Abstract
The lateral organization of biological membranes is thought to take place on the nanometer length scale. However, this length scale and the dynamic nature of small lipid and protein domains have made characterization of such organization in biological membranes and model systems difficult. Here we introduce a new method for measuring the colocalization of lipids in monolayers and bilayers using stable isotope labeling. We take advantage of a process that occurs in dynamic SIMS called atomic recombination, in which atoms on different molecules combine to form diatomic ions that are detected with a NanoSIMS instrument. This process is highly sensitive to the distance between molecules. By measuring the efficiency of the formation of 13C15N− ions from 13C and 15N atoms on different lipid molecules, we measure variations in the lateral organization of bilayers even though these heterogeneities occur on a length scale of only a few nm, well below the diameter of the primary ion beam of the NanoSIMS instrument or even the best super-resolution fluorescence methods. Using this technique, we provide direct evidence for nanoscale phase separation in a model membrane, which may provide a better model for the organization of biological membranes than lipid mixtures with microscale phase separation. We expect this technique to be broadly applicable to any assembly where very short scale proximity is of interest or unknown, both in chemical and biological systems.
Graphical Abstract
INTRODUCTION
Super-resolution methods based on fluorescence have revolutionized the imaging of biological samples.1–3 Fluorescence is a highly sensitive method but, of course, requires a fluorescent label, which can be problematic, especially for lipid components or small molecule metabolites.4,5 Furthermore, one is often interested in the spatial relationship of different molecules, for example, the lateral organization of components in a biological membrane, which is the focus of our work. When studying the spatial distribution of molecules, Förster resonance energy transfer (FRET) and multicolor fluorescence correlation spectroscopy (FCS) offer the sensitivity of fluorescence, again with the caveat of large labels, while NMR, small angle neutron scattering (SANS), and ESR methods can sample short distances, albeit with much less sensitivity.6–16 Imaging mass spectrometry offers an alternative imaging method, with secondary ion mass spectrometry (SIMS) and MALDI each offering advantages and trade-offs. We have used a NanoSIMS for imaging of model membranes, using isotopic (or atom, e.g., F) labels that avoid bulky fluorophores. The NanoSIMS achieves an ultimate lateral resolution of about 50 nm and allows imaging with the multiplex advantage of mass spectrometry.17
In the NanoSIMS experiment, a primary Cs+ ion beam is rastered across the sample, generating monatomic and small polyatomic secondary ions, such as 12C−, 13C−, 13C1H−, 12C2H−12C14N−, 12C15N−, and 13C15N−, and allowing imaging based on isotopic labeling of the parent molecules. The interaction between the Cs+ beam and the organic and underlying solid surface is complicated. Atomic and polyatomic ions are generated, and most of the diatomic ions result from inter- and intramolecular atomic recombination, either between atoms on the same (not necessarily covalently bonded) molecule or between atoms on different molecules.18–22 This recombination offers the possibility of a different type of imaging on a length scale much shorter than the 50 nm primary beam diameter, as atomic recombination has a very strong distance dependence.23 As depicted in Figure 1A and B, if two molecules, one labeled with 13C (blue) and another with 15N (red), are very close to each other, 13C15N− will be generated at a level that is higher than that due to the natural abundance of these isotopes (1.1% for 13C, 0.36% for 15N, joint probability 0.0040%). The 13C15N− yield, normalized to 12C15N−, can be taken as a measure of the distance between isotopically labeled molecules, and, as shown in the following, on a length scale less than about 3 nm. For simplicity, the ratio 13C15N−/(13C15N− + 12C15N−) will be referred to as ℜ(13C15N−). As ℜ is already normalized to the concentration of 15N-lipid (by dividing by 12C15N−), it is only a function of the distance between 13C and 15N atoms and the concentration of 13C-labeled lipid. Note that 13C15N− normalized to 13C14N− could also provide similar information; however, this ratio and the 15N-lipid concentration (from 12C15N−) cannot be measured simultaneously (see Supporting Information section 1 for further discussion of the choice of secondary ions). The NanoSIMS instrument is particularly well suited for high precision isotope ratio measurements, with precision in the ppm range for many samples. Atomic recombination has been reported in earlier work but little developed.18,23 Here, by using well-defined model membrane monolayer and bilayer samples, we show that this approach can provide unique information on lateral heterogeneity on a length scale far below the Cs+ beam diameter.
Figure 1.
Schematic of the atomic recombination experiment and sitespecifically labeled lipids. (A) When the 13C and 15N labels are far apart during Cs+ bombardment (beam shown schematically—actual diameter on the order of 100 nm), 12C15N− and 13C14N− are primarily observed, with only natural abundance 13C15N−. (B) When the 13C and 15N labels are close together, higher counts of 13C15N− are observed, with less 12C15N− and 13C14N−. The ratio ℜ ≡ 13C15N−/(12C15N− + 13C15N−) is, therefore, related to the average distance between 13C and 15N labels in the sample under Cs+ bombardment. (C) Isotope-labeled lipids used in this study, from left to right: 13C3-DOPC, 15N-DOPC, 2,3,4-13C3-CHOL, and 25,26,27-13C3-CHOL and cartoon representations (no color = natural abundance). 13C3- and 15N-labeled DSPC and POPC follow the same headgroup labeling pattern as labeled DOPC; DSPC is shown with straight fatty acid tails in the schematics.
MATERIALS AND METHODS
Phospholipids and unlabeled cholesterol were from Avanti Polar Lipids. 13C-Methyl iodide, 15N-choline chloride, 2,3,4-13C3-cholesterol, and 25,26,27-13C3-cholesterol were from Sigma. Texas Red 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (TR-DHPE) was from Thermo Fisher Scientific. Four inch 〈100〉 p-type silicon wafers were from Silicon Quest International. Solvents and other chemicals were from Fisher and used as supplied.
