Abstract
Background:
H-cadherin (CDH13) is commonly downregulated through promoter methylation in various cancers. However, the role of CDH13 promoter methylation status in patients with colorectal cancer (CRC) remains to be clarified.
Methods:
Eligible articles were identified from online electronic database based on the preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement criteria. The pooled odds ratio (OR) and the corresponding 95% confidence interval (95% CI) were calculated and analyzed.
Results:
Eventually, a total of nine studies were included in this meta-analysis, including 488 CRC, 298 adjacent, 144 normal, 68 premalignant tissues. The results demonstrated that CDH13 promoter methylation was notably higher in CRC than in normal, adjacent, and premalignant tissues (cancer tissues vs normal tissues: OR = 16.94, P < 0.001; cancer tissues vs adjacent tissues: OR = 20.06, P < 0.001; cancer tissues vs premalignant tissues: OR = 2.23, P = 0.038). CDH13 promoter methylation had a significantly increased risk for poorly differentiated CRC (OR = 4.07, P = 0.001). CDH13 promoter methylation was not associated with sex status, tumor stage, and lymph node status (all P > 0.05). One study with 85 CRC patients reported that CDH13 promoter methylation was correlated with poor prognosis in overall survival (OS).
Conclusions:
CDH13 promoter methylation may play an important role in the initiation and progression of CRC, and may be correlated with OS of patients with CRC. Additional studies with large sample sizes are needed to further confirm our findings in the future.
Keywords: CDH13, CRC, methylation, premalignant lesions, progression
1. Introduction
Colorectal cancer (CRC) is the most frequently digestive malignancy and the fourth leading cause of death of malignant tumors worldwide.[1] According to global cancer statistics, approximately 1,360,600 new cases with CRC were diagnosed, leading to killing an estimated 693,900 people in the word in 2012.[1] Despite the recent and main improvements in diagnostic and therapeutic opportunities, more than 50% of the patients with CRC easily metastasize to liver, lung, and lymph nodes, and these cases are called as metastatic CRC.[2] Thus, the prognosis and survival rate for advanced CRC is very poor.[3]
CRC is a multifactorial disease associated with genetic and epigenetic alterations, and develops from normal colonic epithelial cells into colon adenocarcinoma cells.[4,5] The hypermethylation of tumor suppressor genes (TSGs) of the promoter region may be correlated with cell cycle control, DNA repair, metabolism of carcinogens, cell–cell interaction, apoptosis, and angiogenesis, and leads to gene silencing, which may facilitate cancer initiation and progression.[6,7]CDH13 as an atypical member of the cadherin family, a TSG, also named as H-cadherin, T-cadherin, or cadherin-13, is located on 16q24 and plays an important role in cell–cell adhesion.[8,9] In most malignant tumor cell lines, CDH13 expression has been showed to be involved in the inhibition of cell invasion and cell proliferation, and the reduction of tumor cell growth.[9–11]
However, the relationship between CDH13 promoter methylation and CRC remains to be assessed. Therefore, the present meta-analysis was carried out to evaluate whether CDH13 promoter methylation was significantly associated with an increased risk of CRC in cancer versus precancerous, adjacent, and normal tissue samples. In addition, we also determined whether CDH13 promoter methylation was correlated with clinicopathological features such as sex status, tumor differentiation, tumor stage, and lymph node status in cancer.
2. Materials and methods
2.1. Search strategy
The PubMed, Embase, EBSCO, and Wanfang databases were systematically searched to achieve eligible studies without any language restriction before July 18, 2016. We used the following search terms: (CDH13 OR cadherin 13 OR H-cadherin OR T-cadherin) AND (methylation OR epigene∗) AND (colorectal cancer OR colorectal tumor OR colorectal carcinoma OR colorectal neoplasm OR CRC). To get other additional studies, we also scanned reference lists from the initially identified articles.
2.2. Inclusion criteria
To identify the eligibility of the included studies, the following inclusion criteria was applied: the patients were limited to primary CRC by histopathological examination; premalignant lesions included adenoma and aberrant crypt foci (ACF); articles were case–control or cohort studies; articles must provide sufficient data with regard to CDH13 promoter methylation in cancer versus premalignant, adjacent, or normal tissues; cohort studies must have sufficient information to evaluate the association between CDH13 promoter methylation and sex status, tumor differentiation, tumor stage, and lymph node status in CRC. In addition, if articles using the same data were published more than once, only paper with the most complete or up-to-date information was included in this meta-analysis.
