Abstract
Acinetobacter sp. strain ADP1 is a nutritionally versatile soil bacterium closely related to representatives of the well-characterized Pseudomonas aeruginosa and Pseudomonas putida. Unlike these bacteria, the Acinetobacter ADP1 is highly competent for natural transformation which affords extraordinary convenience for genetic manipulation. The circular chromosome of the Acinetobacter ADP1, presented here, encodes 3325 predicted coding sequences, of which 60% have been classified based on sequence similarity to other documented proteins. The close evolutionary proximity of Acinetobacter and Pseudomonas species, as judged by the sequences of their 16S RNA genes and by the highest level of bidirectional best hits, contrasts with the extensive divergence in the GC content of their DNA (40 versus 62%). The chromosomes also differ significantly in size, with the Acinetobacter ADP1 chromosome <60% of the length of the Pseudomonas counterparts. Genome analysis of the Acinetobacter ADP1 revealed genes for metabolic pathways involved in utilization of a large variety of compounds. Almost all of these genes, with orthologs that are scattered in other species, are located in five major ‘islands of catabolic diversity’, now an apparent ‘archipelago of catabolic diversity’, within one-quarter of the overall genome. Acinetobacter ADP1 displays many features of other aerobic soil bacteria with metabolism oriented toward the degradation of organic compounds found in their natural habitat. A distinguishing feature of this genome is the absence of a gene corresponding to pyruvate kinase, the enzyme that generally catalyzes the terminal step in conversion of carbohydrates to pyruvate for respiration by the citric acid cycle. This finding supports the view that the cycle itself is centrally geared to the catabolic capabilities of this exceptionally versatile organism.
INTRODUCTION
Acinetobacter spp. are widespread in nature and can be obtained from water, soil and living organisms. These gram-negative bacteria are oxidase-negative, non-motile, strictly aerobic and tend to be paired non-motile cocci, rather than the typical monoflagellate rods. They can use various carbon sources for growth and can be cultured on relatively simple media. Using cell shape, absence of flagella, G+C content of DNA and nutritional properties, Baumann (1) classified these organisms in the genus Moraxella, now known as Acinetobacter, and designed procedures for specific selection of Acinetobacter strains from the environment (2). The members of the Acinetobacter group are nutritionally versatile chemoheterotrophs: the range of substrates used as sole carbon and energy sources parallels that of the aerobic Pseudomonads (3). Species of Acinetobacter have been attractingly growing interest in both environmental and biotechnological applications: they are known to be involved in biodegradation of a number of different pollutants and in the extra- and intracellular production of a number of economically valuable products (4).
Some representatives of the group, characterized by a relatively narrow nutritional spectrum, have achieved recent notoriety as agents of nosocomial infections (5,6). Others possess a rich genetic repertoire adapting them to the natural environment where they are found in abundance. Much of this biochemical biodiversity is generated by plants that introduce a large variety of chemicals into the environment. Ecological balance is achieved by these soil microorganisms which remove the chemicals by using them as growth substrates.
Unique among this highly heterogeneous bacterial genus (7) is strain ADP1, a soil bacterium, exceptionally competent for natural transformation (8). From a strain called BD4 (BD stands for butanediol) isolated by Taylor and Juni (9–11) on a mineral medium with meso 2,3-butanediol as the sole carbon source, a mutant called BD413 was obtained by ultraviolet irradiation which was later called ADP1. The ease with which it can be transformed offers specific advantages for genetic analysis (90). In addition, laboratory investigations are facilitated by the rapid growth of strain ADP1 with doubling times <1 h allowing a single bacterium to grow to a colony overnight using a variety of substrates. This Acinetobacter ADP1 system makes it possible to easily recover genes with biodegradative functions from other microorganisms.
Robust physiological properties and simple genetic manipulation led us to choose Acinetobacter sp. ADP1 as a good candidate for the determination of the complete genome sequence. In addition to its interest for comparative genomics with closely related bacteria, such as Pseudomonas species and Escherichia coli, it affords an opportunity for analysis and manipulation of genes and gene products, and paves the way for a future analysis of metabolic transformations in the environment.
MATERIALS AND METHODS
Sequencing, assembly and quality control
Two libraries of 3–4 kb inserts (A, obtained by mechanical shearing) and 20 kb inserts (B, obtained by enzymatic digestion) were constructed into pcDNA2.1 (INVITROGEN) and pBeloBAC11 (CALTECH) (both modified), respectively. Plasmid DNAs were purified and end-sequenced using dye-primer [25974 (A), 5.3-fold coverage] and dye-terminator chemistries [18286 (B), 2.5-fold coverage] on LICOR4200L and ABI3700 sequencers (12). The Phred/Phrap/Consed software package (www.phrap.com) (13–15) was used for sequence assembly and quality assessment. Seven large gap closures were obtained by long-range PCRs and subcloning. A total of 4610 additional reactions were necessary to close gaps and to raise the quality of the sequence to finished standards. The integrity of the assembly was confirmed by comparing the restriction profile of the sequence with a physical map (16).
Sequence annotation and comparative genomics
The first set of potential CoDing Sequences (CDSs) was identified using the AMIGene software (Annotation of MIcrobial Genes) (17) trained with a set of CDSs >500 bp from the genomic sequence. The multivariate statistical technique of factorial correspondence analysis and clustering methods were then applied to this set of predicted CDSs in order to derive multiple models taking into account the compositional diversity of genes within the Acinetobacter ADP1 genome. Three gene models were then subsequently used together in the core of AMIGene with minimum CDSs length set to 60 bp. This second set of putative genes (made of 3683 CDSs) was submitted to functional annotation: exhaustive BLAST (18) searches against the UniProt databank (19) were performed to determine homology. Protein motifs and domains were documented using the InterPro databank (20). In addition, genes coding for enzymes were classified using PRIAM software (21). TMHMM vs2.0 was used to identify transmembrane domains (22), and SignalP vs2.0 was used to predict signal peptide regions (23). Finally, tRNAs were revealed out by tRNAscan-SE (24). Intrachromosomal repeats were detected using the method described by Achaz et al. (25).
Sequence data for comparative analyses were obtained from NCBI databanks (ftp://ftp.ncbi.nlm.nih.gov/genbank/genomes/Bacteria). A total of 148 sequenced genomes were used in the analyses (Supplementary Table 1). Putative orthologs between Acinetobacter ADP1 and the 148 other genomes were defined as genes showing a minimum of 30% identity and a ratio of 0.8 of the length of the smallest protein. Orthology relations were strengthened by synteny detection (i.e. conservation of the chromosomal co-localization between pairs of orthologous genes from different genomes) using the Syntonizer software (L. Labarre et al., manuscript in preparation). Our method is not restricted to the bidirectional best hits (BBHs) definition, and thus allows for multiple correspondences between genes (fusion/fission, duplication). In addition, all possible kinds of chromosomal rearrangements are allowed (inversion, insertion/deletion). A ‘gap’ parameter, representing the maximum number of consecutive genes not involved in a synteny group, was set to five genes.
From these comparison results, we defined a specific gene as a gene having no ortholog in the compared specie (significant similarities were not detected in the compared specie and this gene is not in a synteny group, see above). This allowed us to compute the list of Acinetobacter ADP1 specific regions in comparison with the complete genomes of Pseudomonas species (Supplementary Table 2). Such regions are defined by at least two consecutive specific genes. Insertion of genes which have homologies in the compared specie is allowed in a specific region. A ‘gap’ parameter, representing the maximum number of consecutive genes with homologies, was set to two genes.
All the data (i.e. syntaxic and functional annotations, and results of comparative analysis) are stored in a relational database (using MySQL SGBDR software). Each predicted gene was assigned a unique numeric identifier prefixed with ‘ACIAD’. The first CDS from the origin of replication, the putative dnaA gene, was assigned as ACIAD0001, and each following CDS was numbered consecutively in a clockwise direction. Manual validation of the automatic annotation was performed using our web interface MaGe (Magnifying Genomes), which allows graphic visualization of the Acinetobacter ADP1 annotations enhanced by a synchronized representation of synteny groups found with compared genomes (D. Vallenet et al., manuscript in preparation). This cartographic representation was helpful to annotate genes taking into account their genomic neighborhood information. Predicted genes were examined visually and, in some cases, removed (‘Artefact’ status in the database) or clearly labeled as ‘doubtful CDS’ (small CDS, low coding probability and no BLAST hits). Translational start codons were corrected based on protein homology, proximity of ribosome-binding site and relative position to predicted signal peptides if these were present. For this purpose, the useful Artemis sequence Viewer (26) was used (an Artemis JAVA applet is directly launched from MaGe). The Acinetobacter ADP1 nucleotide sequence and annotation data have been deposited at EMBL databank under accession number CR543861. In addition, all these data are also available at (http://www.genoscope.cns.fr/agc/mage/wwwpkgdb/Login/log.php?dir=acinetopublic&login=guest).
RESULTS
General features of the genome
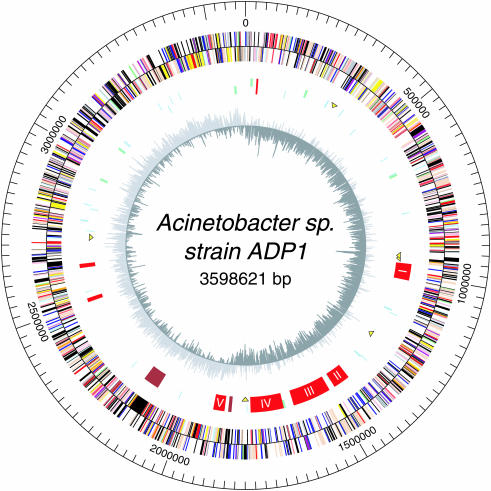
Acinetobacter ADP1 has a single, circular chromosome of 3 598 621 bp in length with an average G+C content of 40.3% (Figure 1 and Table 1). Interestingly, the close proximity of Acinetobacter and Pseudomonas species, as judged by the sequences of their 16S RNA genes (Supplementary Figure 3), contrasted with the extensive divergence in the GC content of their DNA (40 versus 62%). On the basis of the G+C skew analysis, we estimated that the site of termination of replication (terC) is ∼1.8 Mb from the probable origin of replication (oriC), which was mapped between the ribosomal protein rpmH (ACIAD3684) and dnaA (ACIAD0001) genes. The chromosome of strain ADP1 contains 3325 CDSs, on average 930 pb in size, which range from 69 bp (ACIAD2503, similar to pyrroloquinoline quinone biosynthesis protein A of the pqqABCDE operon) to 11 133 bp (ACIAD0940, similar to hemagglutinin/hemolysin-related protein). These CDSs cover 88.8% of the chromosome and show an asymmetry in their distribution between the leading strand (60%) and the lagging strand (40%). Seventy six tRNA species, representing all 20 amino acids, were identified. Of these tRNAs, two are organized in association with the seven annotated rRNA operons whose copies are strictly identical (16S-tRNA-Ile-Ala-23S-5S) (Figure 1 and Table 1). In addition, three other RNA motifs have been found using the Rfam databank (see Methods): a tmRNA at position 914 849 bp (known to act as a mRNA template for tagging proteins resulting from premature transcription termination in E.coli), the RNA component of RNase P at position 2 831 879 bp (rnpB) and a component of Signal Recognition Particle at position 1 380 047 bp [4.5S RNA (ffs) component of the ribonucleoprotein particle].