Synthesis of Isotopically Labeled Lipids
15N-Phospholipids were synthesized as reported earlier by phosphoesterifying the appropriate phosphatidic acid with 15N-choline (Figure 1C).24,25 13C3-Phospholipds were synthesized as before by methylating the appropriate phosphatidylethanolamine with 13C-methyl iodide (Figure 1C).24 Abbreviations used: DOPC = 1,2-dioleoyl-sn-glycero-3-phosphocholine; POPC = 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; DSPC = 1,2-distearoyl-sn-glycero-3-phosphocholine; CHOL = cholesterol. Unless specifically indicated, all lipids have natural isotopic abundance and are not colored in schematic diagrams.
Preparation of Samples for NanoSIMS
Oxidized silicon wafers were prepared as described before by thermally growing 10 nm of SiO2 on silicon wafers and dicing them to 5 × 5 mm2 to fit the NanoSIMS sample holder.24,25
Langmuir–Blodgett (LB) monolayers were deposited onto the diced silicon wafers using standard techniques.26 Briefly, a KSV NIMA KN 2002 (Biolin Scientific, Stockholm, Sweden) Langmuir trough equipped with a 273 cm2 Teflon trough and symmetric Delrin barriers was used to form and deposit the LB monolayers. A Wilhelmy plate made of Whatman filter paper was used to monitor changes in surface pressure. Water (> 18 MΩ; from a Milli-Q system) was added to the clean trough. The desired mixtures of lipids in chloroform were spread onto the surface of the water slowly, and solvent was allowed to evaporate for 10 min. The barriers were compressed at a rate of 10 mm/min until the surface pressure reached 32 mN/m. The plasma-cleaned 5 × 5 mm2 NanoSIMS substrates were glued to microscope coverslips and pulled through the air–water interface at 2 mm/min while maintaining a surface pressure of 32 mN/m (lipid monolayers are stable in air). NanoSIMS substrates with supported monolayers were then stored in a vacuum desiccator until NanoSIMS analysis.
In order to form asymmetric supported bilayers with isotopically labeled lipids in different leaflets, standard Langmuir–Schaefer (LS) techniques were used.27 Briefly, LB monolayers were formed as described. Then, the LB trough was cleaned thoroughly, and a new monolayer of a different lipid mixture was compressed to 32 mN/m. The substrate with the supported monolayer was then slowly passed through the air–water interface to transfer the second leaflet. In contrast to supported monolayers, supported bilayers are not stable in air, and the substrates with supported bilayers tend to dewet when lifted out of the LB trough.28 In order to preserve the bilayer, the substrates were lifted out of the LB trough upside down with tweezers. This leaves a drop of water hanging from the substrate to keep the bilayer hydrated. The substrate is then plunged into liquid nitrogen to flash freeze it. Previous work has shown that flash freezing preserves the lateral organization of lipid bilayers.24,25 The substrates with vitreous ice on them were then transferred to a chamber at liquid nitrogen temperature, and the ice was sublimed overnight at 50 µbar with an oil-free scroll pump with a liquid nitrogen trap.
Supported lipid bilayer (SLB) patches were formed by fusion of giant unilamellar vesicles (GUVs) as previously reported.25 Briefly, GUVs were formed by the gentle hydration method. A 100 nmol portion of lipids was mixed in the desired ratios in chloroform in glass vials, and the chloroform was evaporated under a gentle stream of nitrogen while rotating the vial. 0.1% TR-DHPE was included in the lipid mixtures to aid in visualization of SLB formation. These lipid films were placed under a vacuum for at least 4 h to remove residual solvent. A 1 mL portion of submicron filtered 500 mM sucrose was gently added to the vials. Lipid films were left to swell overnight at either 37 or 70 °C, depending on the melting temperature (Tm) of the lipids. GUVs were routinely analyzed with ESI-MS to confirm the composition and to check for lipid breakdown. NanoSIMS substrates were plasma cleaned for 15 min. The substrates were then placed in PDMS wells with 150 µL of phosphate buffer (240 mM NaCl, 10 mM NaH2PO4, pH 7.4), and 10 µL of the GUV solution was gently added. SLB formation was monitored with fluorescence microscopy. Once 15–25% of the area of the substrates was covered with SLB patches, the substrates were rinsed thoroughly with water. Substrates were then flash frozen and freeze-dried in a manner similar to the asymmetric bilayers. GUVs were used instead of extrusion of small vesicles because they use much less isotopically labeled lipids; SLB patch formation and rinsing removes any inclusions in the GUVs that result from the gentle hydration method, leaving clean planar bilayers. Also, much of the literature on the phase behavior of lipid mixtures uses GUVs, and domains observed by dye partitioning in GUVs are captured in a planar format by rupturing to form an SLB.
NanoSIMS Analysis
SIMS analysis was performed on the Cameca NanoSIMS 50L at Stanford University. Images were collected with a 2 pA 133Cs+ primary ion beam focused to a 100 nm spot. Fifteen 20 µm × 20 µm frames of 256 × 256 pixels were collected with a dwell time of 1 ms/pixel. The ion detectors were set to 12C−, 13C−, 13C1H−, 12C14N−, 12C15N− (or 13C14N−), and 13C15N−, and secondary electrons were detected simultaneously (see Supporting Information section 2 for details on peak identification). Imaging was necessary to calculate isotope ratios within specific domains. Imaging is not strictly necessary for the samples with nanoscale phase separation, as one can measure ion ratios without using the imaging mode, but imaging proved to be useful for avoiding the occasional piece of debris on the samples. Special care was taken to ensure the accuracy of isotope ratios. NMR regulation was used to stabilize the magnetic field of the mass spectrometer, and secondary ion peaks were recentered frequently from the high mass resolution spectra in order to correct for magnet drift. Standard samples were also used to ensure that natural abundance isotope ratios were accurate and reproducible from day to day.
