Abstract
RNA polymerase II carboxy terminal domain (CTD) kinases are key elements in the control of mRNA synthesis. Yeast CTD kinase I (CTDK-I), is a non-essential complex involved in the regulation of mRNA synthesis at the level of transcription elongation, pre-mRNA 3′ formation and nuclear export. Here, we report that CTDK-I is also involved in ribosomal RNA synthesis. We show that CTDK-I is localized in part in the nucleolus. In its absence, nucleolar structure and RNA polymerase I transcription are affected. In vitro experiments show an impairment of the Pol I transcription machinery. Remarkably, RNA polymerase I co-precipitates from cellular extracts with Ctk1, the kinase subunit of the CTDK-I complex. In vitro analysis further demonstrates a direct interaction between RNA polymerase I and Ctk1. The results suggest that CTDK-I might participate in the regulation of distinct nuclear transcriptional machineries, thus playing a role in the adaptation of the global transcriptional response to growth signalling.
INTRODUCTION
Eukaryotic cells contain three nuclear RNA polymerases (Pol I, II and III) that can be distinguished by their polypeptide composition, gene specificity and sub-cellular localization (1). Pol I is localized in the nucleolus where it transcribes the 35S rDNA to generate the precursor transcript of the mature 25, 18 and 5.8S rRNA species. Pol I transcription accounts for ∼60% of the total transcription activity of exponentially growing yeast cells (2). Specific initiation requires four general initiation factors: the upstream activating factor complex (UAF), the TATA binding protein (TBP), the core factor complex (CF) and Rrn3 (3). Assembled onto the 35S rDNA promoter, UAF, TBP and CF form a pre-initiation complex that recruits the Pol I–Rrn3 complex (4–6), the only form of Pol I competent for initiation. Phosphorylation of Pol I is necessary for its interaction with Rrn3, suggesting that one way to regulate rDNA transcription is at the level of Pol I–Rrn3 complex formation (7). Strong functional and structural homologies can be drawn from yeast to mammals, and in both systems, rRNA synthesis rate is under the control of growth signalling (8–10).
Pol II produces mRNA precursors, and the majority of the snRNA implicated in splicing. A key element of transcription by Pol II is the carboxy terminal domain (CTD) of its largest subunit, Rpb1. The CTD is an essential domain specific to Pol II, composed of numerous repeats of the consensus heptapeptide YSPTSPS (11). This domain involved in the major steps of mRNA synthesis, is present under two distinct forms: a highly phosphorylated form (IIo) and a poorly phosphorylated form (IIa) (12). Only the Pol IIA is recruited to the promoter to participate in the formation of the pre-initiation complex, while it needs to be converted to the Pol IIO form in order to enter transcription elongation. A transcription cycle thus involves the cyclical phosphorylation and dephosphorylation of the CTD. In addition, the phosphorylation pattern of the Pol II CTD changes during the transcription process (13), and varies in response to growth conditions and environmental stress (14).
Four CTD kinases have been described in the yeast Saccharomyces cerevisiae, all of them being cyclin-dependent kinases: Kin28, Srb10, Bur1 and Ctk1. Kin28 is required for transcription of most class II genes (15). Like its human counterpart, Cdk7, it is an essential subunit of the general transcription factor TFIIH. Its activity is required for transcription initiation (16), and for recruitment of capping enzymes (17). Srb10 (18) is structurally related to mammalian Cdk8. This non-essential kinase phosphorylates the CTD prior to the formation of the pre-initiation complex, thus preventing its formation (16). BUR1/SGV1 encodes an essential CTD kinase that might be implicated in elongation (19,20). The non-essential CTD kinase I complex (CTDK-I) comprises three specific subunits (21): Ctk1, the catalytic subunit (22), Ctk2, the cyclin subunit (23), and Ctk3 a co-cyclin factor (24). Deletion of the CTK1 gene results in defects in the pattern of the CTD phosphorylation, and correlated defects in transcriptional induction of sets of genes, suggesting a role for CTDK-I in response to nutrient depletion (25). Several studies have implicated CTDK-I in the control of transcription elongation: (i) CTDK-I can modulate the elongation efficiency of Pol II in HeLa extracts (26), (ii) ctk mutants are synthetic lethal with mutants in various genes implicated in elongation control (27,28) and (iii), a recent study has revealed that Ctk1 might be the major kinase involved in CTD phosphorylation during elongation (29). Ctk1 has also been connected to 3′ end processing (30,31), splicing (32), histone methylation (33), DNA damage-induced transcription (34) and nuclear export of mRNA (35). In human, the closest characterized kinase to Ctk1 is Cdk9, the catalytic subunit of the positive elongation factor P-TEFb in metazoans (36). P-TEFb/Cdk9 is a key cellular factor that supports Tat trans-activation and HIV replication (37).
