Abstract
Ribosomes from the extreme thermophile Thermus thermophilus are capable of translation in a coupled transcription–translation system derived from Escherichia coli. At 45°C, T.thermophilus ribosomes translate at ∼25–30% of the maximal rate of E.coli ribosomes, and synthesize full-length protein. T.thermophilus and E.coli subunits can be combined to effect translation, with the spectrum of proteins produced depending upon the source of the 30S subunit. In this system, T.thermophilus ribosomes function in concert with E.coli translational factors and tRNAs, with elongation and release factors being supplied from the E.coli extract, and purified initiation factors (IFs) being added exogenously. Cloned and purified T.thermophilus IF1, IF2 and IF3 supported the synthesis of the same products in vitro as the E.coli factors, although the relative levels of some polypeptides were factor dependent. We conclude that, at least between these two phylogenetically distant species, translational factor function and subunit–subunit interactions are conserved. This functional compatibility is remarkable given the extreme and highly divergent environments to which these species have adapted.
INTRODUCTION
There is little controversy over the universality of the key elements of the bacterial translational apparatus; the widely accepted view is that ribosomal RNA (rRNA) is the key catalytic element in translation, reinforced by the extraordinarily high degree of conservation existing within the crucial domains in each of the rRNAs (1). Concomitantly, there has been the increasingly tacit view that, given this high degree of conservation, all bacterial ribosomes must function in very much the same way. Thus, the genetic and biochemical data, acquired principally from Escherichia coli, which have been used to develop functional models of protein synthesis, are being interpreted in the light of crystal structures from other bacteria (2,3) and even from an archaeon (4). One way to address the validity of this extrapolation, is to explore the exchangeability, or otherwise, of components of the translational machinery from two diverse species. In this work, we have chosen to examine the ribosomes and initiation factors (IFs) from E.coli and the extreme thermophile Thermus thermophilus in an in vitro coupled transcription–translation system derived from E.coli.
There are a number of reasons to have chosen T.thermophilus, aside from the fact that many of the relevant crystal structures are from this organism (2,3,5,6). The evolutionary distance between E.coli and T.thermophilus spans the divergence of Gram-negative from Gram-positive organisms, with T.thermophilus being one of the most deeply branching bacterial species known (7), so questions concerning the conservation of the translational machinery over great evolutionary distances can be addressed. The use of T.thermophilus also allows us to ascertain whether adaptation to extreme temperature requires alteration in conserved structural elements involved in inter-subunit communication or in translational factor interactions with the ribosome. Specifically in this work, we have asked whether there is compatibility between the subunits and between the IFs.
In vitro coupled transcription–translation seems, at first sight, an extremely demanding assay system; however, the arguments for its use are compelling. Provided the production of full-length protein products can be achieved, then the consequences for the polypeptides formed of using heterologous factors and subunits can be examined. We previously developed the system employed here for coupled transcription– translation in Streptomyces lividans (8). This in turn was derived from an efficient E.coli system (9) developed from early work by Zubay (10). All these systems employed crude cellular fractions that contained all the enzymes required for transcription and translation. Subsequently (11), the crude extract was depleted of ribosomes, providing a system that was now dependent on the addition of exogenous ribosomes, although only those derived from other Streptomyces spp. were tested in that study. Here, we took advantage of such a depleted system to examine whether ribosomes from T.thermophilus can translate mRNA transcribed in vitro from a DNA template using only components produced from E.coli.
We report here that translation is indeed possible in such an in vitro chimeric system, albeit at a temperature (45°C) between those required for optimal growth of T.thermophilus (72°C) and E.coli (37°C). This finding opens the potential for exploring other cross-species compatibilities. Additionally, as this system is absolutely dependent upon added IFs, translation by ribosomes from the mesophilic E.coli in concert with translational factors from the thermophilic T.thermophilus could be explored.
MATERIALS AND METHODS
Preparation of coupled transcription–translation extracts from E.coli strain MRE600
Cultures of E.coli strain MRE600 were grown in Luria–Bertani (LB) medium at 37°C to OD600 0.4–0.5. Cells were harvested (Sorvall RC5 centrifuge; GSA rotor; 5000 r.p.m., 10 min, 4°C), washed twice with 0.5 vol buffer containing 10 mM HEPES–KOH pH 7.6, 10 mM MgCl2, 1 M NH4Cl, 5 mM β-mercaptoethanol, and once with 0.25 vol cracking buffer (CB) (10 mM HEPES–KOH pH 7.6, 10 mM MgCl2, 50 mM NH4Cl, 10% v/v glycerol, 5 mM β-mercaptoethanol). Cells were resuspended in CB (1.5 ml/g cells) and pressed once in a French pressure cell at 3500–4000 psi. Cell debris and high molecular weight DNA were pelleted by centrifugation (Beckman L8, Ty65 rotor, 30 000 r.p.m., 30 min, 4°C). Ribosomes were removed from the supernatant by further centrifugation (Ty65 rotor, 40 000 r.p.m., 4 h, 4°C). The supernatant was depleted of lower molecular weight DNA by treatment with CaCl2 (1 mM final) and staphylococcal (S7) nuclease (750 U/ml, 20 min, 30°C), and the nuclease was silenced by the addition of EGTA (2 mM final). Finally, the extract (S100) was dialyzed against 2 × 1000 vol CB for 1 h at 4°C, and stored at −80°C. Activity was stable over at least 6 months, provided extracts were not refrozen.
