Abstract
Here we report that transcriptional effects of the insertion of a retrotransposon can occur simultaneously both upstream and downstream of the insertion site. We have identified an intra-cisternal A particle (IAP) retrotransposon in intron 6 of a gene that we have named Cabp (CDK5 activator binding protein). The presence of the IAP is associated with an aberrant transcript initiating from a cryptic promoter in the IAP, reading out into the adjacent Cabp gene sequence. The expression of this transcript is highly variable among isogenic mice within the C57BL/6J strain and so CabpIAP can be classified as a metastable epiallele. As expected, the presence or absence of the transcript correlates with differential DNA methylation of the 5′ LTR of the IAP. More surprisingly, in mice where the retrotransposon is unmethylated and presumably transcriptionally active, we find a number of short Cabp transcripts which initiate at the normal 5′ end of the gene but terminate prematurely, just 5′ of the retrotransposon. This is the first report of a retrotransposon having both upstream and downstream effects on transcription at the site of insertion and it suggests that alternative polyadenylation may sometimes be caused by a downstream convergent transcription unit.
INTRODUCTION
There is increasing evidence that transposon-like elements, which are scattered throughout the genomes of higher organisms, can influence the expression of adjacent genes (1–3). Although the majority of these elements are inactivated by genetic and epigenetic mechanisms, there is a small proportion of transcriptionally active retrotransposons that escape silencing in somatic cells.
In mammals, a number of mutant alleles have been identified which are associated with the insertion of an intra-cisternal A particle (IAP) retrotransposon, where the activity of the retrotransposon affects the expression of the adjacent gene. These mutant alleles, which include agouti viable yellow (Avy), agouti hypervariable yellow (Ahvy), agouti intra-cisternal A particle yellow (Aiapy) and axin fused (AxinFu), have been termed metastable epialleles and show a variety of unusual characteristics including variable expressivity among genetically identical individuals (4). In the case of AxinFu and Avy, the insertion of the IAP retrotransposon in the opposite orientation influences transcription as a result of a powerful bi-directional promoter in the 5′ long terminal repeat (LTR) of the IAP. The cryptic promoter reads out into the adjacent genomic DNA producing aberrant transcripts (5,6). The stochastic nature of the establishment of the epigenetic state of the 5′ LTR leads to the variable expressivity of the adjacent coding exons among isogenic littermates (7,8).
Here we report the identification of a novel metastable epiallele, which we call CabpIAP, containing an IAP inserted in the reverse orientation into intron 6 of the murine homologue of the rat Cabp (CDK5 activator binding protein) gene. We demonstrate that when the 5′ LTR of the IAP is hypomethylated and active, aberrant transcription occurs not only downstream, but also upstream of the insertion site. Some transcripts, which initiate at the wild-type Cabp promoter, terminate prematurely, 5′ of the IAP at alternative poly(A) sites. In addition, transcripts initiate at a cryptic promoter in the 5′ LTR of the IAP and read out into and through the remaining exons of the Cabp gene. When the IAP is inactive and hypermethylated, no aberrant transcription is observed either upstream or downstream. In theory, both of these aberrant transcripts could produce novel, truncated forms of the protein CABP. These studies highlight the complex nature of the effects of active retrotransposons on transcription at adjacent loci. To our knowledge, this is the first report of a retrotransposon causing alternative polyadenylation at a site outside of the retrotransposon itself. The stochastic nature of the epigenetic state of the IAP has enabled us to observe what appears to be transcriptional interference (9–11). These observations raise the interesting possibility that alternative polyadenylation may sometimes be the result of differential expression of downstream transcription units.
MATERIALS AND METHODS
Inbred mouse strains
Inbred mouse strains used in this study: C57BL/6J (Oak Ridge National Laboratory, Oak Ridge, TN); 129P4/RrRk (The Jackson Laboratory); FVB/NJ (Dr G Robertson, Westmead Hospital, Sydney, Australia); and C3H/HeJ (Australian Research Council, Perth, Western Australia).
Expression analysis
Kidneys were harvested and total RNA was isolated using Tri-Reagent (Sigma-Aldrich). Poly(A)+ mRNA was extracted using the PolyATtract® mRNA Isolation System (Promega Corporation) and 10–50 μg was separated on a 1.5% formaldehyde agarose gel and analysed by northern transfer and hybridization with either the 3′ or the 5′ probe. The 3′ probe (249 bp of intron 6, exons 7 and 8 of CabpIAP) was amplified from C57BL/6J kidney cDNA using intron6-up (5′-gcaccattgcccacttgtta-3′) and exon8-down (5′-ccctgcattattgaactgga-3′) oligonucleotides. The 5′ probe (347 bp of exons 2 and 3 of CabpIAP) was amplified using exon2-up (5′-atgtgctgggtgttgctta-3′) and exon3-down (5′-tcttggaggttgctggtc-3′) oligonucleotides. The membranes were incubated overnight at 68°C with a radiolabelled probe in ExpressHyb hybridization solution (Clontech Laboratories Inc.) and exposed to a PhosphorImager storage phosphor screen (Molecular Dynamics). The image was visualized using PhosphorImager special performance hardware and ImageQuant (v5.1) software (Molecular Dynamics). Membranes were then stripped and rehybridized as indicated in the Results (Figures 1B and 3A).