In higher eukaryotes, CTD kinases have been implicated in phosphorylation of additional substrates (38–42). Similarly, in yeast, both Kin28 and Srb10 have been shown to phosphorylate substrates other than the CTD. However, these substrates are all implicated in Pol II transcription. Kin28 phosphorylates subunits of Mediator (43,44), while Srb10 phosphorylates subunits of TFIID (43) and several Pol II specific transcription factors (45–48).
In this report, we show that, in addition to its role as a Pol II CTD kinase, CTDK-I is implicated in Pol I transcription.
MATERIALS AND METHODS
Yeast strains
The yeast strains used in this study were derived from W303-1B (MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1) and grown in either YPD or SD medium supplemented with casamino-acids and appropriate nutrients (SD-casa). The CTK tagged-strains were constructed by fusion of sequences encoding several epitope-tags (49) at the 3′ end of the CTK coding sequence: CTK1–HA (3-HA::Schizosaccharomyces pombe HIS5), CTK1–Myc (13-Myc::S.pombe HIS5) and CTK2–Myc (13-Myc::Escherichia coli KanR). Δctk1 and Δctk3 null mutants are described in (23). The isogenic Δpdr13 strain (pdr13::S.pombe HIS5) was constructed using the pFA6a-His3MX6 plasmid (49).
Ctk1–Myc purification
Wild-type and CTK1–Myc strains were grown in 12 l of YPD to an OD600 of 2, and pelleted by centrifugation. Fifty gram of cells were typically obtained and resuspended in 50 ml of buffer B [25 mM HEPES, pH 7.5, 10 mM MgCl2, 1 mM EDTA, 1 mM DTT, 20% glycerol (v/v) with 1 mM phenylmethylsulphonyl fluoride (PMSF) and complete protease inhibitors mixture (Roche)] containing 130 mM potassium acetate (B130). Cell suspensions were frozen at −70°C and disrupted in an Eaton press. After ultracentrifugation at 100 000 g for 1 h in a 45 Ti rotor (Beckman), crude extract was loaded on a SP-Sepharose column (10 × 2.5 cm, Amersham Biosciences) equilibrated in buffer B130 at a flow rate of 2 ml/min. Elution of bound proteins was performed by a 300 ml linear gradient of potassium acetate (0.13–1.2 M) in buffer B. Fractions (5 ml) containing Ctk1 were pooled, dialysed for 14 h against 2 l of buffer B100 (0.1 M potassium acetate), and loaded at a flow rate of 1 ml/min on a Poros 10 HQ column (0.8 ml, Perkin Elmer) equilibrated in B100. Ctk1 was eluted with a 13.4 ml linear gradient of potassium acetate (0.1–1M) in buffer B. Fractions (0.8 ml) were analysed by SDS–PAGE and western blot.
Purification of Ctk1–HA/Pol I complexes
Wild-type and CTK1–HA strains were submitted to the purification procedure described above except for the second chromatography step. The pooled solution obtained from SP-Sepharose chromatography was dialysed for 14 h against 2 l of buffer B containing 0.3 M potassium acetate and loaded at a flow rate of 0.5 ml/min on a MonoQ column (1 ml, Amersham Biosciences) equilibrated in buffer B300. Ctk1 was eluted with a 12 ml linear gradient of potassium acetate (0.3–1.6 M) in buffer B. Fractions (0.3 ml) containing Ctk1 were pooled and further loaded on an immuno-affinity column. Protein G-Sepharose (200 μl, Amersham Biosciences) crosslinked to HA-12CA5 antibodies (0.03 mg/ml) was packed in micro-tips and equilibrated with buffer B1000. Proteins were loaded by gravity flow, recycling the flow through eight times. After intensive washing with buffer B1000, the resin was re-equilibrated in buffer B130. Three successive elutions were performed in batch by adding 300 μl of buffer B130 containing 0.1 mg/ml of a synthetic haemagglutinin (HA) peptide. Elutions were pooled and analysed by SDS–PAGE and western blot with anti-HA, anti-Pol I and anti-Rrn3 (50) antibodies.
