the major gene families of Plasmodium species can be categorized into similarity groups. Based on copy number, intra-family sequence conservation, genome location, expression pattern, and gene size, three main groups emerge: one group comprised of rif/stevor, vir, cir, bir, kir and yir, whereas sicavar and var comprise separate groups each. Our test for homology was based on the presence or absence of unique sequence motifs, in conjunction with similarity of the gene structure and predicted protein structure levels. We define unique sequence motifs as motifs that are found in all genes belonging to a family, but not found in any other genes represented in the NR (ftp://ftp.ncbi.nlm.nih.gov/blast/db/) or in any other ORFs found in the Plasmodium genomes. Based on these criteria, we came to the conclusion that both var and sicavar are unique to their respective genomes in the context of those Plasmodium species that were subject of the present study. However, the other gene families must be considered to be related.
The hypothesis of homology of rif/stevor genes and cir genes (and the other rodent malaria homologous genes) is largely based on the key finding of shared conserved unique amino acid sequence motif modules that are not present in any other gene families within the genomes of P.falciparum or P.chabaudi and the other rodent malarias, nor are found in any other taxa outside of the genus Plasmodium represented in the NR. The alignment of rif and cir genes establishes a topographical identity of individual amino acid sites, the first step in assessing homology (1). This hypothesis was tested using phylogenetic inference including genes from vir and kir. The resulting tree gave a hypothetical lineage congruent with the previously hypothesized evolutionary relationships of these species (41,42) (Figure 2). Although some of the amino acid motifs conserved between RIFINS and their rodent malaria homologues described in Figure 1 overlap, they are the products of independent statistical analyses using different window sizes for motif identification, and represent independent and different probability matrices (with the exception of motifs number 4 and 8, which significantly overlap both quantitatively and qualitatively). The four classes of motifs conserved between rif and cir genes (Figure 1 and Table 1) form modules (43) in each family of genes. These modules display different connectivity relationships in CIRS and RIFINS (Figure 1), but are conserved within a family. This difference in the relationships of amino acid module connectivities between CIRS and RIFINS, as well as the difference in number of introns, indicate that these gene families were probably expanded after changes in progenitor genes, which were inherited from a common ancestor, became fixed in the genomes post speciation.
Table 1. Amino acid motifs conserved amongst products of the cir and rif related multi-gene families.
| Motif according to Figure 1 | Best possible match consensus against NR | Best-fit motif from training set in PROSITE notation |
|---|---|---|
| Motif 2 | SIVAILVPVLVM | S-[IV]-[VI]-A-I-[LV]-[IV]-I-[VI]-L-[IV]-[MI] |
| Motif 3 (supplement PIR superfamily motif 2) | VLCQYAYIWL | [KV]-L-C-I-[YF]-[LA]-[YI]-[LIY]-W-L |
| Motif 4 | CDKRIQKIILRD | [CF]-D-K-[ED]-I-Q-K-[IQ]-[IY]-L-[KR]-[DE] |
| Motif 5 | LLFAFPLNIL | L-L-F-A-L-P-L-N-I-L |
| Motif 7 | SVVAILIPVL | S-[IV]-[VA]-A-I-[LV]-[IV]-[IP]-[VI]-L |
Simplified consensus sequences of the amino acid motifs conserved amongst products of the cir and rif-related multi-gene families. The table shows the best-fit output from the PSPMs for the motifs described in the text, shown in Figure 1 and the Supplementary Material. The ‘best possible match against NR’ column shows the consensus of the most common position-specific residues found in significantly matching sequences in the NR database. The ‘best-fit motif from training set’ column shows the most probable position-specific residues found in significantly matching sequences found in the motif-discovery sequence training set. Any annotations of these motifs on sequences should only be carried out with the full original PSPMs that are supplied in the Supplementary Material.
The conservation of key sequence motifs (see above) and the conservation of variant domain distribution within the protein families indicate that the molecules may perform the same function in both P.falciparum and P.chabaudi (as well as in the other species that have genes belonging to this superfamily). This is further supported by the conservation of predicted secondary structure among RIFINS and CIRS. Variable and conserved regions are found in the same topographical positions in both CIRS and RIFINS, and the hypervariable region is positionally conserved. The sequence variation pattern is also topographically conserved in terms of predicted secondary structure (Figure 1), which is a good indication that RIFINS and CIRS share some similar tertiary structure. Conservation of the distribution of variation and sequence conservation between RIFINS and CIRS indicates that both protein families may be under similar selection pressure. Further support for this can be found in the fact that the average amino acid sequence identity is at 30%, the same for both RIFINS and CIRS. Given that both families of proteins are postulated to be under immune selection, the above conservation of structure and variable domain position indicates that both RIFINS and CIRS may well interact with the host immune system in the same way. Further, should CIR and RIFIN proteins perform a biological function other than antigenic variation, the observed patterns of structural conservation provide good evidence that this function is conserved between both families of proteins. The localization of CIR protein within the infected erythrocyte and timing of protein expression in the trophozoite stage further underline the similarity to RIFINS. Although the IFAT localization of CIR shows only an association with the infected erythrocyte membrane but does not deliver evidence of exposure on the surface, it does place the protein within the same organellar area as RIFINS and VIRS. Further experiments will clarify the exact relationship of the CIR localization with the infected erythrocyte surface.