Protein synthesis in coupled extracts
Protein synthesis was totally dependent upon exogenous ribosomes (or subunits), template DNA and IFs. Typically, reaction mixes (15 μl) contained 4–6 pmol ribosomes (70S ribosomes or equimolar amounts of 50S plus 30S subunits); IFs (optimized input of 10 pmol each of IF1, IF2 and IF3); S100 (optimized input, but typically 5 μl); synthesis mix (4 μl; see below); [35S]-methionine (1 μl; specific activity 1000 Ci/mol, 15 mCi/ml for gel analysis; diluted to 6000 c.p.m./pmol, 88 μM with cold methionine for time courses); MgCl2 (12 mM final); and reactions were started by the addition of template plasmid pUC18 (1 μg). Incubation temperatures and times are as indicated.
For time courses, samples were removed directly into 1 ml trichloroacetic acid (TCA; 10% w/v), boiled for 10 min, filtered through Whatman GF/A 2.5-cm diameter discs, and washed with 10 ml TCA (5% w/v). Discs were dried and radioactivity was estimated by liquid scintillation spectrometry. All time courses were repeated at least four times with at least two different preparations of ribosomes or subunits.
For gel electrophoresis, reaction mixes were incubated for 20 min at the temperature indicated, 1 μl was removed for estimation of [35S]-methionine incorporation as described above, unlabelled methionine (1 μl of a 0.44 mg/ml solution) was added, and the chase carried out for a further 10 min. Proteins were either loaded directly in the reaction mix (Figures 1C and 2B) or were precipitated from reaction samples by addition of an equal volume of ice-cold 20% (w/v) TCA, recovered by centrifugation and washed with acetone prior to dissolving in loading buffer (Figure 4C). SDS–PAGE (15% acrylamide) (12) was carried out in a BioRad mini-Protean II gel system at 150 V. Gels were fixed for 15 min in methanol:glacial acetic acid:water (30:10:60), dried, and exposed to Kodak X-Omat film. Gel analysis of products under each experimental condition was carried out at least twice and with different preparations of ribosomes or subunits.
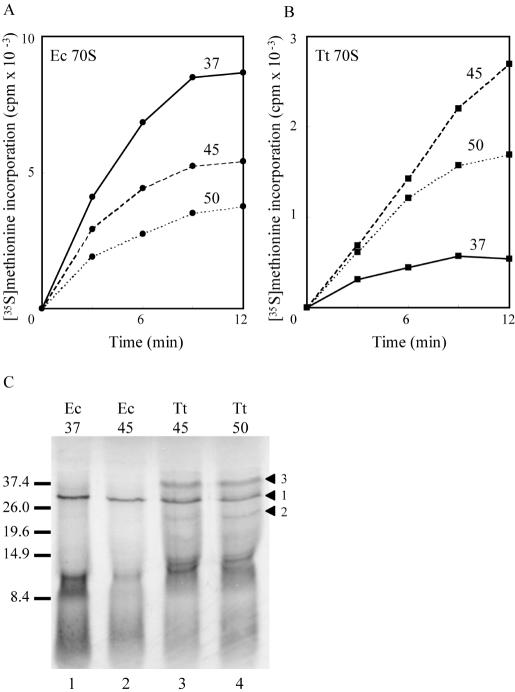
Figure 1.
Coupled transcription–translation directed by plasmid pUC18 in extracts of E.coli at 37°C (solid lines), 45°C (dashed lines) and 50°C (dotted lines). Time courses (A and B) measure the incorporation of [35S]-methionine into TCA-precipitable material directed by ribosomes from E.coli (Ec 70S) (A) or T.thermophilus (Tt 70S) (B). Background incorporation in the absence of ribosomes (usually <2%,) has been subtracted. (C) SDS–PAGE (12) of the products of translation terminated after 10 min. Samples containing ∼105 c.p.m. of TCA-precipitable material were loaded on a 15% acrylamide gel and electrophoresed at 150 V until just after the bromophenol blue had migrated out of the bottom of the gel. Following brief fixing and drying, the gel was exposed to film overnight. Arrowhead 1 marks the position of β-lactamase (31.5 kDa); arrowheads 2 and 3 mark apparently discrete products synthesized by T.thermophilus ribosomes. Size markers (kDa) are indicated at the left. Lanes 1 and 2: products from E.coli 70S at 37°C (lane 1) and 45°C (lane 2). Lanes 3 and 4: products from T.thermophilus 70S at 45°C (lane 3) and 50°C (lane 4).
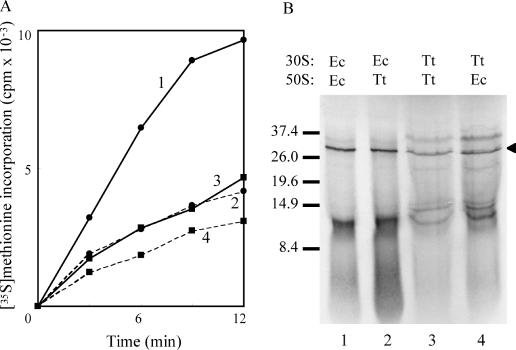
Figure 2.