Figure 1.
Circular representation of the Acinetobacter ADP1 genome. Circles display (from the outside): (i) predicted coding regions transcribed in the clockwise direction. (ii) Predicted coding regions transcribed in the counterclockwise direction. Genes displayed in (i) and (ii) are color coded according to different functional categories: salmon, amino acid biosynthesis; light blue, biosynthesis of cofactors, prosthetic groups and carriers; light green, cell envelope; red, cellular processes; brown, central intermediary metabolism; yellow, DNA metabolism; green, energy metabolism; purple, fatty acid and phospholipid metabolism; pink, protein fate/synthesis; orange, purines, pyrimidines, nucleosides, nucleotides; blue, regulatory functions; grey, transcription; teal, transport and binding proteins; black, hypothetical and CHPs. (iii) Transposable elements (yellow triangles), phage regions (brown), genes involved in catabolic pathways (red); the five catabolic islands (>20 kb) are numbered from I to V. (iv) tRNAs (light blue) and rRNA (light green). (v) GC bias (G+C/G-C).
Table 1. Comparison of general features and functional categories between Acinetobacter sp. ADP1 and selected free-living organismsa.
| Acinetobacter ADP1 | PSAE | PSPU | PSSY | RALTO | ESCO | |
|---|---|---|---|---|---|---|
| General featuresb | ||||||
| Size (Mb) | 3.6 | 6.3 | 6.4 | 5.8 | 5.8 | 4.6 |
| GC% | 40.3 | 66.6 | 61.6 | 58.4 | 66.9 | 50.8 |
| Nb CDS | 3325 | 5567 | 5420 | 5615 | 5129 | 4273 |
| % Coding | 88.8 | 89 | 87.7 | 86.8 | 87.3 | 92 |
| rRNA operon | 7 | 4 | 7 | 5 | 4 | 7 |
| tRNA | 76 | 63 | 63 | 63 | 58 | 82 |
| Known and putative proteins (%) | 62.6 | 65.9 | 62.0 | 61.0 | 44.1 | 80.5 |
| CHP (%) | 20.3 | 22.0 | 23.0 | 28.0 | 41.3 | 12.3 |
| No homology (%) | 13.9 | 11.1 | 15.0 | 11.0 | 14.6 | 7.2 |
| TIGR categories (%)c | ||||||
| Amino acid biosynthesis | 3.52 | 1.65 | 2.16 | 1.81 | 1.01 | 2.09 |
| Biosynthesis of cofactors, prosthetic groups and carriers | 3.61 | 2.02 | 2.57 | 2.17 | 1.00 | 1.85 |
| Cell envelope | 3.97 | 2.54 | 5.58 | 6.55 | 6.73 | 3.16 |
| Cellular processes | 6.16 | 3.49 | 6.14 | 8.01 | 2.62 | 3.47 |
| Central intermediary metabolism | 3.49 | 2.08 | 1.34 | 1.49 | 1.64 | 1.34 |
| DNA metabolism | 2.16 | 1.33 | 2.01 | 2.27 | 2.42 | 1.87 |
| Energy metabolism | 2.97 | 5.29 | 7.81 | 5.47 | 5.73 | 6.78 |
| Fatty acid and phospholipid metabolism | 1.47 | 1.27 | 1.91 | 1.51 | 0.73 | 1.23 |
| Protein fate | 2.25 | 2.17 | 3.07 | 3.40 | 2.40 | 2.13 |
| Protein synthesis | 4.03 | 1.94 | 2.25 | 2.04 | 2.34 | 2.24 |
| Purines, pyrimidines, nucleosides and nucleotides | 0.96 | 0.95 | 1.10 | 0.97 | 0.58 | 1.41 |
| Regulatory functions | 6.97 | 4.69 | 9.13 | 7.45 | 4.55 | 3.23 |
| Transcription | 2.19 | 0.73 | 1.12 | 0.85 | 0.65 | 0.75 |
| Transport and binding proteins | 10.43 | 4.68 | 11.20 | 10.03 | 3.87 | 5.79 |
PSAE, Pseudomonas aeruginosa; PSPU, Pseudomonas putida; PSSY, Pseudomonas syringae; RALTO, Ralstonia solanacearum; ESCO, Escherichia coli.
aThese bacteria have been selected for their close proximity to Acinetobacter sp. ADP1 in terms of lineage or in proportion of BBHs.
bThe data have been extracted from the original papers as far as P.putida (30), P.syringae (31) and R.solanacearum (86) are concerned, and from the re-annotation papers for E.coli (87) and P.aeruginosa (88).
cOnly TIGR's categories which are common to the six species are listed. Values have been extracted from the Comprehensive Microbial Resource database (89) for PSAE, PSPU, PSSY, RALSO and ESCO. As far as Acinetobacter sp. ADP1 is concerned, we made a manual correspondence between Monica Riley's classification (Multifun) and the TIGR's one.
Genes for the primary replication machinery, the DNA polymerase III complex in Acinetobacter ADP1, are similar to those of comparable components in the Pseudomonas lineage. Interestingly, the gene encoding dnaQ function (i.e. the N-terminal part of the protein encoded by ACIAD1137) is also highly similar, in the first part of its sequence, to RNase HI which is known to remove RNA primers from the Okazaki fragments of lagging strand synthesis (50% identity with the rhnA gene from Pseudomonas species). The dnaQ gene codes for the epsilon-subunit of the DNA polymerase III complex and plays a crucial role in fidelity of replication as it encodes the 3′→5′ proofreading exonuclease activity. Although these two genes, dnaQ and rnhA, are very often co-localized in a larger synteny group (mainly in proteobacteria), such a gene fusion is, to date, unique to Acinetobacter ADP1. Three RNA polymerase core-enzyme (rpoA, ACIAD3194, rpoB, ACIAD0307 and rpoC, ACIAD0308) and a gene that encoded polymerase subunit omega (rpoZ, ACIAD3325) are readily defined by sequence homology. Fifteen sigma factors and additional transcription-related factors, such as elongation factors (gre genes), the rho factor and termination factors (nus genes), are all unambiguously recognized. In addition, Acinetobacter ADP1 has about 180 transcriptional regulators acting as activators or repressors involved in many physiological and metabolic pathways. Finally, all translation-related genes are highly conserved as seen in other procaryotes and shared by both Eubacteria and Archea. Twenty genes that encode twenty essential tRNA synthetases are predicted together with the three genes for the Gln/Asn transaminases (ACIAD0822 to 0824).
The Acinetobacter ADP1 genome has a small fraction of repetitive sequences (1.6%) ranging from a few short repeats to several complex ones such as clusters encoding transposases or strong paralogs. Six copies of Insertion Sequence IS1236, a member of the IS3 family, have been identified on the chromosome of Acinetobacter ADP1 (27). The longest repeated sequence is composed of two IS1236 copies flanking a Hypothetical Protein (HP, unique to this bacterium to date), and is known as transposon Tn5613 (27). It is found at position 941 306 not far from the vanillate operon (starting with ACIAD0978, vanR, at position 964 806) where the transposon and the insertion sequence cause much genetic scrambling (28). The G+C content of IS1236 and Tn5613 is in the 36–39% range, making them very Acinetobacter-like, and suggesting that they do not contribute greatly to Horizontal Gene Transfer (HGT). The proportion of ADP1 Insertion Sequences (0.5%) is close to that found in Pseudomonas aeruginosa (0.2%) (29), but contrasts with those found in P.putida (9.4%) (30) and P.syringae (7%) (31). Most of the other repeats are localized close to putative phage-associated genes. Genome analysis revealed two main prophage regions. The longest one (53 kb in size), located at 2117 kb, is flanked by a repeat (485 bp in length) on the left side, and by a transposase and a tRNA gene on the right side. Among the 64 identified genes in this prophage, 70.3% are unique to Acinetobacter ADP1 and 18.7% are similar to genes of unknown function [Conserved Hypothetical Proteins (CHPs) mainly from bacteriophage VT2-Sa]. The remaining genes (11%) share functional annotation with phages from Xylella fastidiosa and Pseudomonas putida, in particular to proteins involved in DNA replication. In addition, one syntenic region is shared with the X.fastidiosa prophage region, and includes two genes orthologous to ‘proteic killer’ proteins (ACIAD2181 and ACIAD2182). Whether or not this Acinetobacter genomic region is a functional prophage is not known. The second prophage region, located at 1.8 Mb, is 9 kb in size and its sequence is similar to that of a filamentous bacteriophage of P.aeruginosa (Pf3). This region is delimited by two complex repeats involving five CDSs; the longest one is similar to a phage replication initiation factor. The Acinetobacter ADP1 prophagic regions are included in the two longest genomic regions that are highly specific to ADP1 compared with genomes of the three complete Pseudomonas species (Supplementary Table 2). The first one (ACIAD2108 to 2200) is 71 412 bp in length and contains genes which have no similarity in Pseudomonas species, apart from the two genes orthologous to ‘protein killer’ proteins (a similarity has been found in P.syringae and P.putida). The second one (ACIAD1835 to 1865) is 18 831 bp in length and is entirely specific to Acinetobacter ADP1 (Supplementary Table 2).