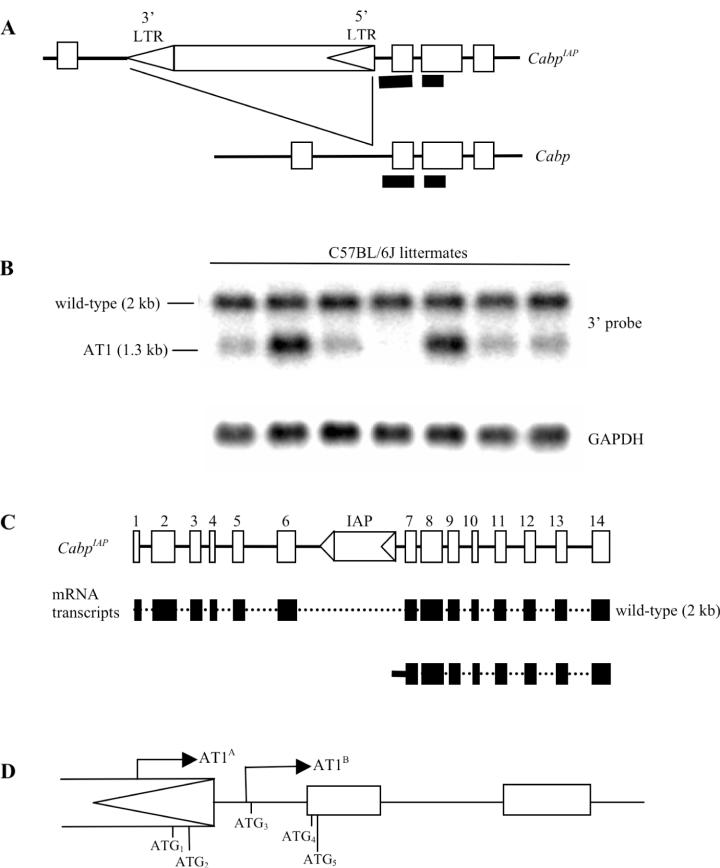
Figure 1.
Variable expressivity of an aberrant transcript at CabpIAP. (A) The CabpIAP allele has an IAP (subtype IΔ1) in intron 6 of the gene, with the direction of transcription from the 5′ LTR (arrowhead) opposite to that of the Cabp gene. The IAP insertion site is 52 bp upstream of exon 7. The 3′ probe consists of intron 6, exons 7 and 8 (thick black lines). (B) Upper panel: Poly(A)+ northern hybridization of seven C57BL/6J mice using the 3′ probe. The 2 kb band, representing the wild-type Cabp transcript, is expressed equally amongst the littermates, whereas the aberrant transcript (AT1) is variably expressed amongst the littermates. Lower panel: GAPDH control. (C) Schematic diagram of the CabpIAP allele showing the 14 exons. The wild-type transcript contains all 14 exons, whereas AT1 which initiates in the 5′ LTR of the IAP contains LTR and intron 6 sequence and all the remaining exons (7–14). (D) 5′ RLM-RACE identified two aberrant transcripts. AT1A initiates in the 5′ LTR (76 bp upstream of the 5′ LTR/intron 6 junction) and AT1B initiates in intron 6 (22 bp downstream of the 5′ LTR/intron 6 junction). Five ATG sites are present in AT1A and three ATG sites are present in AT1B. ATG4 and ATG5 are in-frame with the Cabp coding exons and in vitro transcription/translation assays revealed that both can initiate translation.
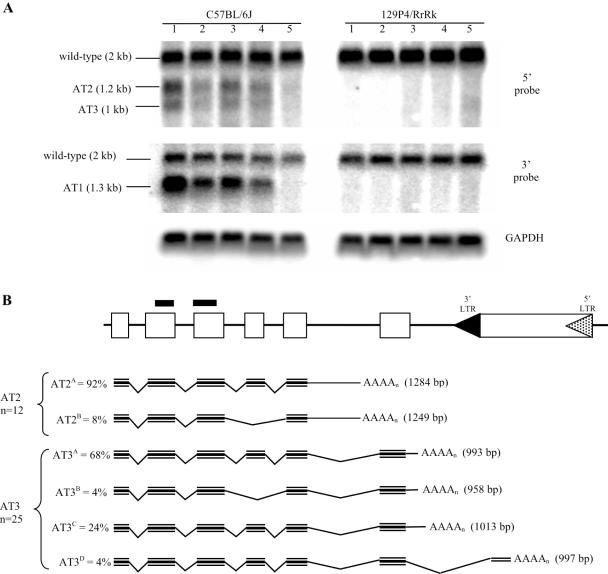
Figure 3.