In vitro translation by subunit combinations (5 pmol each) at 45°C. (A) Time course of incorporation of [35S]-methionine into TCA-precipitable material. Filled circles, solid line (1): E.coli 30S, E.coli 50S. Filled circles, broken line (2): E.coli 30S, T.thermophilus 50S. Filled squares, solid line (3): T.thermophilus 30S, T.thermophilus 50S. Filled squares, broken line (4): T.thermophilus 30S, E.coli 50S. Synthesis by each set of subunits in the absence of the other was negligible, indicating that subunit preparations had very low contamination with other ribosome species. (B) SDS–PAGE of the products of translation. The samples were prepared and gel electrophoresed and processed as described in the legend to Figure 1. Lane 1: E.coli 30S, E.coli 50S; lane 2: E.coli 30S, T.thermophilus 50S; lane 3: T.thermophilus 30S, T.thermophilus 50S; lane 4: T.thermophilus 30S, E.coli 50S. The arrowhead marks the position of β-lactamase (31.5 kDa); size markers (kDa) are indicated on the left.
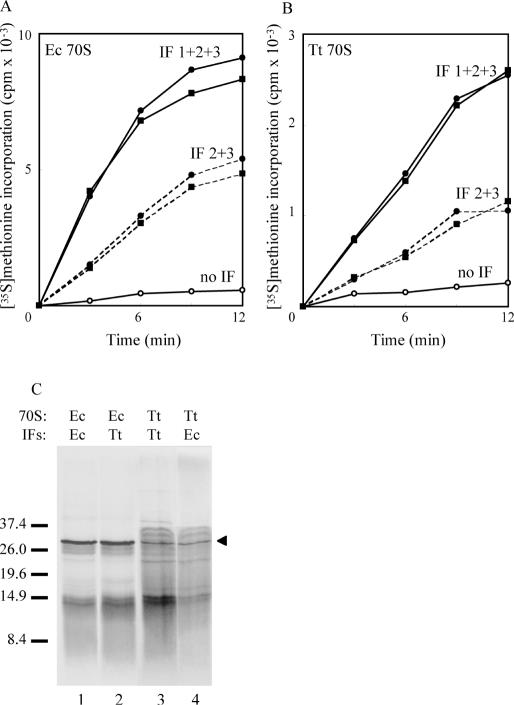
Figure 4.
Effect of added IFs on coupled transcription–translation by (A) E.coli 70S and (B) T.thermophilus 70S at 45°C. Closed circles: E.coli IFs; closed squares: T.thermophilus IFs; solid lines: all three IFs; broken lines: IF2 plus IF3; open circles: no added IFs. In all assays, IFs, where present, were at a 2-fold molar excess over ribosomes. (C) SDS–PAGE of the products of translation. Samples were prepared by TCA precipitation, and gel electrophoresed and processed as described in the legend to Figure 1. Lane 1: E.coli 70S, E.coli IFs; lane 2: E.coli 70S, T.thermophilus IFs; lane 3: T.thermophilus 70S, T.thermophilus IFs; lane 4: T.thermophilus 70S, E.coli IFs. The arrowhead marks the position of β-lactamase (31.5 kDa); size markers (kDa) are indicated on the left.
Synthesis mix
The mix contains energy sources, amino acids, buffer, crowding agents and monovalent cations (concentration in synthesis mix are indicated in parenthesis). Composition: HEPES–KOH pH 8.2 (205 mM); DTT (7 mM); ATP, pH adjusted to 7.0 with KOH (5 mM); CTP, GTP, UTP, pH adjusted as for ATP (each at 3.5 mM); amino acids, excluding methionine (each at 1.5 mM); PEG-6000/8000 (8% w/v); folinic acid (293 μM); pyruvate kinase (250 U in 20% v/v glycerol); phosphoenolpyruvate, pH adjusted to 7.0 with Tris base (107.8 mM); ammonium acetate (143 mM); potassium acetate (286 mM).
Preparation of ribosomes and subunits
Preparative techniques were essentially identical for particles from both E.coli and T.thermophilus cells. Cultures of E.coli strain MRE600 were grown as described above; cultures of T.thermophilus strain HB8 were grown in Thermus Enhanced Medium (ATCC medium 1598) at 72°C with vigorous aeration, and cells were harvested when OD600 readings of 0.5–0.6 were reached. Cells were harvested and washed as described above, but cracked at 20 000 psi in the presence of DNase I (10 μg/ml final; Worthington). Cell debris was pelleted as described above, the S30 was brought to 0.5% w/v with Brij58, incubated for 30 min at 4°C, then layered over an equal volume of buffer containing 10 mM HEPES–KOH pH 7.6, 10 mM MgCl2, 1 M NH4Cl, 10% w/v sucrose, 5 mM β-mercaptoethanol. The ribosomes were pelleted by centrifugation (Ty65 rotor, 40 000 r.p.m., 4 h, 4°C). Ribosomes were resuspended in CB, layered over 10 vol of the same buffer containing 40% w/v sucrose, and sedimented by centrifugation (Ty65 rotor, 20 000 r.p.m., 14 h, 4°C). Finally, ribosomes were resuspended in CB and stored in small aliquots at −80°C.
For subunit preparations, ribosomes were dialyzed against buffer containing 10 mM HEPES–KOH pH 7.6, 1 mM MgCl2, 50 mM NH4Cl, 5 mM β-mercaptoethanol, layered over 36-ml 0–12.5% sucrose gradients in the same buffer (max. 60 A260 units/gradient), and centrifuged (SW28 rotor, 15 000 r.p.m. 16 h, 4°C). Gradients were collected by pumping through an ISCO density gradient fractionator and appropriate fractions pooled conservatively. The MgCl2 concentration was raised to 10 mM, the sucrose was removed by dialysis against CB, and subunits pelleted by centrifugation (Ty65 rotor, 40 000 r.p.m., 14 h, 4°C). Finally, subunits were resuspended in CB and stored in small aliquots at −80°C.