2.3. Ethical review from patients
Although the present study was not primary research involving human samples, our study was a secondary analysis regarding human subject data published in the public domain.
2.4. Data extraction
The following data were independently extracted by 2 reviewers (JL and PY) from the included studies: the surname of first author, publication year, country, ethnic population, detection method of methylation, frequency of methylation, the number of case and control groups, and clinicopathological features such as sex status, tumor differentiation, tumor stage, and lymph node status. Three reviewers discussed (JL, PY, and CN) the disagreements and received the final consensuses.
2.5. Data analysis
This meta-analysis was conducted using the version 12.0 Stata statistical software (Stata Corp, College Station, TX). The pooled odds ratios (ORs) with 95% confidence intervals (CIs) were used to evaluate the strength of the correlation between CDH13 promoter methylation and CRC. Moreover, the pooled ORs with 95% CIs were also calculated to assess the association of CDH13 promoter methylation with sex status, tumor differentiation, tumor stage, and lymph node status in CRC. The assessment of statistical heterogeneity was done based on the Chi-square test.[12] Substantial heterogeneity among studies was detected using the random-effects model (I2 ≥ 50%), a fixed-effects model was used without significant heterogeneity (I2 < 50%).[13,14] We also performed a sensitivity analysis to determine the influence and stability of single study on the results by omitting 1 study.[15] A P-value of less than 0.05 was considered to be statistically significant for the pooled OR.
3. Results
3.1. Characteristics of eligible studies
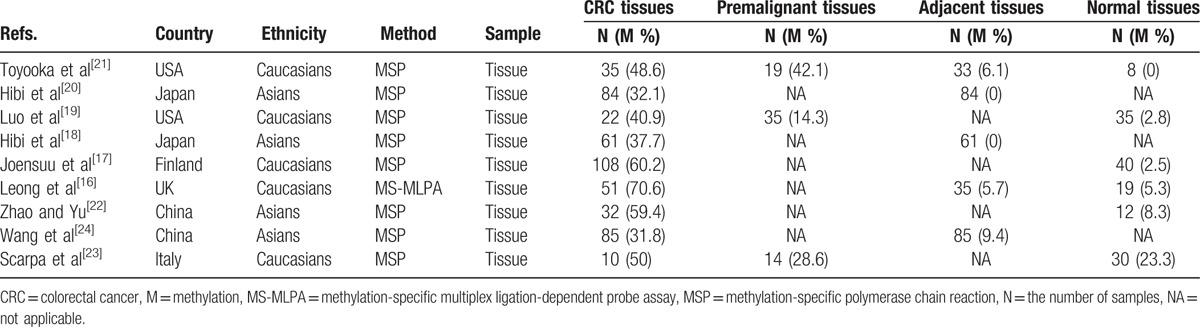
The detailed steps of the systematic search and selection procedures of literature are shown in Fig. 1. According to the above inclusion criteria, finally, 9 case–control studies with 998 tissue samples were included in this study.[16–24] Of these eligible studies, 6 studies with 258 CRC and 144 normal tissue samples assessed the correlation between CDH13 promoter methylation and CRC in CRC versus normal tissues,[16,17,19,21–23] 5 studies with 316 CRC and 298 adjacent tissue samples assessed the correlation between CDH13 promoter methylation and CRC in CRC versus adjacent tissues,[16,18,20,21,24] 3 studies with 67 CRC and 68 premalignant tissue samples assessed the association between CDH13 promoter methylation and CRC in CRC versus premalignant lesions.[19,21,23] Additionally, 3 studies with 191 CRC samples evaluated the association of CDH13 promoter methylation with clinicopathological features.[19,20,24]Table 1 summarizes the main characteristics of the included studies.
Figure 1.
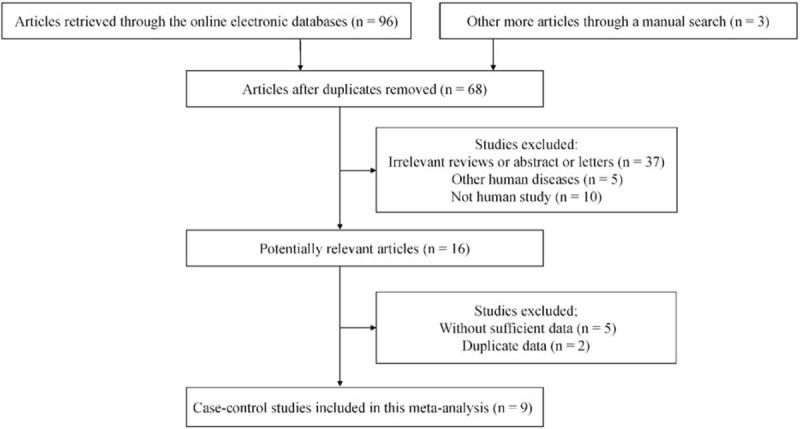
Flow diagram of the literature selection.