Data Analysis
SIMS images were analyzed with ImageJ 4.18 (National Institutes of Health, USA) using the OpenMIMS plugin (National Resource for Imaging Mass Spectrometry, Harvard University, USA). Image planes were summed in OpenMIMS, and regions of interest were selected either within certain domains or for the whole image. Then, the total counts for each ion within the selected region were calculated within the “Tomography” tab of OpenMIMS. Isotope ratios were then calculated as the isotope divided by the sum of stable isotopes for that element so that isotope ratios range from 0 to 1 (e.g., 13C/(13C + 12C) or 12C15N/(12C15N + 12C14N)). The standard deviations of at least three separate isotope ratio determinations for each sample were also calculated. 13C3-Lipid concentrations were determined from calibration curves from monolayers 13C3-DOPC and 15N-DOPC or from dried drops of the two 13C3-cholesterols (see Supporting Information section 3 for details on calibration curves).
RESULTS AND DISCUSSION
Recombination of 13C and 15N to Form 13C15N− Ions
In order to determine whether atomic recombination occurs to a significant extent between atoms in lipid molecules separated from each other by the distances characteristic of lipid packing in leaflets of a bilayer, we first measured the 13C15N− ion yield from monolayers of 13C3-DOPC, 15N-DOPC, and DOPC. As these molecules are chemically identical except for the isotope label, they mix together randomly, and the average distance between the 13C and 15N atoms is governed by the concentrations of 13C3- and 15N-DOPC and the distribution of distances between choline headgroups. Furthermore, the mean molecular area of DOPC molecules in a monolayer at 32 mN/m approximates the lipid packing in a DOPC bilayer where the average molecular area is 75 Å2 with an average center-to-center distance of 8.7 Å.29 The mol % of 15N-DOPC was held constant at 10% for all samples, and 13C3-DOPC varied from 0 to 90% (shown schematically in Figure 2A, all percents and ratios refer to mol %). Figure 2B shows a plot of 13C15N−/(13C15N− + 12C15N−) (hereafter, ℜ(13C15N−)) vs [13C3-DOPC]; there is an apparently linear increase from 0.011, the natural abundance of 13C, to nearly 0.043. ℜ(13C15N−) expresses the amount of 13C15N− observed relative to the amount of 12C15N−, or in other words, the proportion of 15N atoms that have undergone recombination with 13C rather than 12C (all 15N is initially only bonded to 12C and natural abundance 13C). This value is therefore normalized to [15N-lipid] and is a function of both [13C3-lipid] and the distance between labels (probability of atomic recombination).
Figure 2.
Atomic recombination in ideally mixed monolayers of DOPC, 13C3-DOPC, and 15N-DOPC. (A) Increasing the concentration of 13C3-DOPC in monolayers with a fixed percent (10 mol %) of 15N-DOPC decreases the average distance between 13C and 15N labels. (B) The measured ℜ(13C15N−) increases as the mole fraction of 13C3-DOPC increases (black). Simulated values for two limiting cases where there is no recombination (purple) and complete, distance-independent recombination (orange) are plotted. (C) Data for lipids in randomly mixed monolayers can be used as a standard for other compositions that might contain a nonrandom distribution of molecules. Note that the horizontal blue boundary is set by the natural abundance of 13C.
To confirm that this increase in 13C15N− results from intermolecular atomic recombination, not simply from natural abundance 13C15N−, we calculated the ℜ(13C15N−) values that would result from the sum of the natural abundance 15N in the 13C3-DOPC, natural abundance 13C in the 15N-DOPC, and natural abundance 13C and 15N in the DOPC (purple dashed line in Figure 2B, see Supporting Information section 4 for details of these calculations). Another limiting case in which all atoms in the beam spot are scrambled can also be calculated (orange dashed line in Figure 2B). In this case, all C and N atoms in the beam spot have equal probability of combining to form CN− regardless of their distance from each other. The experimental data in black falls between these two limiting cases, indicating that intermolecular recombination is happening in DOPC monolayers and that the probability of recombination depends on the average distance between C and N atoms in different DOPC molecules.23
This data serves as an empirical calibration, where the isotopically labeled lipids are completely randomly distributed laterally, with which other monolayers or bilayers with nonrandom lateral distributions of molecules can be compared. For example, if the two labeled species (at a given mol %) are colocalized in the same phase or by other noncovalent interactions, they will be closer together, on average, than if they were randomly mixed. Similarly, if the two labeled species are in different phases or otherwise not colocalized, they will be farther away from each other on average than if they were randomly mixed. As Figure 2C depicts, if a data point falls above the line for randomly mixed DOPC (in the green shaded region), it indicates that the labeled molecules are colocalized, and if it falls below the line (in the red shaded region), they are noncolocalized. We note that direct comparison of the DOPC monolayer calibration for random mixing to supported bilayers is valid because monolayers at 32 mN/m have the similar lipid packing as bilayers (see Supporting Information sections 5 and 6). For systems with molecular packing that is different than lipid bilayers, a similar calibration would have to be constructed.
Changing the concentrations of 13C3- and 15N-DOPC changes both the mean nearest neighbor distance between labeled lipids and the number of labeled lipids neighboring a labeled lipid. Therefore, the data in Figure 2B results from a convolution of the distance between labels and the number of neighboring labeled lipids. The two factors are difficult to deconvolute, but both are ultimately important in phase separation and do not need to be deconvoluted for most applications. See Supporting Information section 7 for a more detailed discussion of distances between labeled lipids in monolayers.