Cloning of T.thermophilus IFs
The strategy for cloning each of the IFs was identical. Oligonucleotides were designed to be complimentary to up- and downstream regions of genomic DNA immediately flanking the factor of interest. An NdeI site was incorporated in the upstream oligonucleotide and an EcoRI site in the downstream oligonucleotide. PCR amplification from genomic T.thermophilus DNA was carried out, and products were digested with NdeI and EcoRI and ligated into the similarly digested pET30b vector (Novagen, Wis.). Importantly, expression is tightly regulated by the T7 promoter, avoiding problems normally associated with lethality due to overexpression of translational factors. Ligation mixes were transformed into competent E.coli DH5α cells and appropriate regions of plasmids recovered from transformants were analyzed by complete sequencing of both strands (UC Davis, CA). Sources of the genomic DNA sequences and sequences of the oligonucleotide PCR primers were as follows: IF1 (13) (accession number AJ495839) upstream: 5′-GATATACATATGGCGAAGGAGAAGGACACCATTCGG-3′, downstream: 5′-GCTCGAATTCACTTGCGGTAAACGATCCGGCCCCGCG-3′; IF2 (14) (accession number Z48001) upstream: 5′-GATATACATATGGCCAAGGTAAGGATCTACC-3′, downstream: 5′-GCTCGAATTCAGGCGGGGACCTCCACCATCTGGAAGGCC-3′; IF3 (13) (accession number AJ495840) upstream: 5′-GATATACATATGAAGGAGTACCTGACCAACGAACGC-3′, downstream: 5′-GCTCGAATTCAGGCGGAGACCTTCACCGGGGCG-3′.
As a consequence of the cloning strategy, silent mutations were introduced into start and stop codons in some of the factors. Termination codons in IF1 and IF3, which in the wild-type gene are TAG, were changed to TGA. The initiation codon of the native protein IF3 is ATA (13) and was changed to ATG.
Translational factor purification
Cultures of E.coli strain BL21(DE3) carrying each of the pET30b constructs encoding the relevant cloned T.thermophilus translational factor were grown for approximately 2 h at 37°C, then expression of the protein was induced by addition of isopropyl-β-d-thiogalactopyranoside (1 mM final concentration) and growth continued for 2–4 h. Cells were harvested and washed by centrifugation (Sorvall GSA rotor, 5000 r.p.m.), cracked in a French pressure cell (20 000 psi) in CB, and cell debris removed by centrifugation (Beckman L8, Ty65 rotor, 30 000 rpm, 4°C). Many of the contaminating E.coli proteins were denatured by heating at 72°C for 30 min and removed by centrifugation (12 000 g, 10 min). Proteins were then concentrated by centrifugation using Centricon (Amicon) units with appropriate molecular weight cut-offs. Concentrations were determined from the extinction coefficients. Chromosomal DNA fragments were removed by treatment with staphylococcal nuclease as described above.
IFs from E.coli and Bacillus stearothermophilus were purified as previously described (15,16), and were a generous gift from Anna La Teana (University of Ancona, Italy).
Translational factor activity assays
IF2 was assayed for its ability to hydrolyze GTP in a ribosome-dependent manner (17). IF1 produced an 8-fold increase in [35S]fmet-tRNAfmet binding to ribosomes in the presence of an excess of IF2 (18). IF3 was assayed for its ability to dissociate 70S ribosomes into subunits (18). All factors were active in the assays employed, regardless of the source of the ribosomes.
RESULTS
Effect of temperature on in vitro protein synthesis
One of the obvious parameters to address in attempting in vitro protein synthesis by ribosomes from a thermophilic organism in extracts derived from a mesophile is the assay temperature. Optimum growth for E.coli MRE600 cells is ∼37°C, although the upper limit of cell survival is ∼49°C with appropriate nutritional supplements (19). For T.thermophilus, the optimum growth temperature is 72°C, with a minimum temperature for growth of ∼55°C (7). Accordingly, incorporation of [35S]-methionine into TCA-precipitable material using the in vitro transcription–translation system was measured over a range of temperatures (Figure 1A and B). E.coli ribosomes performed optimally at 37°C (Figure 1A), with synthesis levels reducing as the temperature increased. At 37°C, as anticipated, T.thermophilus ribosomes barely synthesized peptide (Figure 1B). At 45°C, however, synthesis was around 25–30% that of E.coli at 37°C (Figure 1B), an unexpected result as this is almost 30°C below the optimum growth temperature for the organism and 10°C below its lowest viable growth temperature. The lower incorporation at 50°C compared with that at 45°C was almost certainly a consequence of heat denaturation of some essential E.coli transcriptional and/or translational component(s) of the reaction; indeed, a precipitate in the reaction mix became visible over the time course of the reaction. When incorporation directed by T.thermophilus ribosomes was examined over shorter time points (data not shown), the initial rate of the reaction was actually faster at 50°C than at 45°C, implying that the major contributory factor to the reduced rate of incorporation relative to that seen with E.coli ribosomes might well be temperature.