Table 1.
General characteristics of the included studies in the report.
3.2. Correlation between CDH13 promoter methylation and CRC in cancer versus controls
The pooled data from 9 studies including 488 cases with CRC, 48 premalignant tissues, 298 adjacent tissues, and 144 normal tissues were included in this study. A random-effects model was used in cancer versus control groups. Our findings showed that the rate of CDH13 promoter methylation was significantly higher in cancer tissues than in normal, adjacent, and premalignant tissues (cancer tissues vs normal tissues: OR = 16.94, 95% CI = 6.10–47.10, P < 0.001; cancer tissues vs adjacent tissues: OR = 20.06, 95% CI = 5.45–73.80, P < 0.001; cancer tissues vs premalignant tissues: OR = 2.23, 95% CI = 1.05–4.74, P = 0.038) (Fig. 2). The above analysis suggested that CDH13 promoter methylation had a significantly increased risk of CRC. However, more studies comparing CRC and premalignant lesions should be essential to further confirm our results in the future.
Figure 2.
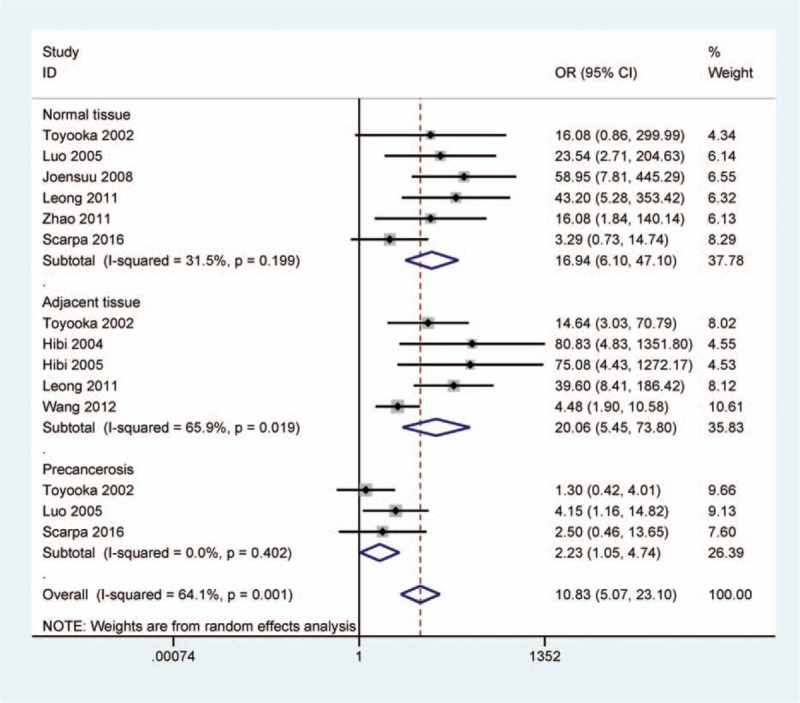
Forest plot of the correlation between CDH13 methylation and CRC, including 9 studies with 488 CRC, 298 adjacent, 144 normal, 68 premalignant tissues (cancer tissues vs normal tissues: OR = 16.94, 95% CI = 6.10–47.10, P < 0.001; cancer tissues vs adjacent tissues: OR = 20.06, 95% CI = 5.45–73.80, P < 0.001; cancer tissues vs premalignant tissues: OR = 2.23, 95% CI = 1.05–4.74, P = 0.038).
3.3. Subgroup and sensitivity analyses in CRC versus adjacent tissues
When cancer tissues were compared to adjacent tissues, significant heterogeneity existed (I2 = 65.9%). Subgroup analysis based on ethnic population was carried out to find the difference. The result demonstrated that CDH13 promoter methylation had significantly increased risk of CRC in Asians and Caucasians (OR = 23.31, 95% CI = 1.79–304.06, P = 0.016; OR = 24.28, 95% CI = 8.04–73.31, P < 0.001, respectively) (Fig. 3).