An alternative way to change the distance between labels without changing the composition of the monolayer is to move the 13C atoms away from the 15N atoms by changing their positions on the 13C-labeled lipid. CHOL with three 13C atoms, the same number per molecule as the 13C3-DOPC, is available at opposite ends of the sterol, so monolayers and asymmetric bilayers of DOPC and DOPC/CHOL were used to systematically vary the distance between 15N and 13C atoms (Figure 3A). Binary mixtures of cholesterol and DOPC do not visibly phase separate and are assumed to be randomly mixed.33,34 When the 13C atoms are in the 2, 3, and 4 positions (see Figure 1C), they are very close to the 15N in the choline headgroup (case 2, Figure 3A) and only slightly farther away than when the 13C atoms are on choline headgroups, as in the earlier example (case 1, Figure 3A). On the other hand, the 13C atoms in 25,26,27-13C3-CHOL are near the edge of the leaflet farthest away from the 15N-choline headgroups (case 3, Figure 3A). Further separation between the 15N and 13C atoms can be achieved by constructing asymmetric bilayers via Langmuir–Blodgett and Langmuir–Schaefer transfers with 15N-DOPC in one leaflet and 13C3-lipid in the other leaflet. Placing 50:50 15NDOPC: DOPC in one leaflet and 50:50 25,26,27-13C3-CHOL:DOPC in the other moves the 13C atoms further from the 15N atoms than in a monolayer of 50:50 15N-DOPC and 25,26,27-13C3-CHOL (case 4, Figure 3A). Similarly, if the second leaflet contains 50:50 2,3,4-13C3-CHOL:DOPC, the separation is even greater (case 5, Figure 3A). Finally, the labels can be separated the most if the second leaflet of the bilayer contains 50:50 13C3-DOPC:DOPC (case 6, Figure 3A). In this last case, ℜ(13C15N−) can be plotted with the data for the randomly mixed monolayer (Figure 2B) in order to see the effect of moving the 13C and 15N labels into different leaflets of a bilayer. As shown in Figure 3B, theℜ(13C15N−) values are far below the line for randomly mixed DOPC monolayers, where it is close to the natural abundance ratio, meaning that very little recombination is occurring between the 13C and 15N atoms at this spacing. The measured ℜ(13C15N−) values for each of the monolayer and bilayer samples are shown in Figure 3C vs the approximate mean separation between the 13C and 15N atoms in order to obtain an estimate of the relationship between CN− recombination efficiency and distance. As seen in Figure 3C, the recombination efficiency drops rapidly from the 13C3-DOPC/15N-DOPC monolayer (case 1) to near 0.011 for the 15N-DOPC/25,26,27-13C3-CHOL asymmetric bilayer (case 4). The other asymmetric bilayers with labels further separated have ℜ(13C15N−) near the natural abundance of 13C. The small differences in recombination efficiency for cases 4–6 are likely due to differences in ionization efficiency for 13C in different chemical environments and are not meaningful. The steep drop in recombination efficiency from a separation of about 1 nm to a separation of about 3 nm indicates that this method is most valuable for measuring very short distances and that recombination signals in the DOPC monolayers likely result primarily from labeled molecules directly neighboring each other. Three nm is somewhat shorter than the Förster radius of most commonly used dye and fluorescent protein pairs, which are generally not useful for measuring changes in distance on this length scale. While FRET efficiency is constant for distances much smaller than the Förster radius (and, of course, requires the attachment of large fluorophores), atomic recombination appears to still be within its dynamic range. Additionally, the distances probed by atomic recombination are similar to those probed by ESR and fluorescence quenching, but these techniques also require potentially disruptive labeling.35,36
Figure 3.
Systematic approaches to separating isotopic labels with leaflet asymmetry and site-specific cholesterol labeling. (A) Compositions (in mol %) are as follows: (1) 50:50 15N-DOPC:13C3-DOPC, (2) 50:50 2,3,4-13C3-CHOL:15N-DOPC, (3) 50:50 25,26,27-13C3-CHOL:15N-DOPC, (4) 50:50 15N-DOPC:DOPC and 50:50 25,26,27-13C3-CHOL:DOPC, (5) 50:50 15N-DOPC:DOPC and 50:50 2,3,4-13C3-CHOL:DOPC, (6) 50:50 15N-DOPC:DOPC and 50:50 13C3-DOPC:DOPC. (B) Data for case 6 demonstrates that when 13C and 15N atoms are separated by approximately 5 nm ℜ(13C15N−) is much smaller. (C) ℜ(13C15N−) decreases as a function of the approximate distance between 13C and 15N atoms for all configurations shown in part A. Mean distances between labels were estimated from X-ray scattering and molecular dynamics models.30–32
Microscale Phase Separating Mixtures
As the recombination of 13C and 15N to form 13C15N− ions depends on the distance between the atoms, it should be possible to detect nonrandom distributions of labeled lipids. We first tested this method with supported bilayers of lipid mixtures that display microscopic phase separation, i.e., detectable qualitatively by dye partitioning or quantitatively by NanoSIMS imaging. Specifically, we chose mixtures of DSPC, CHOL, and DOPC because their phase behavior is well studied in GUVs and due to the high level of immiscibility of these components.37 Lipids were labeled with 15N or 13C atoms on different DSPC molecules only. For ternary compositions within the region of coexisting liquid-ordered (Lo) and liquid-disordered (Ld) phases on the phase diagram (see Supporting Information section 8 for phase diagrams and compositions), phase separation will result in colocalization of the 15N-DSPC and 13C-DSPC in the liquid-ordered phase (see Supporting Information section 9 for examples of NanoSIMS images of phase-separating mixtures). GUVs composed of 20:20:25:35 13C3-DSPC:15N-DSPC:CHOL:DOPC were formed and allowed to rupture on plasma-cleaned oxidized silicon substrates. As shown in Figure 4A, SLB patches resulting from the fusion of a single GUV and imaged by 12C15N− showed clear separation into a bright region on the right where 15N-DSPC accumulates (the liquid ordered domain) and a dark region on the left (the liquid disordered domain) where it is depleted (see schematic diagram). The SLB patches were typically 20–70 µm in diameter with 5–20 µm diameter liquid ordered domains (as visualized by NanoSIMS imaging in Figure 4A), and regions of SLBs containing approximately equal areas of liquid ordered and liquid disordered domains were chosen for NanoSIMS imaging. The concentration of 13C3-lipid in the entire 20 × 20 µm2 field of view was then calculated from calibration curves (see Supporting Information section 3 for details on calibration curves).
Figure 4.