Alternative polyadenylation at CabpIAP. (A) Poly(A)+ northern analysis of five C57BL/6J and five 129P4/RrRk mice. Upper panel: the membrane was hybridized with the 5′ probe [shown as two thick black lines in (B)] and shows the wild-type transcript (2 kb) and two other transcripts of 1.2 and 1.0 kb, labelled AT2 and AT3 respectively. Middle panel: the same membrane was stripped and rehybridized with the 3′ probe (see Figure 1A) and shows the wild-type and AT1 transcripts (see Figure 1B). Lower panel: GAPDH control. (B) Schematic diagram of transcripts resulting from 3′ RLM-RACE analysis of the CabpIAP allele when the 5′ LTR is hypomethylated (dotted arrowhead). 3′ RLM-RACE amplified two major PCR products (data not shown) and sequencing of these products identified six different transcripts. In order to gain insight into the relative abundance of these different transcripts, a number of clones were sequenced. The AT2 band is made up of two splice variants which terminate in intron 5. Ninety two per cent of the clones sequenced (n = 12) contained exons 1–5 and 685 bp of intron 5 (AT2A) and eight per cent of clones lacked exon 4 (AT2B). The AT3 band is made up of four transcripts. The majority of the clones (68%) sequenced (n = 25) contained exons 1–6 and 183 bp of intron 6 (AT3A), 24% of the clones contained exons 1–6 and 203 bp of intron 6 (AT3C) and 4% of clones lacked exon 4 and terminated 183 bp into intron 6 (AT3B). Another 4% of the clones sequenced (AT3D) contained exons 1–6 of the Cabp gene and 187 bp of IAP sequence. The sizes of all transcripts are indicated.
Phenotype classification
The phenotype of each mouse (at three weeks) was determined by kidney poly(A)+ northern analysis using the 3′ probe. A mouse that showed any AT1 expression was classified as penetrant. A mouse that showed no AT1 expression was classified as silent.
5′ and 3′ RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE)
5′ and 3′ RLM-RACE was performed with the FirstChoice RLM-RACE kit (Ambion) using kidney RNA from adult C57BL/6J mice. In 5′ RLM-RACE, the primary PCR was conducted with a reverse outer oligonucleotide (5′-ccctgcattattgaactgga-3′) which annealed to exon 8. Two secondary PCRs were conducted, each with one of the two reverse inner oligonucleotides (5′-cgagaatggaggcaactgg-3′ and 5′-ttaacaagtgggcaatggtg-3′) which annealed to exons 7 and 6, respectively. In 3′ RLM-RACE, the primary PCR was conducted with a reverse outer oligonucleotide (5′-atggtgctgggtgttgctta-3′) which annealed to exon 2. Two secondary PCRs were conducted, each with one of the two reverse inner oligonucleotides (5′-gtgaacgacacagagatagcc-3′ and 5′-tatcatgccagtccagacga-3′) which annealed to exons 3 and 6, respectively. The PCR products were cloned into pGEM®-T Easy vector (Promega Corporation) and sequenced.
Bisulfite sequencing
Ten microgram of kidney DNA was digested with BamHI, ethanol precipitated, and resuspended in 10 μl of Milli-Q. The DNA was embedded in agarose (2%) and treated with sodium bisulfite as described previously (12). The bisulfite treated DNA was resuspended in 30 μl of deionized water and heated at 80°C for 1 min before use in the PCR.
To amplify the 5′ and 3′ LTRs for sequencing analysis, 1 μl (5′ LTR) or 2 μl (3′ LTR) of the bisulfite treated DNA was used in the primary PCR, generating a 879 bp (5′ LTR) and a 512 bp (3′ LTR) product. One microlitre of each product was used in a semi-nested PCR, generating a 410 bp (5′ LTR) and a 378 bp (3′ LTR) fragment which were cloned into pGEM®-T Easy vector for sequencing. Oligonucleotides used were (common, primary PCR, semi-nested PCR): 5′-ggttaggaagaatattatagattagaattttt-3′, 5′-ccaaaaatttcatacttaaatatcttatcc-3′, 5′-aacaccaacatacaattaacaaataaac-3′ for the 5′ LTR and 5′-aacatcctatattctaaattaataaacaaa-3′, 5′-tagtgttataagtgttaagttaggtatatg-3′, 5′-tgattttggtttgagatgtgttaag-3′ for the 3′ LTR.