Ortholog and synteny analysis
Of the 3325 identified protein coding genes (Table 1), a probable biological function, based on a classification scheme adapted from Riley (32,33), has been assigned for more than 62.6% of the CDSs (35% with a definitive assignment and 27.6% with a putative role assignment). In addition, 20.3% were identified as CHPs, of which 28% have domains and/or motifs contained within public databanks. Finally, 13.9% of the CDSs were designated HPs. The remaining CDSs (3.2%) have been annotated as ‘Doubtful CDSs’ and ‘Gene remnant’.
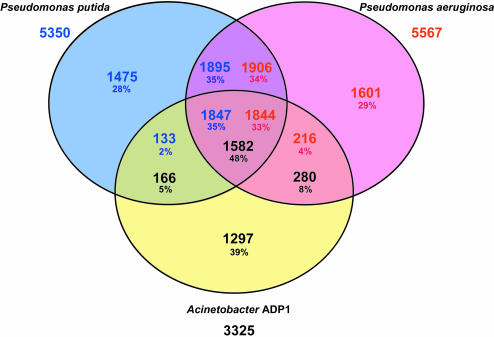
Consistent with 16S rRNA-based phylogeny, the proteomes of Pseudomonas species are the closest to Acinetobacter ADP1 (Table 1). ADP1 CDSs share more than 63% BBHs with P.aeruginosa, a free-living bacterium which causes opportunistic infection in humans (29), and about 35% of ADP1 CDSs have been found in synteny groups with this organism only (Supplementary Figure 1). Transporter functions (Table 1) and enzymatic functions (a total of 34.6%) are clearly the most abundant in the Acinetobacter ADP1 genome, in agreement with the fact that, like Pseudomonads, it is engaged in major metabolic activities related to its natural environment (see below). Gene content comparisons have been performed between the genome of ADP1 and the three complete genomes of Pseudomonas species. Apart from the core of common orthologous genes between Acinetobacter ADP1 and the two well-known Pseudomonas species (Figure 2), few orthologous genes have been found between ADP1 and P.aeruginosa on the one hand, and between ADP1 and P.putida on the other hand (similar results were obtained with P.syringae genome; see Supplementary Figure 2). These genes are mainly involved in uncharacterized catabolic pathways and transport systems in ADP1 (Supplementary Table 2), or in transferred genomic regions shared by Acinetobacter ADP1 and one of the three other Pseudomonas species (mainly P.syringae; see below). Because that two Pseudomonads have been compared in the set of the three species (Figure 2), the number of specific genes is higher in Acinetobacter ADP1 than in P.aeruginosa and P.putida (39% versus 28% and 29%). In ADP1, these genes are mainly involved in the metabolism of various compounds (i.e. ester, nitrile), and in Pseudomonas species several specific secretion systems have been found (Supplementary Table 3). For example, the psc type III export system is specific to P.aeruginosa (PA1715 to PA1725), and the hrp type III secretion system is specific to P.syringae (PSPTO1381 to PSPTO1404).
Figure 2.
Gene content comparisons between Acinetobacter ADP1, P.aeruginosa and P.putida genomes. Putative orthologs are defined as genes showing a minimum of 30% identity and a ratio of 0.8 of the length of the smallest protein, or as two genes included in a synteny group. The intersections between the three circles give the number (and percentage) of genes found in the 2 or 3 compared species. Genes outside these areas are specific to the corresponding organism. The total number of annotated genes is also given under each specie name.
Acinetobacter ADP1, readily modified by natural transformation, is amenable to genetic manipulation because of its relatively small genome, <60% the size of the genomes of P.aeruginosa and P.putida (Table 1). Furthermore, ADP1 appears to be a far more attractive industrial microorganism because it lacks genes known to be associated with pathogenesis: virulence-related genes such as toxins, invasins and secretory systems are absent from the Acinetobacter ADP1 genome. However, it has 10 open reading frames encoding hemolysin-like proteins for which the role still remains puzzling.
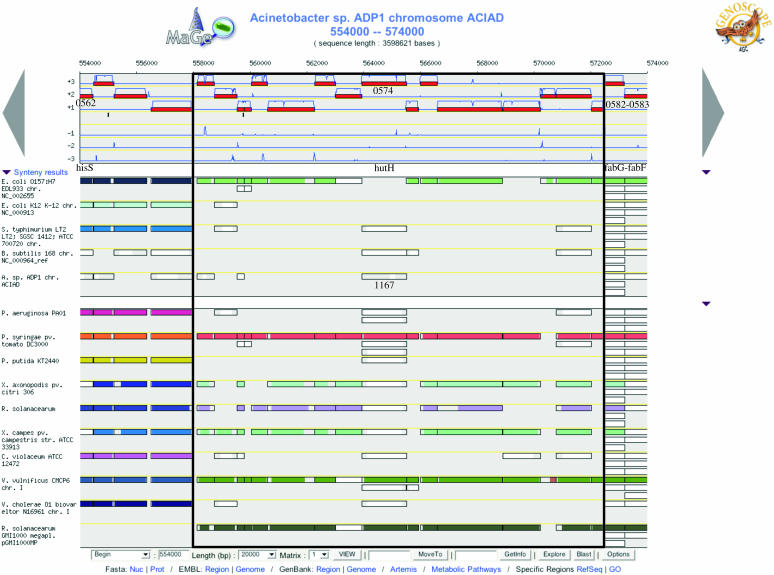
Comparison of the Acinetobacter ADP1 genome with chromosome and megaplasmid of Ralstonia solanacearum shows that this soil-borne phytopathogen is just behind the Pseudomonas species in genomic similarity to Acinetobacter ADP1 (Supplementary Figure 1 and Table 1). The niche occupied by Acinetobacter ADP1, a terrestrial environment associated with different plant sources, provides particularly favorable conditions for HGT. Kay and collaborators (34) have shown that plant environments can provide soil bacteria with opportunities to colonize new niches in which they could particularly have an active metabolism. In the case of Acinetobacter sp., conjugative transfer of a range of plasmids and transformation-mediated transfer of chromosomal genes appear to occur at significant frequencies during R.solanacearum plant colonization. These gene transfers are related to the extensive multiplication of these two bacteria in plants and the development of a competence state in Acinetobacter (35). Evidence that HGT probably occurred is presented in Figure 3, which shows an Acinetobacter ADP1 chromosomal region of 16 kb (from ACIAD0566 to ACIAD0581) delimited by very well conserved synteny groups. Annotated genes from this region are mainly described as CHPs (52%) and acyl carrier proteins (33%). In addition, ACIAD0574 is highly similar to histidine ammonia lyase (hutH gene which is involved in the first step of histidine degradation) from pathogens, such as Vibrio parahaemolyticus and V.vulnificus, Pseudomonas syringae and P.fluorescens, Shewanella oneidensis and the R.solanacearum megaplasmid sequence (60% identity). Although this portion of the Acinetobacter ADP1 chromosome is very well conserved in these bacteria, it is clearly absent in E.coli K12, Bacillus subtilis, Mycobacterium tuberculosis, P.aeruginosa, P.putida and Vibrio cholerae as well as Chromobacterium violaceum, which is a free-living microorganism that populates the soil and water in tropical regions (Figure 3 and Supplementary Figure 3). Intriguingly, ACIAD0574 also shows significant similarities with histidase from Eucaryotes, mainly Homo sapiens, Rattus norvegicus, Mus musculus and Gallus gallus (E-value = e−72; identity = 33%), re-inforcing the probable exogenous nature of this function. In contrast, tiny co-localized fragments of hutH (ACIAD1167), hutU (urocanase hydratase; ACIAD1166) and hutG (formiminoglutamase; ACIAD1169) are highly similar to the corresponding P.aeruginosa genes, suggesting that deletions of large portions of the Acinetobacter ADP1 genes (without loss gene arrangement in synteny) were recent events in the evolutionary history of this organism.
Figure 3.
Part of the Acinetobacter ADP1 genome containing a likely horizontally transferred region. The chromosomal segment, extending between positions 553 000 and 583 000 bp, is represented on this graphical map of the MaGe interface developed on our database. Annotated CDSs are represented in the six reading frames of the sequence by red rectangles, and coding prediction curves are superimposed on the predicted CDSs (blue curves). The synteny maps, calculated on a set of selected genomes in the NCBI databank, are displayed below. In contrast to the graphic interface of the Acinetobacter ADP1 genome, there is no notion of scale on the synteny map: a rectangle has the same size of the CDS which is exactly opposite in the ADP1 genome, and it represents a putative ortholog between one CDS of the compared genome and one CDS of the Acinetobacter ADP1 genome. If, for several CDSs co-localized on the ADP1 genome, there are several co-localized orthologs in the compared genome, the rectangles will all be of the same color; otherwise, the rectangle is white. A group of rectangles of the same color thus indicates synteny between Acinetobacter ADP1 and the compared genome. The compared ADP1 chromosomal region is not found in E.coli K12, P.aeruginosa and P.putida genomes. In contrast, very well conserved syntenies are found with P.syringae, V.parahaemolyticus, V.vulnificus and the R.solanacearum megaplasmid.
Finally, more than 60% of the Acinetobacter ADP1 genes have been found in synteny groups with the 145 genomes with which it was compared (see Methods). This result allowed us to perform functional annotation using gene context and was particularly relevant in cases of low similarity results.
Energy production
Acinetobacter ADP1 can grow on glucose as its sole carbon source. However, genes coding for a glucokinase, a hexokinase and a glucose transporter phosphotransferase system (PTS) were not detected. Presumably, ADP1 cannot directly phosphorylate glucose. Furthermore, in accordance with earlier biochemical studies (9–11), genes encoding 6-phosphofructokinase and pyruvate kinase are missing, whereas the latter enzyme is present in P.aeruginosa. This confirms that ADP1 uses the Entner–Doudoroff (ED) pathway instead of the Embden–Meyerhof–Parnas pathway for glucose dissimilation. In contrast to P.aeruginosa and P.putida, ADP1 does not encode genes for a glucose-6-phosphate dehydrogenase and for a 6-phosphogluconolactonase. The only apparent way this organism can utilize glucose is by a periplasmic oxidation via a membrane-bound glucose dehydrogenase. Glucose oxidation is followed by gluconate or 2-keto-gluconate uptake and metabolism by the ED pathway. This pathway leads to pyruvate and glyceraldehyde 3-phosphate formation. Enzymes for this gluconate conversion are encoded by five genes which are clustered together on the bacterial genome (ACIAD0542 to ACIAD0546).