In vitro co-immunoprecipitation
Pol I, Pol IΔ and Pol I* were purified from yeast cells as previously described (51–53) and analysed by SDS–PAGE and silver staining. Expression of recombinant HA–Ctk1, HA–Ctk2 and HA–Ctk3 was performed as previously described (24) in E.Coli Rosetta (DE3) strain (Novagen). Immunoprecipitation experiments were performed with 5 μg of purified Pol I, and different combinations of bacterial extracts each expressing a single recombinant protein, with 30 μl of Protein G-Sepharose (Amersham Biosciences) coupled with 5 μl of a serum directed against A190 (54), as described in (24). Proteins were eluted with Laemmli buffer and separated by SDS–PAGE prior to western immunoblotting.
Immunofluorescence microscopy
Ten millilitre of cells grown in YPD at an OD600 of 0.5 were fixed by the addition of formaldehyde to a final concentration of 3.7%. Cells were further incubated for 10 min at room temperature, centrifuged and resuspended in KP–Sorbitol buffer (50 mM potassium phosphate, pH 6.5, 0.5 mM MgCl2, 1.2 M Sorbitol). Spheroplasts were prepared by incubation at 37°C for 30 min with zymolyase 100T (0.2 mg/ml, ICN). Cells were washed [once with phosphate-buffered saline (PBS) buffer containing 1% fish gelatine (Teleostan gelatin, Sigma), twice with PBS buffer with 0.1% Triton X-100 (Fluka) and once more with PBS buffer], prior to incubation with the primary antibody in PBS buffer for 1 h at room temperature. Antibodies used were: 10 μg/ml of purified monoclonal anti-Myc 9E10 (Babco), rabbit polyclonal serum anti-A190 at 1:100 dilution (54) and monoclonal anti-Nop1 Mab66 at 1:150 dilution (55). Cells were washed, resuspended in PBS buffer containing either anti-mouse Alexa 594 IgG or anti-rabbit Alexa 488 IgG (1:100 dilution, Molecular Probes) for 1 h at room temperature. After washing, cells were incubated with 0.5 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI, Sigma) in 0.5 ml of PBS buffer for 2 min. Cells were washed once with PBS buffer and observed with a Leica microscope.
In vivo RNA labelling
Ten millilitre of yeast cells were grown in SD-casa at an OD600 of 0.5. Cell labelling was conducted for 30 min at 30°C with 150 μCi of 3H-Uracil (Amersham Biosciences). After centrifugation, cells were washed with water prior to freezing in liquid nitrogen. Total RNA was extracted by hot acid–phenol treatment as described previously (56). RNA was quantified, and 5 μg was separated by electrophoresis on an 8% acrylamide gel containing 7 M urea in TBE. The gel was stained with ethidium bromide, dried and submitted to autoradiography.
Primer extension analyses
Ten millilitre of yeast cells were grown in SD-casa at an OD600 of 1 and 2, respectively. Total RNA was extracted by hot acid–phenol treatment as previously described (56). RNA was quantified by OD260 measurements, and 3 μg was analysed by primer extension [as in (57)] with 0.2 pmol of end-labelled 35S and 25S oligonucleotides (58), plus 5 pmol of cold 25S oligonucleotide. Extension products were separated by electrophoresis on an 8% acrylamide gel containing 7 M urea in TBE. The gel was dried, submitted to autoradiography and analysed with a PhosphorImager (Molecular Dynamics).
Chromatin immunoprecipitation
Wild-type and Δctk1 strains were grown in SD-casa medium to an OD600 of 2. Cross-link, cell lysis, chromatin preparation, cross-link reversal and immunoprecipitation were performed as described previously (59,60). Immunoprecipitation was realized with protein A-Sepharose (Sigma) coupled with polyclonal anti-A190 (54). Immuno-precipitated and total DNA were analysed by real-time PCR with SYBR Green (Applied Biosystems) using two sets of primer amplifying a 122 bp fragment specific of the 35S rDNA promoter region (GTGTGAGGAAAAGTAGTTGGGAGGTA, GACGAGGCCATTTACAAAAACATAAC), and a 236 bp fragment specific of the 5S rDNA (GACGAGGCCATTTACAAAAACATAAC, CATGTCTGGACCCTGCCCTCATA). For each experiment, triplicates of three dilutions were analysed; and five independent experiments were performed. The occupancy level was defined as the ratio of immunoprecipitated DNA over total DNA.