Sequenced clones were excluded from the analysis if they contained more than two non-CpG cytosines that were unconverted by bisulfite treatment, i.e. a conversion efficiency of <97%. Also, clones which were identical from one PCR were discarded to avoid any possible PCR bias.
RESULTS
The CabpIAP allele is the result of a recent IAP insertion into the C57BL/6J inbred mouse strain
In order to identify novel murine alleles where transcription initiates in IAP retrotransposons, we searched C57BL/6J cDNA databases for sequences that contained IAP LTR sequence. One of the novel sequences found (GenBank accession number BB842254) shows homology at its 5′ end to an IAP LTR and reads into intron 6 of a gene, not previously named in the mouse. This gene has homology to the CDK5 activator binding protein (Cabp) in the rat, which is known to inhibit CDK5 kinase activity (13). We have termed the gene Cabp. It lies on chromosome 2 in the mouse genome, contains 14 exons and produces a 2 kb wild-type transcript (Figure 1B and C). In the C57BL/6J strain this gene contains an IΔ1 IAP retrotransposon inserted in the reverse orientation into intron 6. We have called this allele CabpIAP (Figure 1A).
From genomic database searches (ENSEMBL and CELERA), it appeared that the CabpIAP allele is exclusive to the C57BL/6J mouse strain and this was confirmed by Southern transfer analysis. The IAP is always found at the Cabp locus in C57BL/6J mice (n = 81) and is not found in 129P4/RrRk (n = 6), FVB/NJ (n = 6) or C3H/HeJ (n = 2) mice (data not shown), suggesting a relatively recent retrotransposition event. The consistent presence of the IAP amongst mice from the C57BL/6J strain argues that the IAP insertion is stable.
Aberrant transcripts associated with the 5′ LTR of the IAP in CabpIAP are variably expressed
Northern analysis of kidney mRNA, using a probe consisting of intron 6, exons 7 and 8 of CabpIAP(see Figure 1A), revealed the predicted wild-type transcript (2 kb) in all mice and a 1.3 kb aberrant transcript (AT1), the amount of which varied amongst the littermates (Figure 1B). This was also observed in mRNA from brain, liver and lung (data not shown). The size of the 1.3 kb band is consistent with a transcript initiating from a cryptic promoter in the 5′ LTR of the IAP reading out into intron 6 and containing all the remaining exons of Cabp (Figure 1C). Some mice do not express AT1 at all and are classified as silent. Those that do express AT1 are classified as penetrant, with some mice being more penetrant than others. Variable expression of transcripts initiating within IAP retrotransposons has previously been reported at metastable epialleles (5,7,14,15). We can now classify CabpIAP as a metastable epiallele due to its variable expressivity within an inbred strain.
5′ RLM-RACE revealed two transcriptional initiation start sites: AT1A initiated in the 5′ LTR of the IAP (76 bp upstream of the 5′ LTR/intron 6 junction) and AT1B initiated in intron 6 (22 bp downstream of the 5′ LTR/intron 6 junction) (Figure 1D). Multiple transcriptional start sites associated with retrotransposon insertions have been observed previously and sometimes these start sites lie just outside the IAP sequence itself (7,16).
Five putative translational start codons were present in AT1A and three were present in AT1B (Figure 1D). ATG4 and ATG5 are in-frame and in vitro transcription/translation assays revealed that both can initiate translation (data not shown). In theory, these would generate a truncated form of the Cabp protein containing only the carboxy terminal domain encoded by exons 7–14. Site directed mutagenesis of the ATG5 to TTT prevented production of the associated polypeptide (data not shown). Although AT1 has the potential to produce a truncated protein, a phenotype associated with CabpIAP in penetrant mice has not yet been identified.
Level of expression of AT1 correlates with DNA methylation at the 5′ LTR
Previous studies of other metastable epialleles have demonstrated a correlation between transcriptional activity and the DNA methylation state of the 5′ LTR (7,14,15,17). This prompted us to analyse the methylation state of the 5′ LTR of the IAP at the CabpIAP allele.