In contrast to glycolysis, gluconeogenesis is complete and allows the formation of fructose-6-phosphate and glucose-6-phosphate, precursors for cell surface polysaccharide synthesis and for the pentose phosphate pathway.
The genes coding for the enzymes of the tricarboxylic acid cycle (TCA) are all present. Acinetobacter ADP1 possesses two isocitrate dehydrogenases (ACIAD1187 and ACIAD1190), 743 and 436 residues long, respectively, which present no significant similarities. This situation is found in several Pseudomonads. Isocitrate dehydrogenase kinase/phosphatase, which determines the active (dephosphorylated) or inactive (phosphorylated) state of isocitrate dehydrogenase and the deviation of the carbon flow to the anaplerotic glyoxylic acid cycle, could not be detected. Although malate synthase A could not be found, the second characteristic enzyme of the anaplerotic cycle, isocitrate lyase (ACIAD1084), is present as evidenced by the high degree of similarity with the authenticated Hyphomicrobium methylovorum enzyme (36). How the existence of the two differentially regulated isocitrate dehydrogenases (37) is related to the possible entry of acetyl CoA into the glyoxylic acid shunt via malate synthase G (in the absence of malate synthase A) remains an open question.
Aerobic life
As expected, Acinetobacter ADP1, a strict aerobic organism, is devoid of the fnr gene, coding for the global regulator Fnr, which regulates over 70 genes involved in cellular adaptations to growth into an anoxic environment in E.coli. The principal elements of the regulation in aerobically grown E.coli cells, mainly the enzymes of the TCA, are encoded by the arcA and arcB genes (arc stands for aerobic respiration control) and are members of a two-component signal transduction system (38). Most two-component systems have very similar sequences and are difficult to assign to a particular function on the sole basis of their sequence. In ADP1, the exact assignation of arcA and arcB genes is not clear. In any case, the absence of ArcA should not be surprising, since this protein acts primarily as a transcriptional repressor of genes whose products are involved in aerobic metabolism (39).
The toxic superoxide radical and hydrogen peroxide can be respectively eliminated by superoxide dismutase and by catalase. In Acinetobacter ADP1, as in E.coli, three superoxide dismutases are found, a probable periplasmic one from the Fe/Mn family (ACIAD1070), an MnSod (ACIAD2072) and a FeSod (ACIAD1346). Although E.coli is endowed with two catalases, we could find only one in ADP1, corresponding to E.coli hydroperoxidase II (ACIAD0451). On the other hand, equivalents of the two subunits of the E.coli superoxide stress regulon oxyR are present, each as two ‘paralogous’ genes (ahpC as ACIAD0436 and ACIAD2103; ahpF as ACIAD1234 and ACIAD2250).The oxyR gene itself, which activates the ahp genes finds its counterpart in ACIAD1063. The oxyS gene, which codes for a small antisense regulatory RNA which represses rpoS (sigma 58) translation in E.coli in response to oxidative shock (40), has not been found in the Acinetobacter ADP1 genome. The rpoS gene could not be found either.
Biosynthesis
Nitrate and nitrite assimilation
The genes coding for proteins involved in nitrate assimilation (reduction of nitrate to ammonia, biosynthesis of the cofactor molybdopterin guanine dinucleotide, transport and transcription regulation) form a cluster (ACIAD1899 to ACIAD1911), corresponding to one synteny group with P.aeruginosa and P.syringae, and two synteny groups with P.putida. Interestingly, one gene in this region (ACIAD1909) is in synteny with two genes of R.solanacearum megaplasmid. These genes are annotated as nitrite reductase (small subunit) and putative FAD-dependent oxidoreductase. In addition, ACIAD1909 is flanked by genes identified as the large subunits of nitrate and nitrite reductases. Based on this fact, and further sequence analysis, we assume that ACIAD1909 is probably a gene fusion product encoding the small subunits of both nitrate and nitrite reductase. It is finally worth noting that ACIAD1909 has no counterpart in the synteny with P.aeruginosa and P.putida, whereas an orthologous gene with P.syringae, showing similarity only in the N-terminal part of ACIAD1909 (small subunit of nitrate reductase), has been identified.
Sulfate assimilation
Although the genes coding for the two subunits of ATP sulfurylase are present in Acinetobacter ADP1 (ACIAD1072 and ACIAD1073), as in Enterobacteriaceae, we could not detect a gene corresponding to APS kinase. In the absence of PAPS, we assume that APS is directly reduced via the protein coded by ACIAD0833. Although PAPS and APS reductases share 25–30% identical amino acids, ACIAD0833 contains the CCXXRKXXPL and SXGCXXCT motifs characteristic of APS reductases found in plant and other bacterial species (41).
Amino acid biosynthesis
Although Enterobacteriaceae possess three aspartate kinases and two homoserine dehydrogenases which catalyze the same reactions, but differ in the regulation of their activity and synthesis (42,43), Acinetobacter ADP1 has only one gene coding for aspartate kinase (ACIAD1252) and one for homoserine dehydrogenase (ACIAD0264), as in Pseudomonads (44). The details of these regulations deserve to be studied in ADP1. Methionine biosynthesis follows the pattern observed in P.aeruginosa, utilizing O-acetylhomoserine instead of O-succinylhomoserine, and obtaining homocysteine by direct sulfhydrylation of O-acetylhomoserine (45). Apart from these differences, the synthesis of threonine, lysine, methionine and isoleucine follows the pattern of E.coli.
All the enzymes for glutamate and glutamine synthesis, including those ensuring the regulation of glutamine synthetase activity (through covalent modification) and synthesis, are present in Acinetobacter ADP1. The same is true for proline and arginine biosyntheses.
It is interesting to note that S-adenosylmethionine decarboxylase and spermidine synthetase are not found. On the other hand, members of the genus Acinetobacter produce 1,3-diaminopropane in large amounts, via the successive action of aspartate semialdehyde-glutamate transaminase and of 2,4-diaminobutyrate decarboxylase, enzymes that have been characterized in Acinetobacter baumanii (46,47). The genes coding for these two enzymes are present in Acinetobacter ADP1 (ACIAD1210 and ACIAD1211). Also present are agmatine ureohydrolase (ACIAD1299), and putative ornithine and lysine decarboxylases (ACIAD2768 and ACIAD3341), which should produce putrescine and cadaverine. One should, however, note that these diamines are barely detected in A.baumanii grown on a synthetic medium (47).
Although three genes coding for 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase are found in E.coli, only two Acinetobacter ADP1 genes could correspond to this enzyme (ACIAD2330 and ACIAD1878). For each enzyme that has been characterized in E.coli for histidine, phenylalanine, tyrosine and tryptophan biosyntheses, a corresponding gene has been found in ADP1. However, no orthologs to tyrA gene has been found in ADP1. Actually, the dehydrogenase activity of TyrA (a bifunctional enzyme) is fused with AroA, the 3-phosphoshikimate 1-carboxyvinyltransferase (ACIAD2222). Synteny results show that this interesting fusion is also present in P.aeruginosa (PA3164) and P.putida (PP1770) but not in P.syringae in which the ortholog to ACIAD2222 (PSPTO1748) aligns only on the C-terminal part of the protein (TyrA). The chorismate mutase activity of TyrA is probably compensated by the product of pheA (ACIAD2223). Whether the alternative arogenate pathway of phenylalanine and tyrosine is present in ADP1 cannot be determined on the basis of the available data. In P.aeruginosa, the two pathways have been shown to coexist (48).
Nucleic acid building blocks
An equivalent of E.coli pyrA, coding for the regulatory subunit of aspartate carbamoyltransferase, could not be found. This absence may be compensated, as it is in P.aeruginosa (49), by a gene coding for a putative dihydroorotase (ACIAD1419). Interestingly, the catalytic subunit of the aspartate carbamoyltransferase coded by pyrB is present (ACIAD1270). All the enzymes of de novo purine/pyrimidine biosynthesis are also present in Acinetobacter ADP1.
As expected, the strictly aerobic Acinetobacter ADP1 lacks the anaerobic nucleoside triphosphate reductase (nrdD). It obtains its deoxyribonucleotides through a reductase type I similar to that of E.coli, coded by nrdA (ACIAD0724) and nrdB (ACIAD0722), with the necessary glutaredoxin and thioredoxin reductase.
Vitamin and coenzyme biosynthesis
Essentially, all the steps leading to thiamine pyrophosphate are detected in Acinetobacter ADP1, as well as the steps for the syntheses of biotin (and acetyl CoA carboxylase), lipoate, folate and pyridoxine derivatives. As far as riboflavin synthesis is concerned, 3,4-dihydroxy-2-butanone 4-phosphate synthase and GTP cyclohydrolase II are fused in a single bifunctional protein (ACIAD3570). This fusion is also present in the Pseudomonas species (PA4054, PP0516, PSPTO0692 are in BBH with ACIAD3570). In addition, this gene is entirely duplicated in P.syringae and P.putida (PSPTO2668 and PP3813 respectively), whereas this is not the case in P.aeruginosa. Curiously, this bifunctional protein coexists in ADP1 and in P.putida with the corresponding monofunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase (ACIAD2913 and PP0530, respectively). ACIAD3570 also coexists in Acinetobacter ADP1 with GTP cyclohydrolase II (ACIAD3249).
Pantothenate kinase has not been detected, although the phosphorylation of pantothenate is an important step in the biosynthetic pathway leading to coenzyme A. We have found the gene coding for the synthesis of 2-(5′-triphosphoribosyl)-3′-dephosphocoenzyme A (ACIAD1754), precursor of the unique prosthetic group specific for the activity of malonate decarboxylase, an enzyme absent in E.coli but present in P.putida, Acinetobacter calcoaceticus 40902 and Klebsiella pneumoniae (50–52).