In vitro transcription assays
Purification of PA600 fractions from cells grown in YPD to an OD600 of 2 was performed as described previously (61). Specific transcription assays were performed [as in (61)] with 40 ng of YepSIRT template (62). Pol I from PA600 extracts (15 μg) was analysed by western immunoblotting with polyclonal antibodies raised against Pol I.
Immobilized templates assays
DNA templates containing [YepSIRT, (62)], or not containing (Δprom, O. Gadal, unpublished data) the 35S rDNA promoter were amplified by PCR using oligonucleotides 5′pSIRT (CCGCTTCCGCTTCCGCAGT) and biotinylated-3′pSIRT (biotin–CAACCCATCTTTGCAACGA). After purification, PCR fragments were bound to streptavidin-linked magnetic beads (Dynabeads M280 Streptavidin, Dynal) according to the manufacturer's recommendations. The DNA bound-beads were extensively washed and pre-incubated in recruitment buffer (20 mM HEPES, pH 7.8, 350 mM potassium acetate, 10 mM MgCl2, 50 μM EDTA, 2.5 mM DTT, 200 μM ATP, 1 mg/ml BSA). Protein complexes were assembled on immobilized DNA templates by incubation in recruitment buffer of 10 μg of PA600 fractions derived from wild-type and Δctk1 cells. Beads were washed, and bound proteins were eluted after enzymatic digestion (EcoRI) to release the templates. Elution fractions were analysed by western blot with anti-Pol I antibodies. Direct recording of the chemiluminescence corresponding to A49, A43, AC40 and A34.5 subunits was performed using a CCD camera, and quantification was achieved using the FluorChem software (Alpha Innotech).
RESULTS
The Ctk1 kinase interacts with Pol I
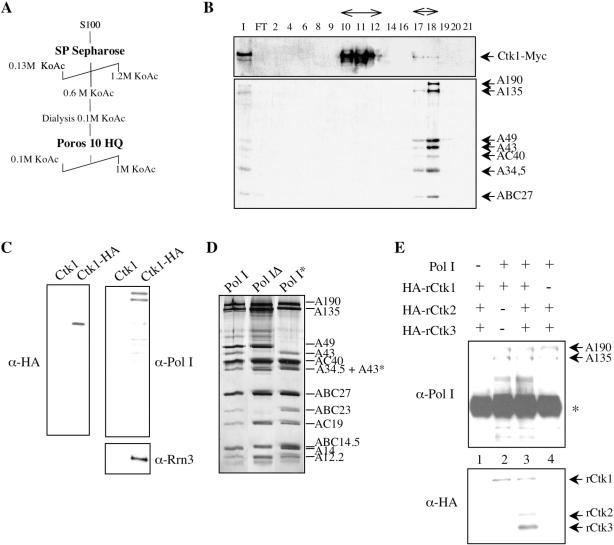
To isolate components interacting with CTDK-I, the Ctk1 kinase subunit was purified from yeast cells under mild salt conditions. After two subsequent ion exchange chromatography columns (Figure 1A), Ctk1–Myc was eluted from the Poros 10 HQ column in two well separated peaks, a major and a minor one (Figure 1B, upper panel). Strikingly, western blot analysis with anti-Pol II antibodies revealed the presence of subunits common to the three nuclear polymerases in the same fractions (data not shown). Further analyses specifically identified the different subunits composing the Pol I enzyme in the minor elution peak of Ctk1 (Figure 1B, lower panel). We did not succeed in immuno-affinity efficient purification of the Ctk1–Myc protein, probably due to a low accessibility of the Myc epitope. We therefore turned to the purification of Ctk1–HA (see Materials and Methods). After the second ion exchange chromatography column, fractions that contained the minor elution peak of Ctk1 were pooled and subjected to HA immuno-affinity purification. A control experiment was also conducted with extracts derived from a wild-type strain. Proteins eluted with HA peptide were analysed by western blot probed with anti-HA (Figure 1C, left panel) and anti-Pol I antibodies (Figure 1C, right upper panel). We observed the presence of Pol I in the fractions derived from the tagged-strain, showing an in vivo interaction between the Ctk1 kinase and Pol I.