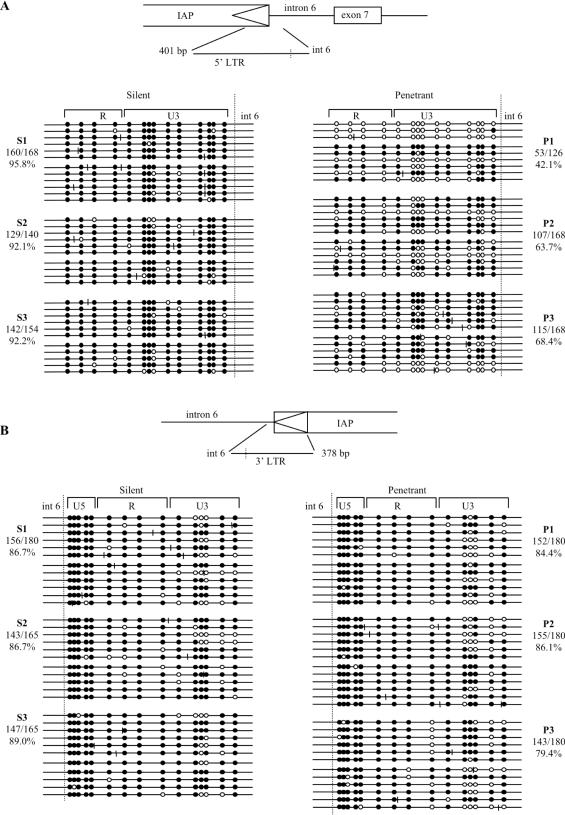
As expected, bisulfite sequencing showed that the 5′ LTR is heavily methylated in somatic tissue of silent mice and less methylated in that of penetrant mice (Figure 2A). In the penetrant mice, there is considerable inter-clone and inter-mouse variation. Therefore, CabpIAP is an epigenetically-sensitive allele where the establishment of the epigenetic mark at the LTR is stochastic and where the epigenetic state correlates, to a degree, with the level of expression of the aberrant transcript. Similar findings have been made at other metastable epialleles (7). The level of methylation at the 3′ LTR was high in both penetrant and silent individuals (Figure 2B), which is interesting, considering that the sequences of the 3′ LTR and the 5′ LTR at this locus are identical (data not shown).
Figure 2.
Methylation profile of the IAP LTRs at CabpIAP. The methylation state of each CpG was obtained by sequencing PCR clones from bisulfite treated genomic DNA. Open and filled circles represent unmethylated and methylated CpGs, respectively. The position of each circle indicates the relative location of each CpG site in the DNA fragment. The short vertical lines represent the location of non-CpG cytosines that were not converted by bisulfite treatment. Each line represents the sequence of one clone. Each block of clones, which is divided into two parts, represents the data from two bisulfite PCRs from the one mouse. Clones which were identical from one PCR were discarded to avoid possible PCR bias. The numbers next to each block of clones represents the proportion of methylated CpG sites relative to the total number of CpG sites and the corresponding percentage of methylation. The data shown are from three silent (S) and three penetrant (P) C57BL/6J mice. (A) The 5′ LTR. A one-tailed, two-sample, unequal variance standard student t-test, showed the three silent mice to be different (P-value <0.05) from the three penetrant mice. (B) The 3′ LTR. A one-tailed, two-sample, unequal variance standard student t-test, showed the three silent mice to be no different (P-value >0.05) from the three penetrant mice.
Alternative polyadenylation at the CabpIAP locus
Northern analysis, using a double-stranded probe consisting of exons 2 and 3 (see Figure 3B), revealed the presence of two additional transcripts, AT2 and AT3 (Figure 3A). The intensity of these bands varied between mice within the C57BL/6J colony and they were absent from mice of the 129P4/RrRk strain. Expression of AT2 and AT3 correlated with expression of AT1, such that a mouse that was highly penetrant with respect to AT1 was also highly penetrant with respect to AT2 and AT3 (Figure 3A). These additional aberrant transcripts suggest that premature polyadenylation is occurring. While it was formally possible that AT2 and AT3 were the result of transcription from the 3′ LTR, northern analysis with a single-stranded sense RNA fragment of exons 2 and 3 of CabpIAP enabled us to rule out this possibility (data not shown). The fact that the 3′ LTR was heavily methylated in both penetrant and silent mice is consistent with a lack of promoter activity (see Figure 2B).
3′ RLM-RACE suggested that AT2 and AT3 were made up of a number of transcripts initiating at the normal start site of the Cabp gene but terminating just 5′ of the IAP. AT2 was made up of two splice variants which terminate in intron 5 and AT3 was made up of four transcripts, three of which terminate in intron 6 and one which terminates in the IAP itself (Figure 3B). AT2A and AT2B contain a consensus polyadenylation signal, AATAAA, whereas AT3A, AT3B and AT3C contain the cryptic polyadenylation signal, ATTAAA (Figure 4), a common variant of the consensus sequence (18). Even though AT3A, AT3B and AT3C, all share the same ATTAAA signal, AT3C contains an extra 20 bp of sequence 3′ of the ATTAAA (Figure 4B). The use of alternative polyadenylation cleavage sites, downstream of a single polyadenylation signal is not uncommon (19). AT3D contains two possible cryptic polyadenylation signals, AGTAAA and ATTTAA and utilizes a cryptic 3′ splice site in the IAP (Figure 4B). All six transcripts contain an in-frame termination codon, TGA (Figure 4).
Figure 4.