The synthesis of isoprenoids by Acinetobacter ADP1 proceeds via the non-mevalonate pathway, leading to the octaprenyl and the undecaprenyl derivatives, necessary for the synthesis of ubiquinone and lipids I and II. The pathway leading to ubiquinone is almost complete, with the notable exception of the absolutely required 3-octaprenyl-4-hydroxybenzoate decarboxylase. None of the enzymes for menaquinone biosynthesis has been detected: this absence may be related to the strict aerobic nature of Acinetobacter ADP1. Menaquinone/menaquinol redox couple has a midpoint potential about 200 mV lower than the quinone/ubiquinol couple, thus making menaquinone more suitable for transferring electrons in an anaerobic electron transfer chain with lower potential electron acceptors.
Tetrapyrroles and vitamin B12 biosynthesis
All the genes coding for tetrapyrrole biosyntheses from glutamyl-tRNA synthetase and reductase to ferrochelatase, validated in comparison with the authentified genes from E.coli or P.aeruginosa, are present in Acinetobacter ADP1. In contrast, a few proteins, only out of the many necessary for the synthesis of adenosylcobalamin in P.denitrificans (53,54), are found in ADP1. Since ADP1 does not require B12 for growth and the gene coding for B12-dependent methionine synthase is present in the ADP1 genome, it is most likely that these enzymes fulfill the expected functions while other unknown enzymes may catalyze the remaining steps.
Polysaccharide metabolism
Cell surface polysaccharides are important for effective colonization, adherence and prevention of desiccation. Acinetobacter ADP1 has a mini-capsule and releases about 50% of its surface polysaccharides into the medium (55). These exo-polysaccharides form a complex with proteins and have emulsifying activities. This feature provides a nutritional advantage to ADP1 because it promotes the uptake of hydrophobic compounds.
We discovered a large region containing genes that encode functions providing activated sugars and other compounds involved in the biosynthesis of capsular or exo-polysaccharides, between two transposase fragments (ACIAD0057 and ACIAD0134). The molecular structure of the Acinetobacter ADP1 exo-polysaccharide has been solved by 13C-NMR studies (56). We identified all genes involved in the synthesis of the monosaccharides of this exo-polysaccharide. Interestingly, the glucose 6-phosphate isomerase (ACIAD0101), an enzyme generally involved in the glycolytic pathway, is located in the cluster of the exo-polysaccharide synthesizing genes. The presence of nine putative glycosyltransferases, three different polysaccharide transport systems and sugar activating enzymes, which are not used in the exo-polysaccharide synthesis, suggests that additional polysaccharides are synthesized in Acinetobacter ADP1. Another surface polymer might be lipopolysaccharide (LPS), the major component of the outer membrane of gram-negative bacteria. It has been reported that ADP1 makes R-type LPS consisting of lipid A and one or two molecule(s) of 3-dehydro-d-manno-octulosonic acid (57). As we found all gene products necessary for lipid A biosynthesis, it was impossible to identify genes involved in the formation of the core and the O-antigen. Besides, no ligase transferring the O-antigen to the lipid A core structure was detected. We therefore conclude that Acinetobacter ADP1 makes no O-antigen, in keeping with its non-pathogenic nature.
Catabolic pathways
Genome analysis of Acinetobacter ADP1 reveals metabolic pathways involved in the transformation of a large variety of compounds (Table 2). For example, malonate can be used as a sole source of carbon and energy. This is due to the existence of the five subunits of malonate decarboxylase, two proteins coding for the synthesis and the utilization of a specific coenzyme A derivative, two specific transport proteins and a transcription factor (51). All these genes are transcribed in the same direction and form a single cluster (ACIAD1753 to ACIAD1762). The decarboxylation of malonate yields acetate. Another typical example is urease, responsible for the utilization of urea as sole nitrogen source, which comprises three main subunits and five accessory proteins (ACIAD1088 to ACIAD1096).
Table 2. Some substrates involved in Acinetobacter sp. ADP1 catabolic pathways.
| Compounds | Final products | Gene name | ACIAD number | Cluster |
|---|---|---|---|---|
| Alkanesulfonates | Sulfite + aldehyde | ssu | 0034–0038 | — |
| Vanillate | Protocatechuate | vana | 0978–0983; 0988 | I |
| Betaine | Betaine aldehyde | bet | 1008–1012 | I |
| Acetoin | Acetate | aco | 1014; 1017–1022 | I |
| Urea | Ammonia | ure | 1088–1096 | — |
| Salicylate esters | Catechol | sala | 1424–1427 | II |
| Aryl esters | Catechol | area | 1428–1431 | II |
| Benzoate | Catechol | bena | 1433–1440 | II |
| Catechol | Succinate + acetyl CoA | cata | 1442–1451 | II |
| Alkanesulfonates | Sulfite + aldehyde | ssu-like | 1505; 1518; 1527; 1535 | III |
| Dibenzothiophene | Hydroxybiphenyl + sulfite | sox | 1510–1512 | III |
| Sulfuric esters | Sulfate + phenol/alcohol | ats | 1586–1588; 1592–1593; 1600–1601 | III |
| Nitriles | Aliphatic amides | nth | 1614–1616; 1622 | III |
| Amidase | Ammoniac + acid | amdA | 1618 | III |
| Dicarboxylic acids | Succinate + acetyl CoA | dcaa | 1684; 1688–1698 | IV |
| Protocatechuate | Succinate + acetyl CoA | pcaa | 1702–1712 | IV |
| Quinate | Protocatechuate | quia | 1713–1716 | IV |
| p-Hydroxybenzoate | Protocatechuate | poba | 1717–1719 | IV |
| Chlorogenate | Quinate + caffeate | hcaa | 1720 | IV |
| Caffeate | Protocatechuate | hcaa | 1722–1728 | IV |
| Malonate | Acetate | mdc | 1753–1762 | IV |
| Nitrate/nitrite | Ammonia | nas | 1908–1914 | V |
| Sarcosine | Glycine + formaldehyde | sox | 2550–2552 | — |
| Anthranilate | Catechol | anta | 2669–2671 | — |
| Alkanesulfonate | Sulfite + aldehyde | msu | 3470–3472 | — |
aGenes already identified in Acinetobacter sp. ADP1.
The utilization of 2,3-butanediol is of historical importance since it was on this compound that the ancestor of the strain under study in this paper was isolated. In P.putida, the 2,3-butanediol dehydrogenase and the acetoin-cleaving system are simultaneously induced during growth on butanediol or acetoin. The aco genes and adh presumably constitute one single operon that encodes all enzymes required for the conversion of 2,3-butanediol to central metabolites (58). Actually, ACIAD1022 (budC) which encodes acetoin dehydrogenase, the four genes encoding the acetoin-cleaving system (ACIAD1017 to ACIAD1020, acoABCD) and a gene coding for a putative butanediol dehydrogenase (ACIAD1021), are adjacent and transcribed in the same direction. Although the genes for acetolactate synthase are present in Acinetobacter ADP1, no gene has been detected that encodes acetolactate decarboxylase, the enzyme giving rise to acetoin. In view of the presence of all the enzymes for butanediol and acetoin metabolism, this may mean that these enzymes are necessary for symbiosis with organisms possessing the acetolactate decarboxylase.
Like Pseudomonas species, Acinetobacter ADP1 is able to modify the diverse structures of many aromatic compounds to common intermediates that can feed into central pathways. About 20% of the genes of the strain ADP1 can be reasonably associated with catabolic functions (Table 2). It is interesting to note that almost all of these genes are located in five major ‘islands of catabolic diversity’ (59), within one-quarter of the overall genome (Figure 1). This situation contrasts with that observed in E.coli or Pseudomonas species in which the aromatic catabolic clusters are spread throughout the chromosome (60,61). Nevertheless, a region of ∼2000 kb in Acinetobacter ADP1 (Figure 1) and 2400 kb in P.putida (61), flanking the replication origin (position 0 kb), is almost devoid of genes related to catabolism of aromatic compounds. To date, the evolutionary basis for the extraordinary clustering of the Acinetobacter ADP1 catabolic islands is unknown. A possible factor contributing to the grouping of the separately regulated genes is the propensity of DNA in this chromosomal quadrant to undergo frequent and extensive tandem duplications (62). These mutations may contribute to elevated expression of the catabolic genes and also may increase their copy number so that they can undergo exchange by natural transformation. Potential for evolutionary variation is present in genes that exist in multiple copies so that a mutation to new function is compensated by a copy of the gene with the original function.
Almost all the characterized genes for aromatic catabolism are located in three of these islands. Island number IV contains the dca-pca-qui-pob-hca gene cluster encoding enzymes associated with utilization of straight chain dicarboxylic acids, aromatic acids and hydroxylated aromatic acids, such as the hydroxycinnamates, caffeate, coumarate and ferulate (Table 2 and Figure 1). These compounds are produced by plants in response to stress and many of them are components of the protective plant polymer suberin (63,64). Notably absent from this island are the van genes required for conversion of vanillate to protocatechuate, a necessary step in ferulate catabolism (Table 2). The van genes are found in the first island (Figure 1), a genetically unstable region ∼720 kb distant from the dca-pca-qui-pob-hca gene cluster (28). The Acinetobacter ADP1 genome sequence extends knowledge of nutritional capabilities associated with these islands of catabolic diversity. Thus, it is evident that genes for malonate catabolism are linked to the dca-pca-qui-pob-hca cluster and genes for 2,3-butanediol utilization are linked to the van genes (Table 2). The second island, the sal-are-ben-cat cluster, positioned between the van genes and the dca-pca-qui-pob-hca cluster (Figure 1), contains genes associated with catabolism through catechol of salicylate, aromatic acids and esters of the latter compounds (65). Compared to the three Pseudomonas species, whose genomes are known, only two catabolic clusters are entirely specific to Acinetobacter ADP1 (nitriles, cluster III and Quinate, cluster IV; Table 2 and Supplementary Table 2). As expected, many of these clusters have not been found in P.syringae (acetoin, benzoate, catechol, protocatechuate and esters) and some of them are also missing in the P.aeruginosa genome, such as caffeate and protocatechuate (Supplementary Table 2). Most of the ADP1 catabolic clusters are shared with the P.putida genome, except the malonate one (cluster IV, Table 2). In addition, other non-identified catabolic clusters, located mainly in cluster III and IV, are specific to Acinetobacter ADP1 (Supplementary Table 2).