Figure 1.
Ctk1 interacts with Pol I in vivo and in vitro. (A and B) Ctk1–Myc purification. (A) Fractionation scheme used to purify Ctk1–Myc from yeast exponential cells by ion exchange chromatographies. (B) The different fractions recovered were analysed by western blot with anti-Myc antibodies (upper panel), and polyclonal antibodies raised against Pol I (lower panel). I and FT indicate input and flow through samples, respectively. (C) Ctk1–HA immunopurification. After the second chromatography, fractions that contained the minor elution peak of Ctk1–HA from CTK1–HA cell extract, and identical fractions from wild-type (Ctk1) cell extract, were pooled and subjected to HA immuno-affinity purification. Eluates were analysed by western blot with anti-HA (left panel), anti-Pol I (right upper panel) and anti-Rrn3 (right lower panel) antibodies. (D and E) In vitro co-immunoprecipitation experiments. (D) Pol I, Pol IΔ and Pol I* purified from yeast cell were analysed by silver staining. A43* corresponds to a degraded form of the A43 subunit. (E) Pol I was incubated with different combinations of recombinant Ctk proteins each tagged with 2 HA epitopes (HA-rCtk). Immunoprecipitation was performed with polyclonal antibodies against the largest subunit of Pol I, A190. Western blot analysis with anti-Pol I (upper panel) and anti-HA (lower panel) antibodies. Asterisk indicates a band corresponding to immunoglobulin heavy chains.
Although only the Pol I–Rrn3 complex is competent for transcription initiation, the majority of Pol I is not associated with Rrn3 during exponential growth. To get an insight into the form of Pol I that was associated with Ctk1, we analysed the eluates of the HA affinity columns by western blotting with antibodies directed against Rrn3. We observed that Rrn3 and Ctk1 co-purify (Figure 1C, right lower panel). It is thus possible that the Pol I co-purified with Ctk1, or at least a substantial fraction, may be the Pol I–Rrn3 form of the enzyme.
To investigate a direct interaction between Ctk1 and Pol I, we performed in vitro co-immunoprecipitation experiments. Three different forms of Pol I were purified from yeast cell and analysed by SDS–PAGE and silver staining (Figure 1D). Pol IΔ is a form of Pol I lacking three subunits: A14, ABC23 and A43 (53), while Pol I* is devoid of A34.5 and A49 subunits (52). HA-tagged versions of the different Ctk subunits were expressed in E.coli cells from the T7 promoter (24), and the bacterial extracts were preincubated independently with each type of polymerase prior to Pol I immunoprecipitation with anti-A190 polyclonal antibodies. When Pol I was incubated with the three distinct Ctk subunits, its precipitation resulted in efficient co-precipitation of the CTDK-I complex (Figure 1E, lane 3). Similarly, after incubation with Ctk1 alone, Pol I precipitation resulted in efficient co-precipitation of the kinase subunit (Figure 1E, lane 2). In contrast, when Pol I was incubated with the Ctk2 and Ctk3 subunits together, neither Ctk2 nor Ctk3 was recovered after Pol I precipitation (Figure 1E, lane 4). Specific co-precipitation of Pol I and Ctk1 was also observed with the Pol IΔ and Pol I* preparations (data not shown).
We conclude that Pol I interacts with the CTDK-I complex as well as with the Ctk1 subunit alone. This interaction does not require the integrity of the Pol I as it does not require the presence of either A14, A34.5, A43, A49 or ABC23 subunit. Furthermore, Pol I does not interact with the Ctk2 cyclin and the Ctk3 co-cyclin subunits, showing that a direct interaction between the CTDK-I complex and Pol I is mediated by the kinase subunit.