Genomic sequence indicating the truncating transcripts AT2A, AT2B, AT3A, AT3B, AT3C and AT3D. (A) Sequence showing the AT2A and AT2B transcripts which terminate at the same site in intron 5. Exon 5 is in bold and shown in uppercase and intron 5 is shown in lowercase. The polyadenylation signal (AATAAA) is double underlined. The sequence of intron 5 found in the AT2A and AT2B transcripts is underlined. AT2B lacks exon 4 (see Figure 3B). The putative termination codon (TGA) is double underlined. (B) Sequence showing the AT3A, AT3B, AT3C and AT3D transcripts. Exon 6 is in bold and shown in uppercase, intron 6 is shown in lowercase and the IAP is shown in italics and bold. In intron 6, the cryptic polyadenylation signal (ATTAAA) for AT3A, AT3B and AT3C is double underlined. The sequence of intron 6 found in the AT3A and AT3B transcripts is underlined. AT3B lacks exon 4 (see Figure 3B). The underlined dotted sequence shows the extra 20 bp present in AT3C. AT3D contains exon 6 and splices into IAP sequence. The sequence of the IAP found in the AT3D is underlined. The two possible cryptic polyadenylation signals, AGTAAA and ATTTAA for AT3D are double underlined. The putative termination codons (TGA) for all transcripts are double underlined.
DISCUSSION
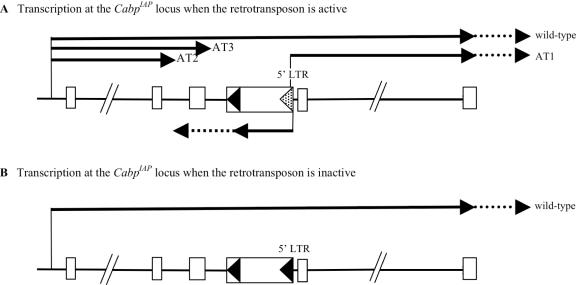
There are ∼1000 copies of the IAP retrotransposon in the mouse genome and a subset of these are transcriptionally active (20). IAP transcripts are detected in most tissues of the mouse (20,21) and their levels are elevated dramatically (50–100 fold) in DNA methyltransferase-1 deficient mice (22). Complex patterns of transcription at the CabpIAP locus only occur when the 5′ LTR of the IAP is hypomethylated. AT2 and AT3 initiate at the endogenous Cabp promoter and terminate 5′ of the IAP, whereas AT1 initiates in the 5′ LTR and reads out into the adjacent Cabp coding exons. As the 5′ LTR of the IAP is a ‘methylation sensitive’ bi-directional promoter, it is expected that the production of AT1 is concomitant with the internal transcription of the IAP itself. However this is difficult to determine directly due to the large number of IAPs in the genome. This raises the possibility that AT2 and AT3 are produced through transcriptional interference. When the 5′ LTR is hypomethylated, the expected internal transcription from the 5′ LTR would occur in an antisense direction with respect to the Cabp gene (Figure 5). The stable IAP pre-mRNA transcripts would terminate at the polyadenylation site in the 3′LTR but the RNA polymerase II complex presumably reads past this and continues into intron 6 or even 5. Read-through transcription is well established in higher eukaryotes, nascent transcripts extending for variable distances (a few hundred nucleotides to several kilobases) into the 3′ flanking region of the gene (23–25). Simultaneously, the endogenous Cabp promoter would drive transcription of Cabp in the opposite direction, establishing a situation of convergent, co-transcribed genes. As transcription proceeds, the RNA polymerase II complexes would collide either in intron 5 or 6, causing one or both to pause or be released from the template. This would result in a failure of splicing and subsequent utilization of alternative polyadenylation signals. Transcriptional interference of this type is thought to underlie the decreased levels of steady-state mRNA reported in cultured cells following transfection of plasmids containing convergent reporter genes (11,26). However, the relevance of this to transcriptional regulation at endogenous loci remains unclear.
Figure 5.
Model of transcriptional interference at CabpIAP. (A) Schematic diagram of the IAP insertion at the CabpIAP allele (not to scale). Transcripts produced when the 5′ LTR of the IAP is hypomethylated (dotted arrowhead). The endogenous Cabp promoter drives expression of the Cabp wild-type transcript and the cryptic promoter in the 5′ LTR drives expression of AT1. The IAP itself is also internally transcribed by its promoter in the 5′ LTR, in an antisense direction to that of Cabp. The stable IAP mRNA transcripts terminate at the polyadenylation site in the 3′ LTR but the RNA polymerase II complex presumably reads past this site and continues into intron 6 or intron 5 (indicated by dotted arrow). Transcriptional interference between the colliding polymerases, causes premature polyadenylation of the wild-type transcripts, producing AT2 and AT3. The solid lines represent the stable mRNA transcripts and the dotted lines represent the presumed nascent transcripts. (B) Only the wild-type Cabp transcript is produced when the 5′ LTR is hypermethylated (black arrowhead).