The link between vir and the cir/bir/yir families has already been described elsewhere (7). The data presented here further strengthens the hypothesis of common ancestry of these gene families by adding data for conserved sequence motifs and structural predictions. The newly described gene family kir resembles vir most closely, which is congruent with the phylogenetic closeness of P.vivax and P.knowlesi.
Based on the evidence presented in this paper, we propose a new gene superfamily named pir found in the genus Plasmodium that comprises the gene families rif/stevor, vir, kir, cir, bir and yir.
The relationship of cir and its rodent malaria homologues to the human parasites' P.falciparum rif and P.vivax vir gene families make them invaluable tools. Results from the study of experimental immunology and genetics with the rodent malaria cir genes will enable predictions to be made with respect to the interactions of P.falciparum and P.vivax with their human host, which can be checked by clinical observations for their direct relevance to the human disease.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR online.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Neil Hall and colleagues for sequencing the P.chabaudi, P.berghei and P.knowlesi genomes. This work was funded by the Wellcome Trust and the European Union. Preliminary genome sequence was made available by the Sanger Institute.
REFERENCES
- 1.Thornton J.W. and DeSalle,R. (2000) Gene family evolution and homology: genomics meets phylogenetics. Annu. Rev. Genomics Hum. Genet., 1, 41–73. [DOI] [PubMed] [Google Scholar]
- 2.Garnham P.C.C. (1966) Malaria Parasites and Other Haemosporidia. Blackwell, Oxford. [Google Scholar]
- 3.Smith J.D., Chitnis,C.E., Craig,A.G., Roberts,D.J., Hudson-Taylor,D.E., Peterson,D.S., Pinches,R., Newbold,C.I. and Miller,L.H. (1995) Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell, 82, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng Q., Cloonan,N., Fischer,K., Thompson,J., Waine,G., Lanzer,M. and Saul,A. (1998) stevor and rif are Plasmodium falciparum multicopy gene families which potentially encode variant antigens. Mol. Biochem. Parasitol., 97, 161–176. [DOI] [PubMed] [Google Scholar]
- 5.del Portillo H.A., Fernandez-Becerra,C., Bowman,S., Oliver,K., Preuss,M., Sanchez,C.P., Schneider,N.K., Villalobos,J.M., Rajandream,M.A., Harris,D. et al. (2001) A superfamily of variant genes encoded in the subtelomeric region of Plasmodium vivax. Nature, 410, 839–842. [DOI] [PubMed] [Google Scholar]
- 6.AlKhedery B., Barnwell,J.W. and Galinski,M.R. (1999) Antigenic variation in malaria: a 3′ genomic alteration associated with the expression of a P.knowlesi variant antigen. Mol. Cell, 3, 131–141. [DOI] [PubMed] [Google Scholar]
- 7.Janssen C.S., Barrett,M.P., Turner,C.M. and Phillips,R.S. (2002) A large gene family for putative variant antigens shared by human and rodent malaria parasites. Proc. R. Soc. Lond. B. Biol. Sci., 269, 431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bahl A., Brunk,B., Coppel,R.L., Crabtree,J., Diskin,S.J., Fraunholz,M.J., Grant,G.R., Gupta,D., Huestis,R.L., Kissinger,J.C. et al. (2002) PlasmoDB: the Plasmodium genome resource. An integrated database providing tools for accessing, analyzing and mapping expression and sequence data (both finished and unfinished). Nucleic Acids Res., 30, 87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su X.Z., Heatwole,V.M., Wertheimer,S.P., Guinet,F., Herrfeldt,J.A., Peterson,D.S., Ravetch,J.A. and Wellems,T.E. (1995) The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell, 82, 89–100. [DOI] [PubMed] [Google Scholar]
- 10.Pearson W.R. (2000) Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol., 132, 185–219. [DOI] [PubMed] [Google Scholar]
- 11.Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics., 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 12.Birney E. and Durbin,R. (2000) Using GeneWise in the Drosophila annotation experiment. Genome Res., 10, 547–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgenstern B. (1999) DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics., 15, 211–218. [DOI] [PubMed] [Google Scholar]
- 14.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L. (2000) The Pfam protein families database. Nucleic Acids Res., 28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailey T.L. and Elkan,C. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol., 2, 28–36. [PubMed] [Google Scholar]
- 16.Bailey T.L., Baker,M.E. and Elkan,C.P. (1997) An artificial intelligence approach to motif discovery in protein sequences: application to steriod dehydrogenases. J. Steroid Biochem. Mol. Biol., 62, 29–44. [DOI] [PubMed] [Google Scholar]