Atomic recombination in microscale and nanoscale phase separating bilayers. (A) An SLB patch of 20:20:25:35 15N-DSPC:13C3-DSPC:CHOL:DOPC formed from the fusion of a single giant unilamellar vesicle shows coexisting liquid phases. In the 15N-DSPC quantitative ion image of the SLB patch, the ordered phase with more 13C3-DSPC and 15N-DSPC is clearly visible as a region of higher 15N (seen as 12C15N−). (B) An SLB patch of 20:20:25:35 15N-DSPC:13C3-DSPC:CHOL:POPC shows a uniform 12C15N− signal in quantitative ion images. Scale bars are 5 µm. (C) When ℜ(13C15N−) within each phase or for the whole field of view is plotted vs the concentration of 13C3-DSPC, deviations from random mixing are observed (solid black squares; note that the vertical axis is expanded compared with Figures 2 and 3). As described in the text, the composition of SLBs with nanoscale Lo/Ld domains and isotope labels in different phases is 20:20:25:20:15 13C3-DSPC:DSPC:CHOL:15N-POPC: POPC. The (dis)ordered domains refer to data from regions in panel A; the nanodomains refer to panel B. Error bars are the standard deviation of NanoSIMS measurements on three different SLB patches with very similar 13C3-DSPC concentrations.
As shown in Figure 4C, ℜ(13C15N−) within both liquid-ordered (green triangle) and liquid-disordered (orange triangle) domains was calculated and plotted vs the 13C-lipid concentration of the whole field of view. If the 15N- and 13C3-lipids were randomly mixed, the ℜ(13C15N−) values for any region of the SLB would be the same. Adding cholesterol and DSPC to DOPC bilayers changes the lipid packing and mean molecular areas of lipids relative to pure DOPC bilayers.38 However, the change in recombination efficiency that results from changes in lipid area are small compared to the changes in local concentration caused by phase separation (see Supporting Information section 6). The ℜ(13C15N−) values within the ordered domains were significantly higher than those for randomly mixed lipids, while ℜ(13C15N−) within the liquid-disordered domains was significantly lower than that for randomly mixed lipids. As the majority of 13C3- and 15N-DSPC is in the ordered domains, these domains dominate the ℜ(13C15N−) value and the ratio for the whole field of view is the same as that of just the liquid-ordered domains (0.0151 for the whole SLB vs 0.0152 for the Lo domain alone). These results are consistent with the published phase diagram of DSPC/CHOL/DOPC.33 As DSPC and CHOL are enriched in liquid-ordered domains while DOPC is depleted, the mean distance between DSPC molecules is smaller than if they were randomly dispersed in the bilayer. As the liquid-disordered domains are enriched in DOPC and depleted in DSPC and CHOL, DSPC molecules have larger mean distances between them than if all of the lipids were uniformly distributed in the SLB.
Nanoscale Phase Separating Compositions
For the microscale phase separating mixtures described above, atomic recombination is not necessary to reveal variations in lateral membrane organization. The domains formed in these mixtures are large enough that their composition can be directly quantified with conventional NanoSIMS imaging, as previously reported and seen in Figure 4A.24,25 What these proof of concept experiments demonstrate and confirm is that NanoSIMS can be used to measure nonrandom mixing of lipid components in lipid bilayers and the lipid packing in phases. While the domains in ternary mixtures of saturated lipids, diunsaturated lipids, and cholesterol have been extensively studied as experimentally tractable models for lipid rafts, their coexisting phases are tens of microns in diameter. Lateral organization, if it is present in the plasma membrane, must exist on a length scale far below the diffraction limit for optical microscopy, and even super-resolution microscopy techniques have failed to directly visualize lateral segregation of lipids. Changing the low-Tm lipid from DOPC to POPC produces bilayers that appear uniform when visualized with fluorescent dyes. Intriguingly, FRET and small angle neutron scattering (SANS) have recently indicated that these mixtures contain nanoscale Lo/Ld phase separation with domains that do not coalesce into microscale domains, explaining why phase separation is not seen under conventional fluorescence microscopy.33,34,39–41 In particular, SANS has revealed the size and physical properties of nanoscale domains in DSPC/CHOL/POPC large unilamellar vesicles.40,43 Due to the size (~15 nm from SANS and FRET) of these domains and the prevalence of POPC in the plasma membrane of mammalian cells, these mixtures may represent a more accurate model for lipid rafts.33,41 However, they remain challenging to study due to the lack of methods for studying nanoscale domains. Despite the small size of these domains, they should create a nonrandom distribution of lipids and should be detectable by atomic recombination in NanoSIMS even if they cannot be directly visualized in composition images.
SLB patches in which POPC replaces DOPC were prepared as before from GUVs with the following compositions: 20:20:25:35 15N-DSPC:13C3-DSPC:CHOL:POPC and 20:20:25:20:15 13C3-DSPC:DSPC:CHOL:15N-POPC:POPC. Fluorescence of TR-DHPE in these GUVs is uniform, confirming that there is no microscale phase separation. As shown in the 15N-DSPC ion image in Figure 4B (shown for 12C15N−), NanoSIMS imaging also revealed uniform ion images, indicating that any phase separation present occurs on a length scale below the spatial resolution of the NanoSIMS (100 nm for the experiments in this work). However, ℜ(13C15N−) for the SLBs with 13C and 15N on separate DSPC molecules is higher than expected for randomly mixed lipids, indicating the labeled DSPC molecules are colocalized (Figure 4C, blue diamond). On the other hand, ℜ(13C15N−) is significantly below the line for the SLBs with 13C3-DSPC and 15N-POPC, indicating that the DSPC and POPC are demixed into different phases (Figure 4C, red circle). The large difference between recombination efficiency for labeled lipids that are in the same vs different phases also proves that the labeled lipids are nonrandomly distributed without the use of the DOPC monolayer calibration. These results are consistent with previous results in which changes in FRET efficiency or ESR are used to find phase boundaries for the same ternary lipid composition.33,39
Other Phase Separating Compositions
The magnitude of the change in recombination efficiency relative to an ideally mixed scenario reports on both the change in proximity of labeled components within a phase, the compositions of coexisting phases, the size of domains, and likely instrumental parameters. While robust theories for FRET correctly predicted the dependence of FRET efficiency on the distance between chromophores, no such theory exists for recombination in dynamic SIMS. Recombination in NanoSIMS is likely to depend on many experimental factors, including choice of labeling site, i.e., chemical context, primary ion beam energy, primary ion beam current, substrate, and pixel dwell time.18 We have also found that there is considerable variation in the compositions of each SLB patch, particularly for phase separating mixtures, making the direct comparison of data for different SLB patches difficult.25 As a result, the displacement of each data point from the line for ideally mixed monolayers makes the data directly comparable (Figure 5). In order to facilitate visualization of the data from many SLB patches of many different compositions, we calculated the difference between each data point and the corresponding point for the calculated 13C3-lipid concentration on the line that fits the ideally mixed monolayer data, and represent the data as shown in Figure 5B. Uncertainty was calculated as the propagated uncertainty from the standard deviation between all of the calculated differences and the mean standard deviation from the points used to determine the ideally mixed curve.