A brief search of the literature and the ENSEMBL human genome database revealed a number of examples of alternative polyadenylation adjacent to a convergent antisense transcription unit. One example is the human excision repair gene, ERCC-1. In addition to the major wild-type transcript (containing exons 1–10), another transcript reads through exon 9 and utilizes a polyadenylation signal in intron 9 (27,28). An antisense transcript of 2.6 kb (ASE-1), which codes for the CD3-epsilon associated protein, overlaps with the ERCC-1 transcription unit. It initiates in a region 3′ of the ERCC-1 gene and terminates in intron 9 of ERCC-1 (29). It is possible that the truncated transcript is produced as a result of transcriptional interference between ERCC-1 and ASE-1.
We have found an IAP retrotransposon insertion at an endogenous gene that generates transcriptional diversity in vivo. Given the abundance of IAP derived elements in the mouse genome, the IAP is likely to influence the regulation of a number of endogenous loci. This suggests a role for the IAP in the evolution of transcriptome complexity. Han et al. recently proposed that the L1 retrotransposon has played an important role in the evolution of the mammalian transcriptome (3), our finding supports and extends this model, by indicating that the IAP retrotransposon may have made a similar contribution.
Our findings raise the possibility that transcriptional interference results in premature polyadenylation elsewhere in the genome. The recent finding in humans that one order of magnitude or more of the genomic sequence is transcribed than remaining as stable mRNA suggests that overlapping transcription units may be more numerous than previously thought (30). Such transcription units would not necessarily produce stable mRNA and may therefore be hard to detect. At CabpIAP, the variable expressivity of the IAP, and the stability of the aberrant transcripts produced, has enabled us to observe what appears to be transcriptional interference in vivo. The approach to understanding alternative polyadenylation has been to look for tissue-specific or stage-specific factors associated with the preferential use of one poly(A) site over another, but a review of the literature reveals that this has been relatively unsuccessful (31,32). A more fruitful approach may be to look for convergent transcription units.
Acknowledgments
ACKNOWLEDGEMENTS
R.D. was supported by an Australian Postgraduate award and this work was funded by a grant from the National Health and Medical Research Council of Australia (to E.W.).
REFERENCES
- 1.Deininger P.L. and Batzer,M.A. (2002) Mammalian retroelements. Genome Res., 12, 1455–1465. [DOI] [PubMed] [Google Scholar]
- 2.Ostertag E.M. and Kazazian,H.H.,Jr (2001) Biology of mammalian L1 retrotransposons. Annu. Rev. Genet., 35, 501–538. [DOI] [PubMed] [Google Scholar]
- 3.Han J.S., Szak,S.T. and Boeke,J.D. (2004) Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature, 429, 268–274. [DOI] [PubMed] [Google Scholar]
- 4.Rakyan V.K., Blewitt,M.E., Druker,R., Preis,J. and Whitelaw,E. (2002) Metastable epialleles in mammals. Trends Genet., 18, 348–351. [DOI] [PubMed] [Google Scholar]
- 5.Duhl D.M., Vrieling,H., Miller,K.A., Wolff,G.L. and Barsh,G.S. (1994) Neomorphic agouti mutations in obese yellow mice. Nature Genet., 8, 59–65. [DOI] [PubMed] [Google Scholar]
- 6.Vasicek T.J., Zeng,L., XGuan,J., Zhang,T., Costantini,F. and Tilghman,S.M. (1997) Two dominant mutations in the mouse fused gene are the result of transposon insertions. Genetics, 147, 777–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rakyan V.K., Chong,S., Champ,M.E., Cuthbert,P.C., Morgan,H.D., Luu,K.V.K. and Whitelaw,E. (2003) Transgenerational inheritance of epigenetic states at the murine AxinFu allele occurs after maternal and paternal transmission. Proc. Natl Acad. Sci. USA, 100, 2538–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolff G.L. (1978). Influence of maternal phenotype on metabolic differentiation of agouti locus mutants in the mouse. Genetics, 88, 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Proudfoot N.J. (1986) Transcriptional interference and termination between duplicated α-globin gene constructs suggests a novel mechanism for gene regulation. Nature, 322, 562–565. [DOI] [PubMed] [Google Scholar]
- 10.Whitelaw E. and Martin,D.I.K. (2001) Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nature Genet., 27, 361–365. [DOI] [PubMed] [Google Scholar]