Figure 3.
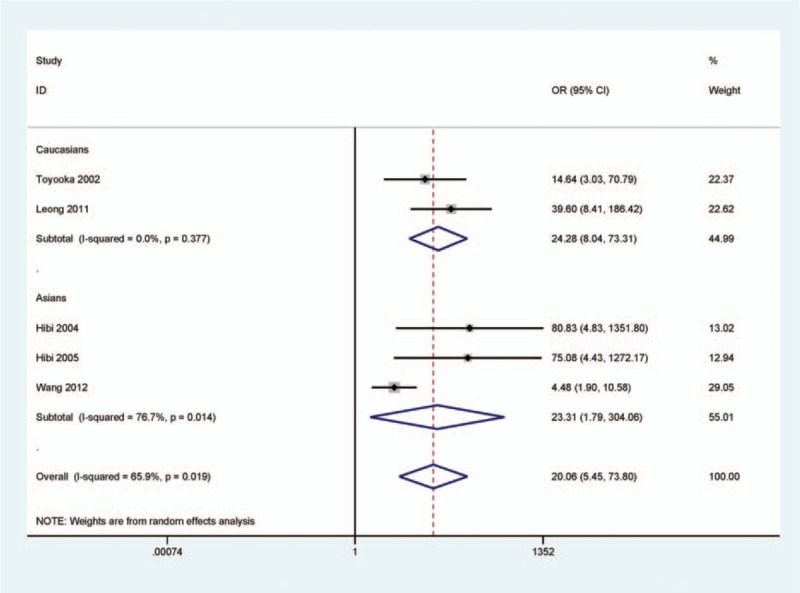
Forest plot of subgroup analysis based on ethnicity for CDH13 promoter methylation in CRC versus adjacent tissues. Asian population: OR = 23.31, 95% CI = 1.79–304.06, P = 0.016; Caucasian population: OR = 24.28, 95% CI = 8.04–73.31, P < 0.001.
Next, a sensitivity analysis was determined to assess the influence and stability by deleting one study. When we removed this study by Wang et al.[24] (Fig. 4), I2 dramatically reduced to 0.0%, indicating no obvious evidence of heterogeneity. A fixed-effects model was used. The pooled OR was not significantly changed (OR = 38.99, 95% CI = 14.69–103.50, P < 0.001), suggesting the stability of our results.
Figure 4.
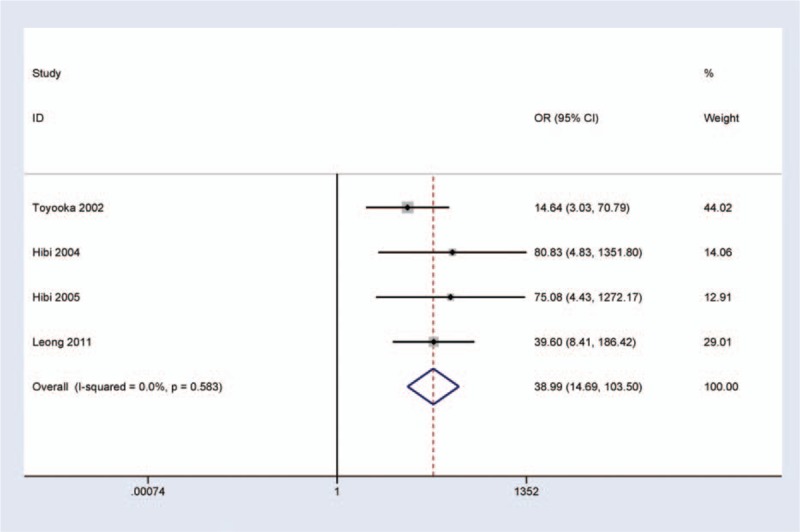
Forest plot of a sensitivity analysis by deleting one study for CDH13 promoter methylation in CRC versus adjacent tissues. I2 = 0.0%, OR = 31.92, 95% CI = 12.14–83.92, P < 0.001.