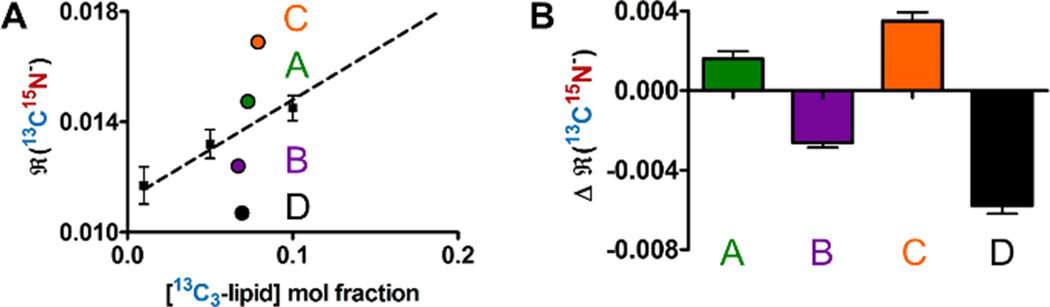
Figure 5.
Comparison of recombination data from micro- and nanoscale phase-separating mixtures. (A) ℜ(13C15N−) values for SLBs of lipid compositions displaying microscale liquid/liquid phase coexistence, microscale liquid/gel phase coexistence, and nanoscale liquid/liquid phase coexistence with isotopic labels either both in the same phase or in different phases. Composition A (nanoscale Lo/Ld): 20:20:25:35 13C3-DSPC, 15N-DSPC, CHOL, POPC. Composition B (nanoscale Lo/Ld): 20:20:25:20:15 13C3-DSPC/DSPC/CHOL/15N-POPC/POPC. Composition C (microscale gel/Ld): 20:20:60 13C3-DSPC/15N-DSPC/DOPC. Composition D (microscale Lo/LS): 20:20:25:20:15 13C3-DSPC/DSPC/CHOL/15N-DOPC/DOPC. (B) ℜ(13C15N−) can be more easily visualized by computing the difference between the measured ℜ(13C15N−) and the fit to the line through the data for ideally mixed monolayers. The error bars represent the propagated uncertainty from the standard deviation of each measurement of a different SLB patch (at least 3 per composition) and the average uncertainty in the monolayer data points (3 per composition) used to calculate the linear fit (see text for more details).
Four different lipid compositions were compared (letters correspond to compositions in Figure 5): (A) 13C3-DSPC:15N-DSPC: CHOL:POPC, which contains nanoscale coexisting Lo/Ld phases; (B) 13C3-DSPC:DSPC:CHOL:15N-POPC, which contains nanoscale coexisting Lo/Ld phases; (C) 13C3-DSPC:15N-DSPC:DOPC, which contains microscale coexisting gel and liquid phases; and (D) 13C3-DSPC:DSPC:CHOL:15N-DOPC, which contains microscale coexisting Lo/Ld phases. These compositions span a range of phase types, domain sizes, and probe locations within phases.33,43,47 The locations of these compositions on phase diagrams can be found in Supporting Information section 8. Compositions A and C localize labeled lipids in the same phase, while compositions B and D localize probes in different phases. The ℜ(13C15N−) values for representative SLB patches of each composition are plotted in Figure 5A. As expected, ℜ(13C15N−) for the composition with 15N- and 13C3-labeled lipids in different phases falls below the line for randomly mixed lipids (Figure 5A, composition D). The ℜ(13C15N−) value for the composition with both labeled lipids colocalized within the gel phase domains falls above the line, consistent with the microscopic colocalization (Figure 5A, composition C).
Next, the magnitudes of the change in recombination for these mixtures was compared (Figure 5B) to examine the effects of phase behavior and choice of lipid labeling scheme on recombination efficiency. Three main conclusions can be drawn from the data, displayed in Figure 5B. First, for both nano- and microscale liquid/liquid phase separation, the magnitude of the change in recombination efficiency is greater when the 13C and 15N atoms are separated into different phases (i.e., 13C3-DSPC and 15N-POPC (composition B) or 15N-DOPC (composition D)) than when they are in the same phase (i.e., 13C3-DSPC and 15N-DSPC (composition A)). Second, the nanoscale phase separating mixtures have smaller magnitudes of changes in recombination efficiency versus the microscale phase separating mixtures (composition B versus D). These mixtures have different domain sizes, and the compositions of the domains vary somewhat. This result is likely a convolution of effects from domain size and domain composition. Reducing the size of the domains from microscale to nanoscale increases the perimeter (of the interface between domains) to the surface area ratio of the liquid-ordered domains. At the interface between the phases, DSPC is close to POPC and recombination can occur between atoms in DSPC and POPC. Thus, we should observe more recombination between 13C3-DSPC and 15N-POPC than between 13C3-DSPC and 15N-DOPC. The Lo phase in compositions A and B contains more POPC than the Lo phase in composition D contains DOPC.39 These compositional differences may also explain the differences in recombination observed for nano- and microscale phase separating mixtures. While nanodomains in vesicles of similar lipid mixtures have previously been measured by SANS and FRET to be ~15 nm in diameter, the size of domains in SLBs is unknown.40 Without more information or simulations, we cannot deconvolute these effects. Finally, the magnitude of the change in recombination efficiency for labeled DSPC molecules in the gel phase is greater than that for DSPC molecules in liquid ordered phases (composition C versus A). This also makes sense because DSPC has a greater local concentration in the gel phases than in liquid ordered phases, which also contain cholesterol. These results provide some guidelines for designing future atomic recombination experiments in lipid bilayers and a framework for interpreting the results.