- 17.Bailey T.L. and Gribskov,M. (1997) Score distributions for simultaneous matching to multiple motifs. J. Comput. Biol., 4, 45–59. [DOI] [PubMed] [Google Scholar]
- 18.Rice P., Longden,I. and Bleasby,A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet., 16, 276–277. [DOI] [PubMed] [Google Scholar]
- 19.Henikoff S. and Henikoff,J.G. (1992) Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA, 89, 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuff J.A. and Barton,G.J. (2000) Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins, 40, 502–511. [DOI] [PubMed] [Google Scholar]
- 21.King R.D., Saqi,M., Sayle,R. and Sternberg,M.J. (1997) DSC: public domain protein secondary structure predication. Comput. Appl. Biosci., 13, 473–474. [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein J. and Churchill,G.A. (1996) A Hidden Markov Model approach to variation among sites in rate of evolution. Mol. Biol. Evol., 13, 93–104. [DOI] [PubMed] [Google Scholar]
- 23.Felsenstein J. (1996) Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol., 266, 418–427. [DOI] [PubMed] [Google Scholar]
- 24.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol., 17, 540–552. [DOI] [PubMed] [Google Scholar]
- 26.Strimmer K. and von Haeseler,A. (1996) Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol., 13, 964–969. [Google Scholar]
- 27.Strimmer K. and von Haeseler,A. (1997) Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl Acad. Sci. USA, 94, 6815–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zmasek C.M. and Eddy,S.R. (2001) ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics., 17, 383–384. [DOI] [PubMed] [Google Scholar]
- 29.Page R.D. (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci., 12, 357–358. [DOI] [PubMed] [Google Scholar]
- 30.Pace T., Ponzi,M., Dore,E. and Frontali,C. (1987) Telomeric motifs are present in a highly repetitive element in the Plasmodium berghei genome. Mol. Biochem. Parasitol., 24, 193–202. [DOI] [PubMed] [Google Scholar]
- 31.Dore E., Pace,T., Ponzi,M., Picci,L. and Fontali,C. (1990) Organization of subtelomeric repeats in Plasmodium berghei. Mol. Cell Biol., 10, 2423–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner C.M. (2002) A perspective on clonal phenotypic (antigenic) variation in protozoan parasites. Parasitology, 125 (Suppl.), 17–S23. [DOI] [PubMed] [Google Scholar]
- 33.Cox H.W. (1962) The behaviour of Plasmodium berghei strains isolated from relapsed infections of white mice. J. Parasitol., 9, 114–118. [Google Scholar]
- 34.Brown K.N. and Brown,I.N. (1965) Immunity to malaria: antigenic variation in chronic infections of Plasmodium knowlesi. Nature, 208, 1286–1288. [DOI] [PubMed] [Google Scholar]
- 35.Voller A. and Rossan,R.N. (1969) Immunological studies with simian malarias. I. Antigenic variants of Plasmodium cynomolgi bastianellii. Trans. R. Soc. Trop. Med. Hyg., 63, 46–56. [DOI] [PubMed] [Google Scholar]
- 36.Handunnetti S.M., Mendis,K.N. and David,P.H. (1987) Antigenic variation of cloned Plasmodium fragile in its natural host Macaca sinica. Sequential appearance of successive variant antigenic types. J. Exp. Med., 165, 1269–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLean S.A., Pearson,C.D. and Phillips,R.S. (1982) Plasmodium chabaudi: antigenic variation during recrudescent parasitaemias in mice. Exp. Parasitol., 54, 296–302. [DOI] [PubMed] [Google Scholar]
- 38.Hommel M., David,P.H. and Oligino,L.D. (1983) Surface alterations of erythrocytes in Plasmodium falciparum malaria. Antigenic variation, antigenic diversity, and the role of the spleen. J. Exp. Med., 157, 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biggs B.A., Anders,R.F., Dillon,H.E., Davern,K.M., Martin,M., Petersen,C. and Brown,G.V. (1992) Adherence of infected erythrocytes to venular endothelium selects for antigenic variants of Plasmodium falciparum. J. Immunol., 149, 2047–2054. [PubMed] [Google Scholar]
- 40.Leech J.H., Barnwell,J.W., Miller,L.H. and Howard,R.J. (1984) Identification of a strain-specific malarial antigen exposed on the surface of Plasmodium falciparum-infected erythrocytes. J. Exp. Med., 159, 1567–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ayala F.J., Escalante,A.A., Lal,A.A. and Rich,S.M. (1998) Evolutionary relationships of human malaria parasites. In Sherman,I.W. (ed.), Malaria: Parasite biology, pathogenesis, and protection. ASM Press, Washington DC., pp. 285–300. [Google Scholar]
- 42.Siddall M.E. and Barta,J.R. (1992) Phylogeny of Plasmodium species: estimation and inference. J. Parasitol., 78, 567–568. [PubMed] [Google Scholar]
- 43.Henikoff S., Greene,E.A., Pietrokovski,S., Bork,P., Attwood,T.K. and Hood,L. (1997) Gene families: the taxonomy of protein paralogs and chimeras. Science, 278, 609–614. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.