Peptides as short as five amino acids are precipitated by TCA; simply measuring radiolabel incorporation, therefore, gives little estimate of the processivity of the reaction. To assess the extent of elongation achievable, we examined the translation products of the reaction by SDS–PAGE (Figure 1C). The principle product formed by E.coli ribosomes (lanes 1 and 2) at both 37 and 45°C is a single polypeptide of a molecular weight corresponding to that of β-lactamase (31 557 Da), the largest protein encoded by plasmid pUC18. Crucially, T.thermophilus ribosomes are capable of synthesizing full-length protein in the E.coli milieu (lanes 3 and 4), with a band produced that also corresponds in size to β-lactamase (arrowhead 1). The temperature at which the reaction is carried out (45 or 50°C) appears to make little difference to the size and range of the polypeptides synthesized. If translation were totally aberrant, with random initiation, termination and frameshifting, it is likely that a complete range of product sizes would be obtained which, upon gel electrophoresis, would produce a smear. Other open reading frames on pUC18 exist, however, and some of these have recognizable, although sub-optimal, ribosome binding sites (20). Notably, one within the β-lactamase gene itself encodes an approximately 24-kDa product (arrowhead 2), and there are a number that would generate polypeptides in the 10–13 kDa range (ExPASy Translate Tool; http://us.expasy.org), consistent with the heavy banding pattern seen in that region of the gel. There is also a product formed of ∼39 kDa (arrowhead 3), ∼7.5 kDa larger than β-lactamase, perhaps a product of, e.g. stop codon readthrough. Whatever the explanation for these extra products, however, there is no doubt that T.thermophilus ribosomes are capable of synthesizing full-length protein in an E.coli background and, therefore, of interacting in an appropriate fashion with all of the E.coli translational factors and tRNAs.
In vitro protein synthesis by heterologous ribosomes
From cryo-electron microscopy studies, it is now clear that there are large relative rotational movements between the subunits during the elongation cycle [for review see (21)]. To address the question of whether ribosomes from species adapted to different growth temperatures might have developed strongly temperature-dependent movement, we examined protein synthesis with homologous and heterologous combinations of subunits. Broadly, any combination involving a T.thermophilus component has reduced synthesis capacity (Figure 2A), although both heterologous combinations are capable of protein synthesis. Closer examination of the translation products (Figure 2B) was more informative. With E.coli 30S subunits, regardless of the source of the 50S subunits (lanes 1 and 2), the product banding patterns are identical and closely similar to the products of E.coli 70S (cf. lane 1; Figure 1C, lanes 1 and 2). Conversely, with T.thermophilus 30S, again regardless of the 50S source, the banding patterns are also identical (lanes 3 and 4) but are now the same as those produced by T.thermophilus 70S (Figure 1C, lanes 3 and 4). Clearly, the source of the 30S subunit is critical in determining the range of products synthesized. Presumably, with T.thermophilus 30S subunits, the sub-optimal temperature may permit the formation of initiation complexes on mRNA sites with lower affinity than the correct ribosome binding sites, resulting in aberrant translation initiation and hence erroneous products. The overall conclusion, however, is that T.thermophilus subunits in combinations with E.coli subunits are able to synthesize full-length polypeptides, suggesting that there must be a high level of conservation of appropriate inter-subunit bridging.
Effects of adding T.thermophilus IFs to the coupled transcription–translation system
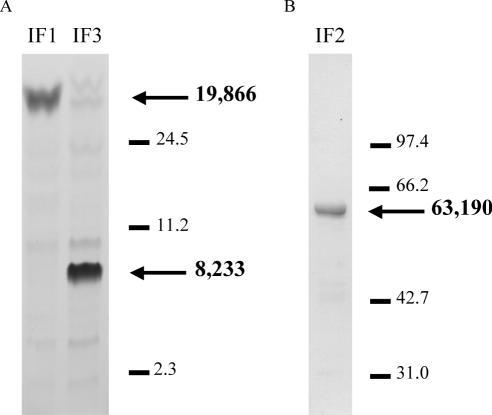
The coupled transcription–translation system is almost totally dependent upon the addition of all three IFs, which in the experiments described thus far have been supplied from E.coli. We have, therefore, the opportunity to examine whether the products of translation are affected by the source of the factors. All three T.thermophilus IFs have previously been cloned (13,14), but with histidine tags to aid purification. Although the presence of tags has not been reported in the case of IFs to cause problems, for elongation factor G, at least, there is evidence that histidine tagging may inhibit translocation (22). For use in the coupled transcription–translation system, purification of the factors to homogeneity was a lesser requirement than the need to use proteins of native sequence, so we cloned each of the factors without histidine tags so that only completely native protein would be expressed. Purification of each of the factors relied largely upon the thermostability of the T.thermophilus proteins. Thus, a single heat denaturation step of the E.coli S30 resulted in >90% purification (Figure 3) (12,23).
Figure 3.
Gel electrophoresis of cloned T.thermophilus translational factors (∼100 pmol/lane). (A) IF1 and IF3 analyzed in the gel system of Schägger and von Jagow (23) for low molecular weight proteins. (B) IF2 electrophoresed in a standard 15% SDS gel (12). Both gels were stained with Coomassie blue. Size markers (kDa) are indicated with lines; arrows indicate the respective IFs with molecular weight (Da) calculated from the amino acid sequence.