Detection of Nonrandom Lipid Distribution below the Imaging Resolution of NanoSIMS
In this work, we establish the distance range over which recombination occurs in model membranes and establish a framework for using atomic recombination to assess the homogeneity or heterogeneity of the lateral distribution of lipids in model membranes. Figure 3C demonstrates that atomic recombination under these experimental conditions occurs when 13C and 15N atoms are within approximately 3 nm of each other, and the resulting changes in ℜ(13C15N−) are easily resolvable with the good signal/noise of the NanoSIMS. See Supporting Information section 10 for a comparison of the distance dependence of atomic recombination in different systems.
There has been much debate about the nanoscale lateral organization of the plasma membrane of cells, limited by methods for directly studying organization on that length scale. FRET, diffusion, NMR, SAXS, SANS, and ESR have been used to provide evidence for the existence of nanodomains in model membranes.39,41–45 The experiments reported here are analogous in design to the FRET experiments of Heberle et al., in which FRET probes that partition into different phases are used to reveal compositions with nanoscale phase separation.39 An important difference is that we are detecting the interactions between the molecules of interest, not between dye molecules that may behave unexpectedly in membranes.5 We note that this method does not distinguish between true phase behavior and nonideal mixing; however, we refer to the nanodomains as phases because they appear to be true thermodynamic phases in FRET experiments.33,46 By varying the identity of the lipids to which the 13C and 15N atoms are attached, we show that nanodomains are enriched in DSPC, while POPC is excluded. These results are consistent with published phase diagrams inferred from FRET experiments but can easily be extended to more complex lipid mixtures for which there are no phase diagrams. Similarly, atomic recombination is complementary to SANS studies and will likely find unique applications, as no solvent contrast matching or complex data fitting is necessary. Additionally, atomic recombination need not rely on published phase diagrams for modeling. Since only two components need to be labeled (e.g., DSPC and POPC), other unlabeled lipid or protein components could be added to the bilayers to determine their effect on nanoscale phase separation or, in the case of a protein, the proximity of a lipid to the protein.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institutes of Health (GM069630 and GM118044), the National Science Foundation Biophysics Program MCB-1408785, and Center for Molecular Analysis and Design (graduate fellowship to F.R.M.). The Cameca NanoSIMS 50L at the Stanford Nano Shared Facilities is supported by the National Science Foundation (Award 0922648). We thank Charles Hitzman of the Stanford Nano Shared Facilities for assistance with the NanoSIMS 50L. Mass spectrometry was performed at the Vincent Coates Foundation Mass Spectrometry Laboratory, Stanford University Mass Spectrometry (http://mass-spec.stanford.edu).
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Choice of secondary ions as a chemical ruler; NanoSIMS high resolution mass spectra; compositional analysis of supported monolayers and bilayers; calculating limiting cases for recombination in monolayers; comparison of recombination efficiency in DOPC monolayers and bilayers; recombination in DOPC monolayers deposited at different surface pressures; modeling intermolecular distances in monolayers; choice of lipid compositions; fluorescence and NanoSIMS images of supported monolayers and bilayers; comparison of recombination in monolayers and bilayers (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 2.Huang B, Babcock H, Zhuang X. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sydor AM, Czymmek KJ, Puchner EM, Mennella V. Trends Cell Biol. 2015;25:730–748. doi: 10.1016/j.tcb.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Swulius MT, Jensen GJ. J. Bacteriol. 2012;194:6382–6386. doi: 10.1128/JB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes LD, Rawle RJ, Boxer SG. PLoS One. 2014;9:e87649. doi: 10.1371/journal.pone.0087649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eggeling C, Ringemann C, Medda R, Schwarzmann G, Sandhoff K, Polyakova S, Belov VN, Hein B, von Middendorff C, Schonle A, Hell SW. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 7.Honigmann A, Mueller V, Hell SW, Eggeling C. Faraday Discuss. 2013;161:77–89. doi: 10.1039/c2fd20107k. [DOI] [PubMed] [Google Scholar]
- 8.Aussenac F, Tavares M, Dufourc EJ. Biochemistry. 2003;42:1383–1390. doi: 10.1021/bi026717b. [DOI] [PubMed] [Google Scholar]
- 9.Veatch SL, Polozov IV, Gawrisch K, Keller SL. Biophys. J. 2004;86:2910–2922. doi: 10.1016/S0006-3495(04)74342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobson K, Mouritsen OG, Anderson RG. Nat. Cell Biol. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- 11.Bae W, Choi MG, Hyeon C, Shin YK, Yoon TY. J. Am. Chem. Soc. 2013;135:10254–10257. doi: 10.1021/ja404276g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rasnik I, McKinney SA, Ha T. Acc. Chem. Res. 2005;38:542–548. doi: 10.1021/ar040138c. [DOI] [PubMed] [Google Scholar]