- 11.Eszterhas S.K., Bouhassira,E.E., Martin,D.I.K. and Fiering,S. (2002) Transcriptional interference by independently regulated genes occurs in any relative arrangement of the genes and is influenced by chromosomal integration position. Mol. Cell. Biol., 22, 469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark S.J., Harrison,J., Paul,C.L. and Frommer,M. (1994) High sensitivity mapping of methylated cytosines. Nucleic Acids Res., 22, 2990–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ching Y.P., Pang,A.S., Lam,W.H., Qi,R.Z. and Wang,J.H. (2002) Identification of a neuronal Cdk5 activator-binding protein as Cdk5 inhibitor. J. Biol. Chem., 277,15237–15240. [DOI] [PubMed] [Google Scholar]
- 14.Argeson A.C., Nelson,K.K. and Siracusa,L.D. (1996) Molecular basis of the pleiotropic phenotype of mice carrying the hypervariable yellow (Ahvy) at the agouti locus. Genetics, 142, 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michaud E.J., van Vugt,M.J., Bultman,S.J., Sweet,H.O., Davisson,M.T. and Woychik,R.P. (1994) Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev., 8, 1463–1472. [DOI] [PubMed] [Google Scholar]
- 16.Christy R.J. and Huang,R.C. (1988) Functional analysis of the long terminal repeats of intracisternal A-particle genes: sequences within the U3 region determine both the efficiency and the direction of promoter activity. Mol. Cell. Biol., 8, 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan H.D., Sutherland,H.E., Martin,D.I.K. and Whitelaw,E. (1999) Epigenetic inheritance at the agouti locus in the mouse. Nature Genet., 23, 314–318. [DOI] [PubMed] [Google Scholar]
- 18.Proudfoot N.J. and Whitelaw,E. (1988) Termination and 3′ end processing of eukaryotic RNA. In Hames,B.D. and Glover,D.M. (eds), Transcription and Splicing. Frontiers in Molecular Biology. IRL Press, Oxford, Washington DC. pp. 97–129. [Google Scholar]
- 19.Pauws E., van Kampen,A.H.C., van de Graaf,S.A.R., de Vijlder,J.J.M. and Ris-Stalpers,C. (2001) Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids Res., 29, 1690–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuff E.L. and Lueders,K.K. (1988) The intracisternal A particle gene family: structure and functional analysis. Adv. Cancer Res., 51, 183–276. [DOI] [PubMed] [Google Scholar]
- 21.Dupressoir A. and Heidmann,T. (1996) Germ line-specific expression of intracisternal A-particle retrotransposons in transgenic mice. Mol. Cell. Biol., 16, 4495–4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh C.P., Chaillet,R. and Bestor,T.H. (1998) Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nature Genet., 20, 116–117. [DOI] [PubMed] [Google Scholar]
- 23.Amara S.G., Evans,R.M. and Rosenfeld,M.G. (1984) Calcitonin/calcitonin gene-related peptide transcription unit: tissue-specific expression involves selective use of alternative polyadenylation sites. Mol. Cell. Biol., 4, 2151–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weintraub H., Larsen,A. and Groudine,M. (1981) α-Globin-gene switching during the development of chicken embryos: expression and chromosome structure. Cell, 24, 333–344. [DOI] [PubMed] [Google Scholar]
- 25.Whitelaw E. and Proudfoot,N.J. (1986) α-Thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human alpha 2 globin gene. EMBO J., 5, 2915–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prescott E.M. and Proudfoot,N.J. (2002) Transcriptional collision between convergent genes in budding yeast. Proc. Natl Acad. Sci. USA, 99, 8796–8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Duin M., Koken,M.H., van Den Tol,J., ten Dijke,P., Odijk,H., Westerveld,A., Bootsma,D. and Hoeijmakers,J.H. (1987) Genomic characterization of the human DNA excision repair gene ERCC-1. Nucleic Acids Res., 15, 9195–9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson M.D., Ruttan,C.C., Koop,B.F. and Glickman,B.W. (2001) ERCC1: a comparative genomic perspective. Environ. Mol. Mutagen., 38, 209–215.. [DOI] [PubMed] [Google Scholar]
- 29.van Duin M., van Den Tol,J., Hoeijmakers,J.H., Bootsma,D., Rupp,I.P., Reynolds,P., Prakash,L. and Prakash,S. (1989) Conserved pattern of antisense overlapping transcription in the homologous human ERCC-1 and yeast RAD10 DNA repair gene regions. Mol. Cell. Biol., 9, 1794–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapranov P., Cawley,S.E., Drenkow,J., Bekiranov,S., Strausberg,R.L., Fodor,S.P.A. and Gingeras,T.R. (2002) Large-scale transcriptional activity in chromosomes 21 and 22. Science, 296, 916–919. [DOI] [PubMed] [Google Scholar]
- 31.Edwalds-Gilbert G., Veraldi,K.L. and Milcarek,C. (1997) Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res., 25, 2547–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao J., Hyman,L. and Moore,C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]