3.4. Correlation of CDH13 promoter methylation with clinicopathological features
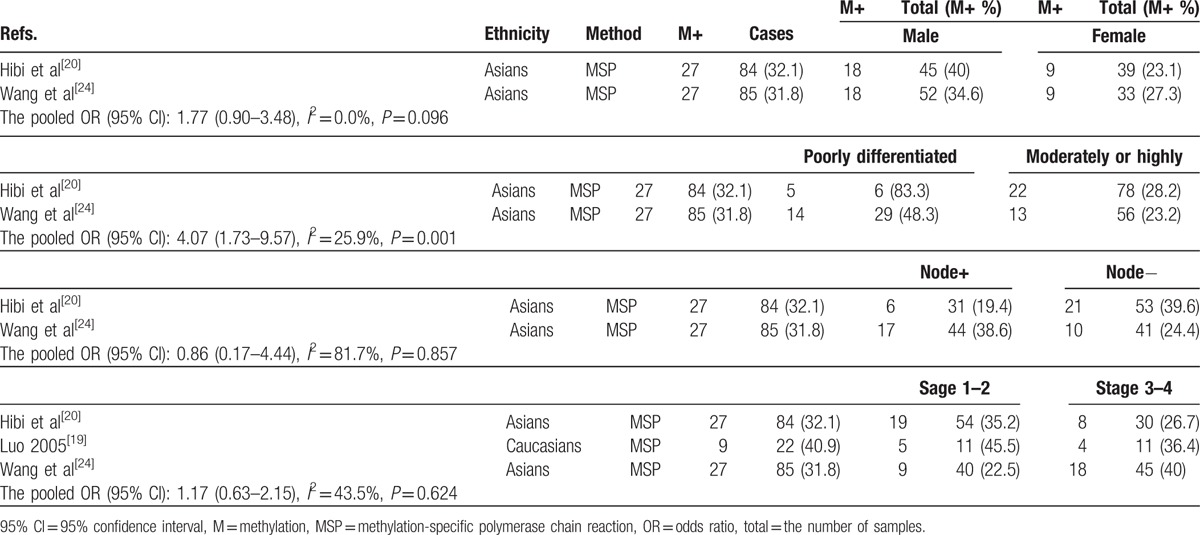
Table 2 shows that the association between CDH13 promoter methylation and clinicopathological characteristics in cancer, including 3 studies with 191 CRC patients. The results of CDH13 promoter methylation demonstrated that no significant association was observed in relation to sex status, tumor stage, and lymph node status in CRC (OR = 1.77, 95% CI = 0.90–3.48, P = 0.096; OR = 1.17, 95% CI = 0.63–2.15, P = 0.624; OR = 0.86, 95% CI = 0.17–4.44, P = 0.857, respectively). The result from 2 studies demonstrated that CDH13 promoter methylation was significantly higher in poorly differentiated CRC than in moderately or highly differentiated CRC (OR = 4.07, 95% CI = 1.73–9.57, P = 0.001). However, the relationships between CDH13 promoter methylation and clinicopathological characteristics should be cautious as only smaller cases were analyzed in this study.
Table 2.
Association of CDH13 promoter methylation with clinicopathological features.
3.5. Prognosis of CDH13 promoter methylation
Only Wang et al[24] reported that CDH13 promoter methylation was associated with poor prognosis in 5-year overall survival (OS), including 85 patients with CRC. More studies with larger sample sizes are necessary to future validate the prognostic value of CDH13 promoter methylation in the future.
4. Discussion
The reduction of CDH13 expression has been reported to be correlated with poor prognosis in several types of human cancers.[8]CDH13 is frequently downregulated by promoter methylation in the CpG islands in various carcinomas, including endometrial carcinoma,[25] breast cancer,[26] cervical cancer,[27] and CRC.[20] The site of the CDH13 methylation was located in the promoter of its 5′-flanking region in this study. However, there were contradictory results regarding the frequency of CDH13 promoter methylation in CRC, precancerous, adjacent, and normal tissues. The methylation frequency of CDH13 promoter was inconsistent in CRC, with a range from 31.8%[24] to 70.6%.[16] In addition, Toyooka et al[21] reported that CDH13 promoter had different methylation rates in CRC, precancerous, adjacent, and normal tissues (CRC: 48.6%; precancerous tissues: 42.1%; adjacent tissues: 6.1%; normal tissues: 0.0%). While Luo et al[19] reported that CDH13 promoter had different methylation frequencies in CRC, precancerous, and normal tissues (CRC: 40.9%; precancerous tissues: 14.3%; normal tissues: 2.8%). Thus, the present study was carried out to assess the correlation between CDH13 promoter methylation and CRC risk.