- 13.Nietlispach D, Mott HR, Stott KM, Nielsen PR, Thiru A, Laue ED. Methods Mol. Biol. 2004;278:255–288. doi: 10.1385/1-59259-809-9:255. [DOI] [PubMed] [Google Scholar]
- 14.Hong M, Schmidt-Rohr K. Acc. Chem. Res. 2013;46:2154–2163. doi: 10.1021/ar300294x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacques DA, Trewhella J. Protein Sci. 2010;19:642–657. doi: 10.1002/pro.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanchet CE, Svergun DI. Annu. Rev. Phys. Chem. 2013;64:37–54. doi: 10.1146/annurev-physchem-040412-110132. [DOI] [PubMed] [Google Scholar]
- 17.Boxer SG, Kraft ML, Weber PK. Annu. Rev. Biophys. 2009;38:53–74. doi: 10.1146/annurev.biophys.050708.133634. [DOI] [PubMed] [Google Scholar]
- 18.McMahon G, Saint-Cyr HF, Lechene C, Unkefer CJ. J. Am. Soc. Mass Spectrom. 2006;17:1181–1187. doi: 10.1016/j.jasms.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 19.Marletta G. Nucl. Instrum. Methods Phys. Res., Sect. B. 1990;46:295–305. [Google Scholar]
- 20.Venkatesan T. Nucl. Instrum. Methods Phys. Res., Sect. B. 1985;7–8:461–467. [Google Scholar]
- 21.Briggs D, Hearn M. Vacuum. 1986;36:1005–1010. [Google Scholar]
- 22.Taylor JA, Lancaster GM, Ignatiev A, Rabalais JW. J. Chem. Phys. 1978;68:1776. [Google Scholar]
- 23.Legent G, Delaune A, Norris V, Delcorte A, Gibouin D, Lefebvre F, Misevic G, Thellier M, Ripoll CJ. Phys. Chem. B. 2008;112:5534–5546. doi: 10.1021/jp7100489. [DOI] [PubMed] [Google Scholar]
- 24.Kraft ML, Weber PK, Longo ML, Hutcheon ID, Boxer SG. Science. 2006;313:1948–1951. doi: 10.1126/science.1130279. [DOI] [PubMed] [Google Scholar]
- 25.Lozano MM, Liu Z, Sunnick E, Janshoff A, Kumar K, Boxer SG. J. Am. Chem. Soc. 2013;135:5620–5630. doi: 10.1021/ja310831m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girard-Egrot AP, Loïc J. Langmuir-Blodgett Technique for Synthesis of Biomimetic Lipid Membranes. In: Martin DK, editor. Nanobiotechnology of Biomimetic Membranes. Springer: New York: 2007. [Google Scholar]
- 27.Tamm LK, McConnell HM. Biophys. J. 1985;47:105–113. doi: 10.1016/S0006-3495(85)83882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cremer PS, Boxer SG. J. Phys. Chem. B. 1999;103:2554–2559. [Google Scholar]
- 29.Stottrup BL, Stevens DS, Keller SL. Biophys. J. 2005;88:269–276. doi: 10.1529/biophysj.104.048439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiu S, Subramaniam S, Jakobsson E. Biophys. J. 1999;76:1929–1938. doi: 10.1016/S0006-3495(99)77352-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kučerka N, Tristram-Nagle S, Nagle JF. J. Membr. Biol. 2006;208:193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 32.Nagle JF, Tristram-Nagle S. Biochim. Biophys. Acta. Rev. Biomembr. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konyakhina TM, Wu J, Mastroianni JD, Heberle FA, Feigenson GW. Biochim. Biophys. Acta, Biomembr. 2013;1828:2204–2214. doi: 10.1016/j.bbamem.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feigenson GW. Biochim. Biophys. Acta, Biomembr. 2009;1788:47–52. doi: 10.1016/j.bbamem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ionova IV, Livshits VA, Marsh D. Biophys. J. 2012;102:1856–1865. doi: 10.1016/j.bpj.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pathak P, London E. Biophys. J. 2011;101:2417–2425. doi: 10.1016/j.bpj.2011.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao J, Wu J, Heberle FA, Mills TT, Klawitter P, Huang G, Costanza G, Feigenson GW. Biochim. Biophys. Acta, Biomembr. 2007;1768:2764–2776. doi: 10.1016/j.bbamem.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heftberger P, Kollmitzer B, Rieder AA, Amenitsch H, Pabst G. Biophys. J. 2015;108:854–862. doi: 10.1016/j.bpj.2014.11.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heberle FA, Wu J, Goh SL, Petruzielo RS, Feigenson GW. Biophys. J. 2010;99:3309–3318. doi: 10.1016/j.bpj.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heberle FA, Petruzielo RS, Pan J, Drazba P, Kučerka N, Standaert RF, Feigenson GW, Katsaras J. J. Am. Chem. Soc. 2013;135:6853–6859. doi: 10.1021/ja3113615. [DOI] [PubMed] [Google Scholar]
- 41.Goh SL, Amazon JJ, Feigenson GW. Biophys. J. 2013;104:853–862. doi: 10.1016/j.bpj.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown AC, Wrenn SP. Langmuir. 2013;29:9832–9840. doi: 10.1021/la401249m. [DOI] [PubMed] [Google Scholar]
- 43.Nickels JD, Cheng X, Mostofian B, Stanley C, Lindner B, Heberle FA, Perticaroli S, Feygenson M, Egami T, Standaert RF, Smith JC, Myles DA, Ohl M, Katsaras J. J. Am. Chem. Soc. 2015;137:15772–15780. doi: 10.1021/jacs.5b08894. [DOI] [PubMed] [Google Scholar]
- 44.Silvius JR. Biophys. J. 2003;85:1034–1045. doi: 10.1016/S0006-3495(03)74542-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marquardt D, Heberle FA, Nickels JD, Pabst G, Katsaras J. Soft Matter. 2015;11:9055–9072. doi: 10.1039/c5sm01807b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feigenson GW. Nat. Chem. Biol. 2006;2:560–563. doi: 10.1038/nchembio1106-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veatch SL, Keller SL. Biophys. J. 2003;85:3074–3083. doi: 10.1016/S0006-3495(03)74726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.