At a 2-fold molar excess over ribosomes of each of IF1, IF2 and IF3, the activity of E.coli ribosomes was the same, regardless of the source of the factors (Figure 4A). Addition of each of the factors individually did not stimulate translation, nor did the combination of IF1 and IF2 or of IF1 and IF3 (data not shown, but closely similar to that shown for synthesis in the absence of added IFs). The combination of IF2 and IF3, however, did allow translation to about half the level achieved with all three factors (Figure 4A), indicating that a low level of IF1 must be present in the S100. Similarly, when T.thermophilus ribosomes were used, all three IFs were required for maximal synthesis, although again the source of the factors made no difference (Figure 4B). Again, only the IF2 plus IF3 combination promoted synthesis (to ∼50% of the maximal level). At the least, it would appear that IFs from either E.coli or T.thermophilus are equally effective in supporting protein synthesis in the coupled transcription–translation system.
The effects of IFs on the pattern of translation from either E.coli (Figure 4C, lanes 1 and 2) or T.thermophilus (lanes 3 and 4) ribosomes were examined with either E.coli IFs (lanes 1 and 4) or T.thermophilus IFs (lanes 2 and 3). Overall, the products of translation appear independent of the source of IFs, with a polypeptide corresponding in size to β-lactamase being synthesized with all sets of ribosome–factor combinations. Curiously, the products in the 10–13 kDa range appear much more intense, relative to the 31.5 kDa band in the homologous T.thermophilus combination (lane 3). We currently have no explanation for this result.
DISCUSSION
The notion that protein synthesis is a universal phenomenon achieved by broadly similar mechanisms across each of the three domains of life is compelling, but one that has rarely been tested. We and others have examined the consequences of constructing chimeric ribosomes, asking questions concerning the mechanistic universality of the GTP hydrolysis center on the large subunit (24–26) and of the peptidyl transferase center (27). Recently, the compatibility of protein L7/L12 between mitochondrial and E.coli ribosomes, focussing again on the GTP hydrolysis center, underlined the functional conservation of the proteins, but also differences in detail (28). These differences between organisms can be exploited to advance our understanding of translation at the molecular level. The use of a coupled transcription–translation system in which heterologous ribosomes can be examined is, therefore, an important tool for determining whether differences exist in the details of protein synthesis between organisms. For example, questions concerning the compatibilities of translational factor interactions and of subunit–subunit communication can be addressed. Determining which molecular contacts are critical for function, of course, relies heavily on available atomic-level structures in order to interpret results. While the reasons for choosing T.thermophilus for this study were in part based on the crystal structures available, an important general point is that ribosomes from many other organisms may potentially be studied in this way. Additionally, the in vitro system has been used to compare relative rates of translation between mutant and wild-type ribosomes and to examine antibiotic resistance profiles.
We have shown here that it is indeed possible to examine protein synthesis directed by T.thermophilus ribosomes in an E.coli extract. Despite its low processivity, compared either to calculated in vivo rates (29) or to optimized in vitro systems (30), it appears that the principle reason for the low rates of protein synthesis achieved by T.thermophilus ribosomes and subunits is primarily an effect of temperature. This is not surprising, given the great differences in growth temperature optima of the two organisms (72°C for T.thermophilus, 37°C for E.coli). On the other hand, it may well be possible that even when optimal conditions are achieved, T.thermophilus ribosomes may not be capable of translation at the same rate as those from E.coli; doubling times for T.thermophilus HB8 (39 min) (31) exceed twice that of E.coli MRE600 (18 min; our unpublished measurement). The fact that full-length proteins are synthesized by heterologous combinations of subunits indicates that subunit–subunit interactions are highly conserved. While this might not be an unexpected observation given the known sequence conservation of the bridge elements (1), it is likely that the temperature dependence is due to differences in distal, less well-conserved regions. Equally, that the system is capable of synthesizing a product of the correct size, also implies that each of the E.coli translational factors is capable of appropriate and functional interaction with T.thermophilus ribosomes. While it has not been possible to examine all of the translational factors in detail here, the dependence of the system upon exogenous IFs has at least allowed a preliminary examination of their compatibility at the level of gross peptide synthesis in this in vitro environment. It is worth noting here that IFs from Bacillus stearothermophilus are also able to support translation with E.coli ribosomes (our unpublished observation), so a reasonably high level of conservation in all three IF interactions with the ribosome seems to be maintained.
Initiation of translation involves the assembly of the 30S subunit with an initiation codon at the P site, initiator tRNA and the three IFs [for review see (32)]. The precise roles of the individual IFs in this process are becoming clearer, aided by some crystallographic details. IF1 augments the activity of both IF2 and IF3, and there is a suggestion that it may promote initiation complex formation by occluding the 30S subunit A site. Upon binding, IF1 appears to cause a number of conformational changes in the subunit (5). The protein is highly conserved between E.coli and T.thermophilus (13), and given that the protein structure appears not to change upon binding the 30S subunit and that the conformational changes it exerts are upon highly conserved rRNA regions, the compatibility of IFs from both organisms is not too surprising. IF2, a GTPase protein, is less well conserved, with a substantial size difference between the T.thermophilus and E.coli orthologs. There is, however, reasonable structural conservation in the N-terminal ribosome binding region and the nucleotide binding domain (33). The NMR structure for the N-terminus of the E.coli protein, in particular, indicates that this three-helix structure should be well conserved (34). IF2 appears to function by binding the initiator tRNA to the P site and is important for the recruitment of the 50S subunit to the 30S initiation complex. Clearly, it interacts with the factor binding domain of the 50S subunit (35), although the role of GTP hydrolysis is disputed. Given the large size of this protein and the flexibility of some of its regions, the interchangeability of IF2 between E.coli and T.thermophilus is somewhat surprising. Detailed analysis of the interaction of the IF2s with the subunits from each of these organisms will be the subject of future scrutiny. The last in the trio of initiation factors, IF3, is also well conserved (13). This factor functions to prevent subunit association until initiation complex formation is appropriate, at which time it facilitates the binding of IF1 and IF2. The protein is structurally divided into approximately two equal halves, separated by a lysine-rich linker region that has been proposed to act as a strap but can be of variable length (36). The structures of both the N- and C-terminal halves have been examined crystallographically in 30S subunits of T.thermophilus (6) and reveal that the C-terminal domain binds close to the anti-Shine–Dalgarno region of 16S rRNA, while the N-terminus binds in the vicinity of the P site, thus explaining the requirement for the flexibility of the linker. As with IF1, binding of IF3, and particularly the C-terminal domain, induces conformational changes in the 30S subunit and not in IF3 itself, so again the exchangeability of the factor indicates conservation of the recognition sequences.