Although genes are separate in the different islands of catabolic diversity, their products are capable of physical and physiological interaction. The pcaIFJ and catIJF gene products catalyze the terminal steps in complex convergent metabolic pathways leading through protocatechuate and catechol, respectively (66–68). The genes are nearly identical in nucleotide sequence, and although separated by 263 kb, pcaIJF can serve as a template for repair of catIJF mutations by gene conversion (69). The catIJF and pcaIJF genes have a G+C content of 55.2% (69), much higher than that of other ADP1 genes (70). This evidence suggests that these genes were acquired by horizontal transfer. A remaining mystery is the evolutionary basis for the selection of heterologous genes for the terminal enzymatic steps in pathways otherwise well established in Acinetobacter ADP1. This event, which appears to occur twice (once with pcaIJF and once with catIJF) with donor genes from the same source, contrasts with the intuitively obvious and generally accepted view that selection builds from core catabolic processes (such as the conversion of ketoadipate to citric acid cycle intermediates by pcaIJF and catIJF gene products) to more peripheral catabolic steps (such as those that give rise to beta-ketoadipate). Physiological interactions between products of the sal-are-ben-cat and dca-pca-qui-pob-hca clusters have been indicated by demonstration that utilization of p-hydroxybenzoate is repressed by growth on benzoate (71).
Competence
Bacterial genetic competence for natural transformation is a physiological state that permits uptake and incorporation of naked exogenous DNA. This process, intensively studied in various gram-positive and gram-negative model organisms, is probably the most versatile mechanism of DNA transfer.
Previously identified and/or published Acinetobacter ADP1 genes involved in competence (72–76) have been found to be organized in two main clusters (comFECB at 3314 kb and comQLONM at 3355 kb). In the complete genome of ADP1, the comC gene (ACIAD3316) is 242 amino acid residues larger than the corresponding proteins annotated in the SWALL database (the beginning of the gene is missing in the EMBL entry AF027189, due to a sequencing error). Genome analysis reveals two additional gene clusters which play a potential role in natural transformation (Table 3). The first one consists of three co-localized annotated CDSs similar to pilD, pilC and pilB from Pseudomonas stutzeri. This organism is also a gram-negative soil bacterium which is naturally transformable. The second cluster contains two CDSs showing high similarity with pilU and pilT genes from Pseudomonas stutzeri. These two genes are known to be involved in natural transformation (77). Apart from these two new gene clusters, ACIAD0209, ACIAD0242, ACIAD2898 and ACIAD3236 may also be involved in natural competence (Table 3).
Table 3. Newly identified Acinetobacter sp. ADP1 genes involved in its competence.
| ACIAD number | Gene name | Size (aa) | Most similar to | Identity % (length aa) | AC/ID SWALL | Description |
|---|---|---|---|---|---|---|
| 209 | smf | 383 | Haemophilus influenzae | 41.7 (373) | SMF_HAEIN | Putative protein involved in DNA uptake |
| 242 | comM | 495 | H.influenzae | 49.5 (509) | COMM_HAEIN | Magnesium chelatase, competence-related protein |
| 360 | pilD | 286 | P.stutzeri | 52.3 (289) | LEP4_PSEST | Type 4 prepilin-like proteins leader peptide processing enzyme |
| 361 | pilC | 408 | P.stutzeri | 58.9 (405) | Q9ZEL5 | Type 4 fimbrial assembly protein |
| 362 | pilB | 579 | P.stutzeri | 59.7 (567) | Q9ZEL4 | Type 4 fimbrial biogenesis protein |
| 911 | pilU | 383 | P.stutzeri | 65.3 (382) | Q9RLD5 | Twitching motility protein |
| 912 | pilT | 344 | P.stutzeri | 83.4 (344) | Q9RLD6 | Twitching motility protein |
| 2898 | comL | 360 | Neisseria gonorrhoeae | 34.1 (267) | COML_NEIGO | Putative competence protein |
| 3236 | comF | 213 | H.influenzae | 30.6 (229) | COMF_HAEIN | Putative DNA transformation protein |
Transport
Consistent with the extensive metabolic versatility for the degradation of various components (specifically aromatics), transport is an important function in Acinetobacter ADP1: 10% of genes are involved in transport compared with 3–12% in other gram-negative organisms (Table 1). In fact, about 320 genes encode transport-related proteins for aromatic compounds of plant origin, amino acids, cations/anions, carbohydrates, nucleotides and toxic molecules such as antibiotics.
Acinetobacter ADP1 has a large capacity for amino acid transport: 38% of its ABC systems are involved in amino acid transport. In contrast, it appears to be notably deficient in sugar transporters: only one ABC system involved in sugar transport was found, and it lacks a solute-binding protein (SBP). Most of the transport features of ADP1 are shared with P.putida and P.aeruginosa but not with P.syringae (29–31). In particular, Acinetobacter ADP1, like P.putida, possesses only one PTS. ADP1 has 26 ABC transporter systems which possess at least two of the typical three components for ABC transporters (nucleotide-binding domain, membrane-spanning domain, and SBP). Transport of sulfate, aliphatic sulfonates, phosphates, copper, zinc and molybdate is carried out by ABC systems similar to those in E.coli.
The metabolic versatility in the degradation of aromatic substrates is mediated by a large number of transporters for these compounds, including two benzoate transporters, BenK and BenK-like, and a shikimate transporter. Ctr3 is an integral membrane protein that assembles as a trimer to form a competent copper uptake permease at the plasma membrane. These transporters belong to the Major Facilitator Superfamily, which represents about 14% of the transporters in Acinetobacter ADP1. A closely related PcaK transporter is formed by ADP1, as well as an additional transporter, VanK, that also acts on protocatechuate (78). Intriguingly, PcaK and VanK have diverged substantially, exhibiting only 28% amino acid sequence identity. Acinetobacter ADP1 has four porins implicated in the uptake of aromatic substrates: VanP (28), BenP (79), QuiX (80) and HcaE (59).
Among the outer membrane proteins annotated in Acinetobacter ADP1, we found a putative protein, OmpA-like, which presents about 30% similarity with the beta-barrel membrane protein OmpA of E.coli (81,82). OmpA-like proteins have already been described in other species of Acinetobacter (83). In the case of ADP1, this protein may function to desorb, solubilize and bring water-insoluble substrates into the bacteria when they are deprived of water-soluble carbon compounds (84). Further genome analysis suggests that Acinetobacter ADP1 possesses several systems for iron uptake. We identified 28 putative ferric siderophore outer membrane receptor proteins and three copies of a putative cluster of tonB, exbB, exbD genes. An intriguing feature of the genome is the presence of a cluster encoding genes for the synthesis of a siderophore composed of 2,3-dihydroxybenzoic acid, serine (or threonine or cysteine) and possibly other unidentified elements. This cluster is flanked by transposase fragments (ACIAD2760 and ACIAD2779) suggesting horizontal acquisition. The exact structure of this siderophore is unknown. It is obviously different from acinetobactin, a siderophore identified in A.baumanii ATCC 19606 (85), because Acinetobacter ADP1 does not contain histidine decarboxylase which is necessary for the biosynthesis of acinetobactin.
DISCUSSION
The overall properties of Acinetobacter ADP1 genes for catabolic pathways resemble those of other soil bacteria. In contrast, organization of the ADP1 genes, distinguished by clustering of five ‘islands of catabolic diversity’ within one-quarter of the chromosome, is unique among known bacterial genomes. Paradoxically, the GC content of its chromosome differs substantially from those of bacteria which by other criteria are most closely related. The small size of the Acinetobacter ADP1 genome is unusual. Remarkably, genes for carbohydrate catabolism are largely lacking, and the central pathway for glycolysis appears to be impeded by the lack of an evident gene for pyruvate kinase. It is particularly reassuring that this genome analysis confirms the biochemical characterizations performed some 40 years ago (9–11), which pointed to the absence of a number of basic enzyme activities associated with catabolism of common carbohydrates (see ‘Energy production’ section). This very simplified sugar metabolism as well as other metabolic features revealed by the Acinetobacter ADP1 genome analysis suggest that biotechnological applications with this versatile organism are likely to be most beneficially based upon metabolic processes that lead directly to the citric acid cycle.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Volker Doring for his critical comments and suggestions. We thank Stéphanie Bocs for preparing the Acinetobacter ADP1 gene models, Ronald Breaker and Jeff Barrick for their examination on candidates for riboswitches, and Beate Averhoff and Kaare Nielsen for helpful discussions concerning ADP1 competence. We wish to thank as well Susan Cure for her help in writing the manuscript. This work was supported by the French Centre National de la Recherche Scientifique (CNRS-URA8030) and the French Ministry of Research (grant no. 9950275).