Although most patients with CRC develop as a consequence of tumor progression from adenomas into adenocarcinomas, adenomas are not defined as the only type of precancerous lesions. Additionally, some studies have shown that serrated polyps or aberrant crypt foci (ACF) are also premalignant lesions.[19,28,29] In the present study, CDH13 promoter methylation status was shown to be significantly higher in CRC tissues than in premalignant, normal, and adjacent tissues, suggesting that CDH13 promoter methylation may play an important role in the carcinogenesis of CRC. However, the result comparing CRC and premalignant lesions should be carefully considered as only 67 CRC tissues and 68 precancerous lesions were included in this study.
Significant heterogeneity was detected in the comparison of CRC and adjacent tissues (I2 = 65.9%). Subgroup analysis by ethnicity was analyzed to find the different association. The result showed that CDH13 promoter methylation was significantly correlated with an increased risk of CRC in Asian and Caucasian patients, indicating that Asian and Caucasian populations were susceptible to the promoter methylation of CDH13. Next, a sensitivity analysis was conducted by omitting this study (Wang et al.[24]), the result showed that the pooled OR of CDH13 promoter methylation was not significantly changed, with no substantial heterogeneity, which suggested that the stability of our analyses. The result of this study (Wang et al[24]) was different from other studies, the reason was not very clear. The adjacent tissue samples might have been slightly contaminated by CRC cells, which may lead to bias in cancer versus adjacent tissues.
Furthermore, our findings revealed that CDH13 promoter methylation was not associated with sex status, tumor stage, and lymph node status in cancer. While CDH13 promoter methylation had an increased risk for poorly differentiated CRC, suggesting that CDH13 promoter methylation may play a key role in CRC progression. However, the results of CDH13 promoter methylation with clinicopathological features should be carefully considered as only 191 CRC patients were analyzed.
This meta-analysis had several limitations. First, the main ethnic population were Asians and Caucasians, such as other ethnicities, Africans, were lack. Second, blood or feces samples were insufficient; the studies with large sample sizes are needed to determine whether CDH13 promoter methylation could be a specific noninvasive biomarker for CRC diagnosis. Third, the results from less than 4 studies with small sample sizes were analyzed in CRC versus precancerous lesions, and in relation to clinicopathological features in cancer. Fourth, although studies published in English or Chinese language were included in this meta-analysis, articles published in other languages were missed, which may lead to a selection bias. Finally, only Wang et al[24] reported that CDH13 promoter methylation was correlated with 5-year OS of patients with CRC. Additional studies with large sample sizes are needed to validate the prognostic value as a potential drug target in the future.
In conclusion, our findings showed that CRC had a higher CDH13 promoter methylation than premalignant, normal, and adjacent tissues, and higher in poorly differentiated CRC than in moderately or highly differentiated CRC. Moreover, CDH13 promoter methylation was not correlated with sex status, tumor stage, and lymph node status in cancer. Further well-designed studies with larger sample sizes are very essential to confirm our results in the future.
Footnotes
Abbreviations: 95% CI = 95% confidence interval, ACF = adenoma and aberrant crypt foci, CDH13 = H-cadherin, CRC = colorectal cancer, OR = odds ratio, OS = overall survival, TSGs = tumor suppressor genes.
Funding: The research was supported by the grants from Natural Science Foundation of Zhejiang Province (LY16H160005), Ningbo Natural Science Foundation (2014A610235), and Project of Scientific Innovation Team of Ningbo (2015B11050).
Authors’ contributions: TH and MY contributed to the conception, design, and final approval of the submitted manuscript. JL, CZ, PY, CN, and SC contributed to the completion of article analysis, including data extraction, and the calculation and design of the figures and tables. All the authors approved the final manuscript.
The authors have no conflicts of interest to disclose.