The ribosome itself undergoes a series of conformational changes during translation, in particular large inter-subunit motions (21) that are, of course, subject to kinetic constraints. This may help to explain why T.thermophilus ribosomes function so slowly at sub-optimal temperatures. It would appear, however, that there are no inherent species-specific barriers to inter-subunit communication between ribosomes of the two organisms studied here. The nature of the 30S subunit- and IF-specific differences in the products of translation between T.thermophilus and E.coli currently remain unresolved. Overall, however, we conclude that the conservation in the bacterial protein synthetic machinery is high enough, at the least, to warrant the extrapolation of crystallographic data from T.thermophilus to E.coli for functional considerations.
Acknowledgments
ACKNOWLEDGEMENTS
We thank A. La Teana for purified IFs from E.coli and B.stearothermophilus. Members of the Dahlberg lab, past and present, are thanked for their continuing support and insightful comments. This research was supported by NIH grant GM19756 to A.E.D.
REFERENCES
- 1.Cannone J.J., Subramanian,S., Schnare,M.N., Collett,J.R., D'Souza,L.M., Du,Y., Feng,B., Lin,N., Madabusi,L.V., Muller,K.M., Pande,N., Shang,Z., Yu,N. and Gutell,R.R. (2002) The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics, 3, 2. [Correction: BMC Bioinformatics, 3, 15.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wimberly B.T., Brodersen,D.E., Clemons,W.M.,Jr, Morgan-Warren,R.J., Carter,A.P., Vonrhein,C., Hartsch,T. and Ramakrishnan,V. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–339. [DOI] [PubMed] [Google Scholar]
- 3.Yusupov M.M., Yusupova,G.Z., Baucom,A., Lieberman,K., Earnest,T.N., Cate,J.H. and Noller,H.F. (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science, 292, 883–896. [DOI] [PubMed] [Google Scholar]
- 4.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 5.Carter A.P., Clemons,Jr.,W.M., Brodersen,D.E., Morgan-Warren,R.J., Hartsch,T., Wimberly,B.T. and Ramakrishnan,V. (2001) Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science, 291, 498–501. [DOI] [PubMed] [Google Scholar]
- 6.Pioletti M., Schluenzen,F., Harms,J., Zarivach,R., Gluehmann,M., Avila,H., Bashan,A., Bartels,H., Auerbach,T., Jacobi,C., Hartsch,T., Yonath,A. and Franceschi,F. (2001) Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J., 20, 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams R. and Sharp,R. (1995) The taxonomy and identification of Thermus. In Williams,R. and Sharp,R. (eds), Thermus Species. Plenum Press, New York, pp. 1–39. [Google Scholar]
- 8.Thompson J., Rae,S. and Cundliffe,E. (1984) Coupled transcription–translation in extracts of Streptomyces lividans. Mol. Gen. Genet., 195, 38–43. [DOI] [PubMed] [Google Scholar]
- 9.Pratt J.M., Boulnois,G.J., Darby,V., Orr,E., Wahle,E. and Holland,I.B. (1981) Identification of gene products programmed by restriction endonuclease DNA fragments using an E.coli in vitro system. Nucleic Acids Res., 9, 4459–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zubay G. (1973) In vitro synthesis of protein in microbial systems. Annu. Rev. Genet., 7, 267–287. [DOI] [PubMed] [Google Scholar]
- 11.Calcutt M.J. and Cundliffe,E. (1989) Use of a fractionated coupled transcription–translation system in the study of ribosomal resistance mechanisms in antibiotic-producing Streptomyces. J. Gen. Microbiol., 135, 1071–1081. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 13.Wolfrum A., Brock,S., Mac,T. and Grillenbeck,N. (2003) Expression in E. coli and purification of Thermus thermophilus translation initiation factors IF1 and IF3. Protein Expr. Purif., 29, 15–23. [DOI] [PubMed] [Google Scholar]
- 14.Vornlocher H.P., Scheible,W.R., Faulhammer,H.G. and Sprinzl,M. (1997) Identification and purification of translation initiation factor 2 (IF2) from Thermus thermophilus. Eur. J. Biochem., 243, 66–71. [DOI] [PubMed] [Google Scholar]
- 15.Gualerzi C.O., Spurio,R., La Teana,A., Calogero,R., Celano,B. and Pon,C.L. (1989) Site-directed mutagenesis of Escherichia coli translation initiation factor IF1. Identification of the amino acid involved in its ribosomal binding and recycling. Protein Eng., 3, 133–138. [DOI] [PubMed] [Google Scholar]
- 16.Soffientini A., Lorenzetti,R., Gastaldo,L., Parlett,J.H., Spurio,R., La Teana,A. and Islam,K. (1994) Purification procedure for bacterial translational initiation factors IF2 and IF3. Protein Expr. Purif., 5, 118–124. [DOI] [PubMed] [Google Scholar]