DDBJ/EMBL/GenBank accession no. CR543861
REFERENCES
- 1.Baumann P. (1968) Isolation of Acinetobacter from soil and water. J Bacteriol, 96, 39–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumann P., Doudoroff,M. and Stanier,R.Y. (1968) A study of the Moraxella group. II. Oxidative-negative species (genus Acinetobacter). J. Bacteriol., 95, 1520–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanier R.Y., Palleroni,N.J. and Doudoroff,M. (1966) The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol., 43, 159–271. [DOI] [PubMed] [Google Scholar]
- 4.Abdel-El-Haleem D. (2003) Acintobacter: environmental and biotechnological applications. Afr. J. Biotechnol., 2, 71–74. [Google Scholar]
- 5.Smolyakov R., Borer,A., Riesenberg,K., Schlaeffer,F., Alkan,M., Porath,A., Rimar,D., Almog,Y. and Gilad,J. (2003) Nosocomial multi-drug resistant Acinetobacter baumannii bloodstream infection: risk factors and outcome with ampicillin-sulbactam treatment. J. Hosp. Infect., 54, 32–38. [DOI] [PubMed] [Google Scholar]
- 6.Manfredi R., Nanetti,A., Valentini,R. and Chiodo,F. (2001) Acinetobacter infections in patients with human immunodeficiency virus infection: microbiological and clinical epidemiology. Chemotherapy, 47, 19–28. [DOI] [PubMed] [Google Scholar]
- 7.Ibrahim A., Gerner-Smidt,P. and Liesak,W. (1997) Phylogenetic relationship of the twenty-one DNA groups of the genus Acinetobacter as revealed by 16S ribosomal DNA sequence analysis. Int. J. Syst. Bacteriol., 47, 837–841. [DOI] [PubMed] [Google Scholar]
- 8.Juni E. and Janik,A. (1969) Transformation of Acinetobacter calco-aceticus (Bacterium anitratum). J. Bacteriol., 98, 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor W.H. and Juni,E. (1961) Pathways for biosynthesis of a bacterial capsular polysaccharide.I. Characterization of the organism and polysaccharide. J. Bacteriol., 81, 688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor W.H. and Juni,E. (1961) Pathways for biosynthesis of a bacterial capsular polysaccharide.II. Carbohydrate metabolism and terminal oxidation mechanisms of a capsule-producing coccus. J. Bacteriol., 81, 694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor W.H. and Juni,E. (1961) Pathways for biosynthesis of a bacterial capsular polysaccharide.III. Syntheses from radioactive substrates. J. Biol. Chem., 236, 1231–1234. [PubMed] [Google Scholar]
- 12.Artiguenave F., Wincker,P., Brottier,P., Duprat,S., Jovelin,F., Scarpelli,C., Verdier,J., Vico,V., Weissenbach,J. and Saurin,W. (2000) Genomic exploration of the hemiascomycetous yeasts: 2. Data generation and processing. FEBS Lett., 487, 13–16. [DOI] [PubMed] [Google Scholar]
- 13.Gordon D., Abajian,C. and Green,P. (1998) Consed: a graphical tool for sequence finishing. Genome Res., 8, 195–202. [DOI] [PubMed] [Google Scholar]
- 14.Ewing B. and Green,P. (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res., 8, 186–194. [PubMed] [Google Scholar]
- 15.Ewing B., Hillier,L., Wendl,M.C. and Green,P. (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res., 8, 175–185. [DOI] [PubMed] [Google Scholar]
- 16.Gralton E.M., Campbell,A.L. and Neidle,E.L. (1997) Directed introduction of DNA cleavage sites to produce a high-resolution genetic and physical map of the Acinetobacter sp. strain ADP1 (BD413UE) chromosome. Microbiology, 143 (Pt 4), 1345–1357. [DOI] [PubMed] [Google Scholar]
- 17.Bocs S., Cruveiller,S., Vallenet,D., Nuel,G. and Medigue,C. (2003) AMIGene: Annotation of MIcrobial Genes. Nucleic Acids Res., 31, 3723–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Apweiler R., Bairoch,A., Wu,C.H., Barker,W.C., Boeckmann,B., Ferro,S., Gasteiger,E., Huang,H., Lopez,R., Magrane,M. et al. (2004) UniProt: the Universal Protein knowledgebase. Nucleic Acids Res., 32 (Database issue), D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M.D. et al. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res., 29, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claudel-Renard C., Chevalet,C., Faraut,T. and Kahn,D. (2003) Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res., 31, 6633–6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krogh A., Larsson,B., von Heijne,G. and Sonnhammer,E.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol., 305, 567–580. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen H., Brunak,S. and von Heijne,G. (1999) Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng., 12, 3–9. [DOI] [PubMed] [Google Scholar]
- 24.Lowe T.M. and Eddy,S.R. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res., 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Achaz G., Coissac,E., Viari,A. and Netter,P. (2000) Analysis of intrachromosomal duplications in yeast Saccharomyces cerevisiae: a possible model for their origin. Mol. Biol. Evol., 17, 1268–1275. [DOI] [PubMed] [Google Scholar]
- 26.Rutherford K., Parkhill,J., Crook,J., Horsnell,T., Rice,P., Rajandream,M.A. and Barrell,B. (2000) Artemis: sequence visualization and annotation. Bioinformatics, 16, 944–945. [DOI] [PubMed] [Google Scholar]
- 27.Gerischer U., D'Argenio,D.A. and Ornston,L.N. (1996) IS1236, a newly discovered member of the IS3 family, exhibits varied patterns of insertion into the Acinetobacter calcoaceticus chromosome. Microbiology, 142 (Pt 7), 1825–1831. [DOI] [PubMed] [Google Scholar]
- 28.Segura A., Bunz,P.V., D'Argenio,D.A. and Ornston,L.N. (1999) Genetic analysis of a chromosomal region containing vanA and vanB, genes required for conversion of either ferulate or vanillate to protocatechuate in Acinetobacter. J. Bacteriol., 181, 3494–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stover C.K., Pham,X.Q., Erwin,A.L., Mizoguchi,S.D., Warrener,P., Hickey,M.J., Brinkman,F.S., Hufnagle,W.O., Kowalik,D.J., Lagrou,M. et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature, 406, 959–964. [DOI] [PubMed] [Google Scholar]
- 30.Nelson K.E., Weinel,C., Paulsen,I.T., Dodson,R.J., Hilbert,H., Martins dos Santos,V.A., Fouts,D.E., Gill,S.R., Pop,M., Holmes,M. et al. (2002) Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol., 4, 799–808. [DOI] [PubMed] [Google Scholar]
- 31.Buell C.R., Joardar,V., Lindeberg,M., Selengut,J., Paulsen,I.T., Gwinn,M.L., Dodson,R.J., Deboy,R.T., Durkin,A.S., Kolonay,J.F. et al. (2003) The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl Acad. Sci. USA, 100, 10181–10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riley M. (1993) Functions of the gene products of Escherichia coli. Microbiol. Rev., 57, 862–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serres M.H., Goswami,S. and Riley,M. (2004) GenProtEC: an updated and improved analysis of functions of Escherichia coli K-12 proteins. Nucleic Acids Res., 32 (Database issue), D300–D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kay E., Bertolla,F., Vogel,T.M. and Simonet,P. (2002) Opportunistic colonization of Ralstonia solanacearum-infected plants by Acinetobacter sp. and its natural competence development. Microb. Ecol., 43, 291–297. [DOI] [PubMed] [Google Scholar]
- 35.Kay E., Chabrillat,G., Vogel,T.M. and Simonet,P. (2003) Intergeneric transfer of chromosomal and conjugative plasmid genes between Ralstonia solanacearum and Acinetobacter sp. BD413. Mol. Plant Microbe Interact., 16, 74–82. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka Y., Yoshida,T., Watanabe,K., Izumi,Y. and Mitsunaga,T. (1997) Characterization, gene cloning and expression of isocitrate lyase involved in the assimilation of one-carbon compounds in Hyphomicrobium methylovorum GM2. Eur. J. Biochem., 249, 820–825. [DOI] [PubMed] [Google Scholar]
- 37.Hoyt J.C., Johnson,K.E. and Reeves,H.C. (1991) Purification and characterization of Acinetobacter calcoaceticus isocitrate lyase. J. Bacteriol., 173, 6844–6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lynch A.S. and Lin,E.C. (1996) In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella, 2nd edn. ASM press, Washington, DC, Vol. 1, pp. 1526–1538. [Google Scholar]
- 39.Iuchi S. and Lin,E.C. (1988) arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc. Natl Acad. Sci. USA, 85, 1888–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Repoila F., Majdalani,N. and Gottesman,S. (2003) Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol. Microbiol., 48, 855–861. [DOI] [PubMed] [Google Scholar]
- 41.Kopriva S., Buchert,T., Fritz,G., Suter,M., Benda,R., Schunemann,V., Koprivova,A., Schurmann,P., Trautwein,A.X., Kroneck,P.M. et al. (2002) The presence of an iron–sulfur cluster in adenosine 5′-phosphosulfate reductase separates organisms utilizing adenosine 5′-phosphosulfate and phosphoadenosine 5′-phosphosulfate for sulfate assimilation. J. Biol. Chem., 277, 21786–21791. [DOI] [PubMed] [Google Scholar]
- 42.Stadtman E.R., Cohen,G.N. and Lebras,G. (1961) Feedback inhibition and repression of aspartokinase activity in Escherichia coli. Ann. NY Acad. Sci., 94, 952–959. [DOI] [PubMed] [Google Scholar]
- 43.Patte J.C., Le Bras,G. and Cohen,G.N. (1967) Regulation by methionine of the synthesis of a third aspartokinase and of a second homoserine dehydrogenase in Escherichia coli K 12. Biochim. Biophys. Acta, 136, 245–247. [DOI] [PubMed] [Google Scholar]
- 44.Cohen G.N., Stanier,R.Y. and Le Bras,G. (1969) Regulation of the biosynthesis of amino acids of the aspartate family in Coliform bacteria and Pseudomonads. J. Bacteriol., 99, 791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foglino M., Borne,F., Bally,M., Ball,G. and Patte,J.C. (1995) A direct sulfhydrylation pathway is used for methionine biosynthesis in Pseudomonas aeruginosa. Microbiology, 141 (Pt 2), 431–439. [DOI] [PubMed] [Google Scholar]
- 46.Ikai H. and Yamamoto,S. (1997) Identification and analysis of a gene encoding L-2,4-diaminobutyrate:2-ketoglutarate 4-aminotransferase involved in the 1,3-diaminopropane production pathway in Acinetobacter baumannii. J. Bacteriol., 179, 5118–5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ikai H. and Yamamoto,S. (1996) Sequence analysis of the gene encoding a novel L-2,4-diaminobutyrate decarboxylase of Acinetobacter baumannii: similarity to the group II amino acid decarboxylases. Arch. Microbiol., 166, 128–131. [DOI] [PubMed] [Google Scholar]
- 48.Patel N., Pierson,D.L. and Jensen,R.A. (1977) Dual enzymatic routes to L-tyrosine and L-phenylalanine via pretyrosine in Pseudomonas aeruginosa. J. Biol. Chem., 252, 5839–5846. [PubMed] [Google Scholar]