References
- [1].Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- [2].Segal NH, Saltz LB. Evolving treatment of advanced colon cancer. Annu Rev Med 2009;60:207–19. [DOI] [PubMed] [Google Scholar]
- [3].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5–29. [DOI] [PubMed] [Google Scholar]
- [4].Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med 2009;361:2449–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology 2008;135:1079–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kulis M, Esteller M. DNA methylation and cancer. Adv Genet 2010;70:27–56. [DOI] [PubMed] [Google Scholar]
- [7].Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol 2005;2(suppl 1):S4–11. [DOI] [PubMed] [Google Scholar]
- [8].Andreeva AV, Kutuzov MA. Cadherin 13 in cancer. Genes Chromosomes Cancer 2010;49:775–90. [DOI] [PubMed] [Google Scholar]
- [9].Takeuchi T, Ohtsuki Y. Recent progress in T-cadherin (CDH13, H-cadherin) research. Histol Histopathol 2001;16:1287–93. [DOI] [PubMed] [Google Scholar]
- [10].Lee SW, Reimer CL, Campbell DB, et al. H-cadherin expression inhibits in vitro invasiveness and tumor formation in vivo. Carcinogenesis 1998;19:1157–9. [DOI] [PubMed] [Google Scholar]
- [11].Lee SW. H-cadherin, a novel cadherin with growth inhibitory functions and diminished expression in human breast cancer. Nat Med 1996;2:776–82. [DOI] [PubMed] [Google Scholar]
- [12].Zintzaras E, Ioannidis JP. HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics 2005;21:3672–3. [DOI] [PubMed] [Google Scholar]
- [13].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].DerSimonian R. Meta-analysis in the design and monitoring of clinical trials. Stat Med 1996;15:1237–48. discussion 1249–1252. [DOI] [PubMed] [Google Scholar]
- [15].Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med 1997;127:820–6. [DOI] [PubMed] [Google Scholar]
- [16].Leong KJ, Wei W, Tannahill LA, et al. Methylation profiling of rectal cancer identifies novel markers of early-stage disease. Br J Surg 2011;98:724–34. [DOI] [PubMed] [Google Scholar]
- [17].Joensuu EI, Abdel-Rahman WM, Ollikainen M, et al. Epigenetic signatures of familial cancer are characteristic of tumor type and family category. Cancer Res 2008;68:4597–605. [DOI] [PubMed] [Google Scholar]
- [18].Hibi K, Kodera Y, Ito K, et al. Aberrant methylation of HLTF, SOCS-1, and CDH13 genes is shown in colorectal cancers without lymph node metastasis. Dis Colon Rectum 2005;48:1282–6. [DOI] [PubMed] [Google Scholar]
- [19].Luo L, Chen WD, Pretlow TP. CpG island methylation in aberrant crypt foci and cancers from the same patients. Int J Cancer 2005;115:747–51. [DOI] [PubMed] [Google Scholar]
- [20].Hibi K, Nakayama H, Kodera Y, et al. CDH13 promoter region is specifically methylated in poorly differentiated colorectal cancer. Br J Cancer 2004;90:1030–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Toyooka S, Toyooka KO, Harada K, et al. Aberrant methylation of the CDH13 (H-cadherin) promoter region in colorectal cancers and adenomas. Cancer Res 2002;62:3382–6. [PubMed] [Google Scholar]
- [22].Zhao L, Yu JP. Methylation status of CDH13 gene promoter in colon cancer. Chin J Postgrad Med 2011;34:23–4. [Google Scholar]
- [23].Scarpa M, Scarpa M, Castagliuolo I, et al. Aberrant gene methylation in non-neoplastic mucosa as a predictive marker of ulcerative colitis-associated CRC. Oncotarget 2016;7:10322–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang Z, Yuan X, Jiao N, et al. CDH13 and FLBN3 gene methylation are associated with poor prognosis in colorectal cancer. Pathol Oncol Res 2012;18:263–70. [DOI] [PubMed] [Google Scholar]
- [25].Sheng Y, Wang H, Liu D, et al. Methylation of tumor suppressor gene CDH13 and SHP1 promoters and their epigenetic regulation by the UHRF1/PRMT5 complex in endometrial carcinoma. Gynecol Oncol 2016;140:145–51. [DOI] [PubMed] [Google Scholar]
- [26].Pang JM, Deb S, Takano EA, et al. Methylation profiling of ductal carcinoma in situ and its relationship to histopathological features. Breast Cancer Res 2014;16:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Siegel EM, Riggs BM, Delmas AL, et al. Quantitative DNA methylation analysis of candidate genes in cervical cancer. PLoS ONE 2015;10:e0122495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Okamoto K, Kitamura S, Kimura T, et al. Clinicopathological characteristics of serrated polyps as precursors to colorectal cancer: current status and management. J Gastroenterol Hepatol 2016;doi: 10.1111/jgh.13482. [DOI] [PubMed] [Google Scholar]
- [29].Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759–67. [DOI] [PubMed] [Google Scholar]