- 17.Stark M. and Cundliffe,E. (1979) On the biological role of ribosomal protein BM-L11 of Bacillus megaterium, homologous with Escherichia coli ribosomal protein L11. J. Mol. Biol., 134, 767–779. [DOI] [PubMed] [Google Scholar]
- 18.Wahba A.J., Woodley,C.L. and Dholakia,J.N. (1990) Initiation of protein synthesis. In Spedding,G. (ed.), Ribosomes and Protein Synthesis: A Practical Approach. IRL Press, Oxford, pp. 31–48. [Google Scholar]
- 19.Ingraham J.L. and Marr,A.G. (1996) Effect of temperature, pressure, pH, and osmotic stress on growth. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, 2nd edn. ASM Press, Washington,DC., pp. 1570–1578. [Google Scholar]
- 20.Ma J., Campbell,A. and Karlin,S. (2002) Correlations between Shine–Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bacteriol., 184, 5733–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank J. (2003) Electron microscopy of functional ribosome complexes. Biopolymers, 68, 223–233. [DOI] [PubMed] [Google Scholar]
- 22.Studer S.M., Feinberg,J.S. and Joseph,S. (2003) Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J. Mol. Biol., 327, 369–381. [DOI] [PubMed] [Google Scholar]
- 23.Schägger H. and von Jagow,G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem., 166, 368–379. [DOI] [PubMed] [Google Scholar]
- 24.Musters W., Concalves,P.M., Boon,K., Raue,H.A., van Heerikhuizen,H. and Planta,R.J. (1991) The conserved GTPase center and variable region V9 from Saccharomyces cerevisiae 26S rRNA can be replaced by their equivalents from other prokaryotes or eukaryotes without detectable loss of ribosomal function. Proc. Natl Acad. Sci. USA, 88, 1469–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson J., Musters,W., Cundliffe,E. and Dahlberg,A.E. (1993) Replacement of the L11 binding region within E.coli 23S ribosomal RNA with its homologue from yeast: in vivo and in vitro analysis of hybrid ribosomes altered in the GTPase centre. EMBO J., 12, 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velichutina I.V., Rogers,M.J., McCutchan,T.F. and Liebman,S.W. (1998) Chimeric rRNAs containing the GTPase centers of the developmentally regulated ribosomal rRNAs of Plasmodium falciparum are functionally distinct. RNA, 4, 594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson J., Tapprich,W.E., Munger,C. and Dahlberg,A.E. (2001) Staphylococcus aureus domain V functions in Escherichia coli ribosomes provided a conserved interaction with domain IV is restored. RNA, 7, 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terasaki M., Suzuki,T., Hanada,T. and Watanabe,K. (2004) Functional compatibility of elongation factors between mammalian mitochondrial and bacterial ribosomes: characterization of GTPase activity and translation elongation by hybrid ribosomes bearing heterologous L7/12 proteins. J. Mol. Biol., 336, 331–342. [DOI] [PubMed] [Google Scholar]
- 29.Pedersen S. (1984) Escherichia coli ribosomes translate in vivo with variable rate. EMBO J., 3, 2895–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pavlov M.Y. and Ehrenberg,M. (1996) Rate of translation of natural mRNAs in an optimized in vitro system. Arch. Biochem. Biophys., 328, 9–16. [DOI] [PubMed] [Google Scholar]
- 31.Cameron D.M., Thompson,J., Gregory,S.T., March,P.E. and Dahlberg,A.E. (2004) Thiostrepton-resistant mutants of Thermus thermophilus. Nucleic Acids Res., 32, 3220–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gualerzi C.O., Brandi,L., Caserta,E., La Teana,A.L., Spurio,R., Tomsic,J. and Pon,C.L. (2000) Translation initiation in bacteria. In Garrett,R.A. et al. (eds), The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions. ASM Press, Washington,DC, pp. 477–494. [Google Scholar]
- 33.Vornlocher H.P., Kreutzer,R. and Sprinzl,M. (1997) Organization of the Thermus thermophilus nusA/infB operon and overexpression of the infB gene in Escherichia coli. Biochimie, 79, 195–203. [DOI] [PubMed] [Google Scholar]
- 34.Laursen B.S., Mortensen,K.K., Sperling-Petersen,H.U. and Hoffman,D.W. (2003) A conserved structural motif at the N-terminal of bacterial translation initiation factor IF2. J. Biol. Chem., 278, 16320–16328. [DOI] [PubMed] [Google Scholar]
- 35.La Teana A., Gualerzi,C.O. and Dahlberg,A.E. (2001) Initiation factor IF 2 binds to the alpha-sarcin loop and helix 89 of Escherichia coli 23S ribosomal RNA. RNA, 7, 1173–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Cock E., Springer,M. and Dardel,F. (1999) The interdomain linker of Escherichia coli initiation factor IF3: a possible trigger of translation initiation specificity. Mol. Microbiol., 32, 193–202. [DOI] [PubMed] [Google Scholar]