- 49.Vickrey J.F., Herve,G. and Evans,D.R. (2002) Pseudomonas aeruginosa aspartate transcarbamoylase. Characterization of its catalytic and regulatory properties. J. Biol. Chem., 277, 24490–24498. [DOI] [PubMed] [Google Scholar]
- 50.Chohnan S., Akagi,K. and Takamura,Y. (2003) Functions of malonate decarboxylase subunits from Pseudomonas putida. Biosci. Biotechnol. Biochem., 67, 214–217. [DOI] [PubMed] [Google Scholar]
- 51.Koo J.H. and Kim,Y.S. (1999) Functional evaluation of the genes involved in malonate decarboxylation by Acinetobacter calcoaceticus. Eur. J. Biochem., 266, 683–690. [DOI] [PubMed] [Google Scholar]
- 52.Hoenke S., Wild,M.R. and Dimroth,P. (2000) Biosynthesis of triphosphoribosyl-dephospho-coenzyme A, the precursor of the prosthetic group of malonate decarboxylase. Biochemistry, 39, 13223–13232. [DOI] [PubMed] [Google Scholar]
- 53.Thibaut D., Couder,M., Famechon,A., Debussche,L., Cameron,B., Crouzet,J. and Blanche,F. (1992) The final step in the biosynthesis of hydrogenobyrinic acid is catalyzed by the cobH gene product with precorrin-8x as the substrate. J. Bacteriol., 174, 1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cameron B., Briggs,K., Pridmore,S., Brefort,G. and Crouzet,J. (1989) Cloning and analysis of genes involved in coenzyme B12 biosynthesis in Pseudomonas denitrificans. J. Bacteriol., 171, 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaplan N., Zosim,Z. and Rosenberg,E. (1987) Reconstitution of emulsifying activity of Acinetobacter calcoaceticus BD4 emulsan by using pure polysaccharide and protein. Appl. Environ. Microbiol., 53, 440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaplan N. and Rosenberg,E. (1982) Exopolysaccharide distribution of and bioemulsifier production by Acinetobacter calcoaceticus BD4 and BD413. Appl. Environ. Microbiol., 44, 1335–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Borneleit P., Blechschmidt,B., Blasig,R., Gunther,P. and Kleber,H.P. (1989) Preparation of R-type lipopolysaccharides of Acinetobacter calcoaceticus by EDTA-salt extraction. Curr. Microbiol., 19, 77–81. [Google Scholar]
- 58.Huang M., Oppermann,F.B. and Steinbuchel,A. (1994) Molecular characterization of the Pseudomonas putida 2,3-butanediol catabolic pathway. FEMS Microbiol. Lett., 124, 141–150. [DOI] [PubMed] [Google Scholar]
- 59.Parke D. and Ornston,L.N. (2003) Hydroxycinnamate (hca) catabolic genes from Acinetobacter sp. strain ADP1 are repressed by HcaR and are induced by hydroxycinnamoyl-coenzyme A thioesters. Appl. Environ. Microbiol., 69, 5398–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diaz E., Ferrandez,A., Prieto,M.A. and Garcia,J.L. (2001) Biodegradation of aromatic compounds by Escherichia coli. Microbiol. Mol. Biol. Rev., 65, 523–569, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jimenez J.I., Minambres,B., Garcia,J.L. and Diaz,E. (2002) Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol., 4, 824–841. [DOI] [PubMed] [Google Scholar]
- 62.Reams A.B. and Neidle,E.L. (2003) Genome plasticity in Acinetobacter: new degradative capabilities acquired by the spontaneous amplification of large chromosomal segments. Mol. Microbiol., 47, 1291–1304. [DOI] [PubMed] [Google Scholar]
- 63.Bernards M.A. and Razem,F.A. (2001) The poly(phenolic) domain of potato suberin: a non-lignin cell wall bio-polymer. Phytochemistry, 57, 1115–1122. [DOI] [PubMed] [Google Scholar]
- 64.Lulai E.C. and Corsini,D.L. (1998) Differential deposition of suberin phenolic and aliphatic domains and their roles in resistance to infection during potato tuber (Solanum tuberosum L.) wound-healing. Physiol. Mol. Plant Pathol., 53, 209–222. [Google Scholar]
- 65.Jones R.M., Pagmantidis,V. and Williams,P.A. (2000) sal genes determining the catabolism of salicylate esters are part of a supraoperonic cluster of catabolic genes in Acinetobacter sp. strain ADP1. J. Bacteriol., 182, 2018–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bundy B.M., Campbell,A.L. and Neidle,E.L. (1998) Similarities between the antABC-encoded anthranilate dioxygenase and the benABC-encoded benzoate dioxygenase of Acinetobacter sp. strain ADP1. J. Bacteriol., 180, 4466–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harwood C.S. and Parales,R.E. (1996) The beta-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol., 50, 553–590. [DOI] [PubMed] [Google Scholar]
- 68.Parke D. (1997) Acquisition, reorganisation, and merger of genes—novel management of the beta-ketoadipate pathway in Agrobacterium tumefaciens. FEMS Microbiol. Lett., 146, 3–12. [Google Scholar]
- 69.Kowalchuk G.A., Gregg-Jolly,L.A. and Ornston,L.N. (1995) Nucleotide sequences transferred by gene conversion in the bacterium Acinetobacter calcoaceticus. Gene, 153, 111–115. [DOI] [PubMed] [Google Scholar]
- 70.Shanley M.S., Harrison,A., Parales,R.E., Kowalchuk,G., Mitchell,D.J. and Ornston,L.N. (1994) Unusual G + C content and codon usage in catIJF, a segment of the ben-cat supra-operonic cluster in the Acinetobacter calcoaceticus chromosome. Gene, 138, 59–65. [DOI] [PubMed] [Google Scholar]
- 71.Gaines G.L. III, Smith,L. and Neidle,E.L. (1996) Novel nuclear magnetic resonance spectroscopy methods demonstrate preferential carbon source utilization by Acinetobacter calcoaceticus. J. Bacteriol., 178, 6833–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Link C., Eickernjager,S., Porstendorfer,D. and Averhoff,B. (1998) Identification and characterization of a novel competence gene, comC, required for DNA binding and uptake in Acinetobacter sp. strain BD413. J. Bacteriol., 180, 1592–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Busch S., Rosenplanter,C. and Averhoff,B. (1999) Identification and characterization of ComE and ComF, two novel pilin-like competence factors involved in natural transformation of Acinetobacter sp. strain BD413. Appl. Environ. Microbiol., 65, 4568–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Herzberg C., Friedrich,A. and Averhoff,B. (2000) ComB, a novel competence gene required for natural transformation of Acinetobacter sp. BD413: identification, characterization, and analysis of growth-phase-dependent regulation. Arch. Microbiol., 173, 220–228. [DOI] [PubMed] [Google Scholar]
- 75.Porstendorfer D., Gohl,O., Mayer,F. and Averhoff,B. (2000) ComP, a pilin-like protein essential for natural competence in Acinetobacter sp. Strain BD413: regulation, modification, and cellular localization. J. Bacteriol., 182, 3673–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Friedrich A., Hartsch,T. and Averhoff,B. (2001) Natural transformation in mesophilic and thermophilic bacteria: identification and characterization of novel, closely related competence genes in Acinetobacter sp. strain BD413 and Thermus thermophilus HB27. Appl. Environ. Microbiol., 67, 3140–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Averhoff B. and Friedrich,A. (2003) Type IV pili-related natural transformation systems: DNA transport in mesophilic and thermophilic bacteria. Arch. Microbiol., 180, 385–393. [DOI] [PubMed] [Google Scholar]
- 78.D'Argenio D.A., Segura,A., Coco,W.M., Bunz,P.V. and Ornston,L.N. (1999) The physiological contribution of Acinetobacter PcaK, a transport system that acts upon protocatechuate, can be masked by the overlapping specificity of VanK. J. Bacteriol., 181, 3505–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu Y., Chen,M., Zhang,W. and Lin,M. (2003) Genetic organization of genes encoding phenol hydroxylase, benzoate 1,2-dioxygenase alpha subunit and its regulatory proteins in Acinetobacter calcoaceticus PHEA-2. Curr. Microbiol., 46, 235–240. [DOI] [PubMed] [Google Scholar]
- 80.Elsemore D.A. and Ornston,L.N. (1995) Unusual ancestry of dehydratases associated with quinate catabolism in Acinetobacter calcoaceticus. J. Bacteriol., 177, 5971–5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morona R., Klose,M. and Henning,U. (1984) Escherichia coli K-12 outer membrane protein (OmpA) as a bacteriophage receptor: analysis of mutant genes expressing altered proteins. J. Bacteriol., 159, 570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koebnik R. (1999) Structural and functional roles of the surface-exposed loops of the beta-barrel membrane protein OmpA from Escherichia coli. J. Bacteriol., 181, 3688–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ofori-Darko E., Zavros,Y., Rieder,G., Tarle,S.A., Van Antwerp,M. and Merchant,J.L. (2000) An OmpA-like protein from Acinetobacter spp. stimulates gastrin and interleukin-8 promoters. Infect. Immun., 68, 3657–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Toren A., Orr,E., Paitan,Y., Ron,E.Z. and Rosenberg,E. (2002) The active component of the bioemulsifier alasan from Acinetobacter radioresistens KA53 is an OmpA-like protein. J. Bacteriol., 184, 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yamamoto S., Okujo,N. and Sakakibara,Y. (1994) Isolation and structure elucidation of acinetobactin, a novel siderophore from Acinetobacter baumannii. Arch. Microbiol., 162, 249–254. [DOI] [PubMed] [Google Scholar]
- 86.Salanoubat M., Genin,S., Artiguenave,F., Gouzy,J., Mangenot,S., Arlat,M., Billault,A., Brottier,P., Camus,J.C., Cattolico,L. et al. (2002) Genome sequence of the plant pathogen Ralstonia solanacearum. Nature, 415, 497–502. [DOI] [PubMed] [Google Scholar]
- 87.Serres M.H., Gopal,S., Nahum,L.A., Liang,P., Gaasterland,T. and Riley,M. (2001) A functional update of the Escherichia coli K-12 genome. Genome Biol., 2, RESEARCH0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weinel C., Ermolaeva,M.D. and Ouzounis,C. (2003) PseuRECA: genome annotation and gene context analysis for Pseudomonas aeruginosa PAO1. Bioinformatics, 19, 1457–1460. [DOI] [PubMed] [Google Scholar]
- 89.Peterson J.D., Umayam,L.A., Dickinson,T., Hickey,E.K. and White,O. (2001) The Comprehensive Microbial Resource. Nucleic Acids Res., 29, 123–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Metzgar D., Bacher, J.M., Pezo,V., Reader,J., Döring,V., Schimmel,P., Marlière,P. and de Crécy-Lagard,V. (2004) Acinetobacter sp. ADP1: an ideal model organism for genetic analysis and genome engineering. Nucleic Acids Res., 32, 5780–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.