Sub-cellular localization of the CTDK-I complex
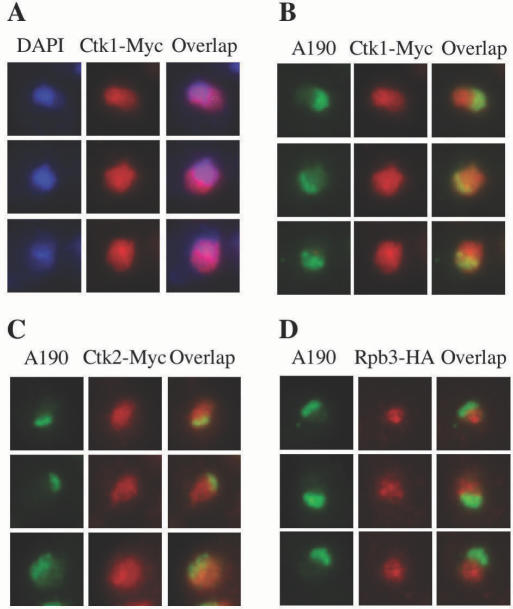
The observation that Ctk1 interacts with a nucleolar component prompted us to study CTDK-I sub-cellular localization. For this purpose, we have epitope-tagged each of its three subunits (see Materials and Methods). The cellular localization of Ctk1–Myc was analysed by immunofluorescence microscopy using monoclonal anti-Myc antibodies. As expected, we observed a nuclear localization in cells grown to early exponential phase (Figure 2A). Myc staining appeared to overflow the DAPI staining that preferentially reveals the nucleoplasmic DNA, suggesting that Ctk1 might be partly localized in the nucleolar compartment. To confirm the presence of Ctk1 in the nucleolus, we next used polyclonal antibodies raised against the largest subunit of Pol I, A190. These antibodies allow the specific staining of the nucleolus that appears as a crescent-shaped structure making extensive contacts with the nuclear envelope (Figure 2, A190). When both anti-Myc and anti-A190 antibodies were combined, we observed some Myc staining overlapping the Pol I staining (Figure 2B). Similar results were obtained when we analysed staining derived from Ctk2–Myc (Figure 2C) and HA–Ctk3 (data not shown) proteins. As a control, we analysed an RPB3-HA strain where the Pol II Rpb3 subunit is tagged at its C-terminus with three HA epitopes (C.B., unpublished data). Unlike that observed with Ctk proteins, Rpb3 staining was restricted to the nucleoplasmic compartment, showing no overlap with A190 staining (Figure 2D).
Figure 2.
Immunolocalization of the Ctk proteins. Indirect immunofluorescence was performed with: (A and B) CTK1–Myc, (C) CTK2–Myc and (D) RPB3-HA early exponential cells. DNA was stained with DAPI (A), while Ctk1 (A and B), Ctk2 (C) and Rpb3 (D) were detected with mouse monoclonal anti-Myc (A, B and C) or anti-HA (D) antibodies, and A190 (B, C and D) was detected using a rabbit antibody. Three distinct representative nuclei are shown in each case.
We conclude that the three subunits composing the CTDK-I complex, co-localize in the nucleoplasm as well as in the nucleolus. This is consistent with the fact that in vivo, a subpopulation of Ctk1 interacts with Pol I, and suggests that CTDK-I might play a role that is distinct from its function in Pol II transcription.
Δctk strains display defects in nucleolar structure
In an attempt to determine whether CTDK-I plays a role in the nucleolar compartment, we analysed the nucleolus structure in Δctk cells by immunofluorescence microscopy. Deletion of either CTK gene generates a slow growth at 30°C and no growth at 15°C. As previously observed, Δctk cells grown in rich medium to early exponential phase, were much larger than wild-type cells (21). Remarkably, antibodies against A190 did not reveal in Δctk1 cells the characteristic crescent shape of the nucleolus observed in wild-type cells (Figure 3A). On the contrary, we observed a more diffuse signal overlapping part of the DAPI staining. A similar result was obtained with the Δctk3 strain (Figure 3A). To confirm these observations, experiments were repeated using antibodies directed against Nop1, a nucleolar protein involved in rRNA maturation (63). Again, instead of the crescent-shaped nucleoli seen in wild-type cells, isogenic Δctk1 cells exhibited a more diffuse Nop1 staining (Figure 3B). Because CTDK-I is involved in Pol II transcription, we were interested to also analyse nucleoli in a Pol II mutant strain. We used a mutant in Rpb3, the third-largest subunit of Pol II. At 30°C, rpb3-2 is a slow growth mutant that exhibits defects in Pol II transcription (64). Microscopy analysis of rpb3-2 cells using antibodies against A190 revealed nucleoli similar to that observed in wild-type cells (Figure 3A).
Figure 3.
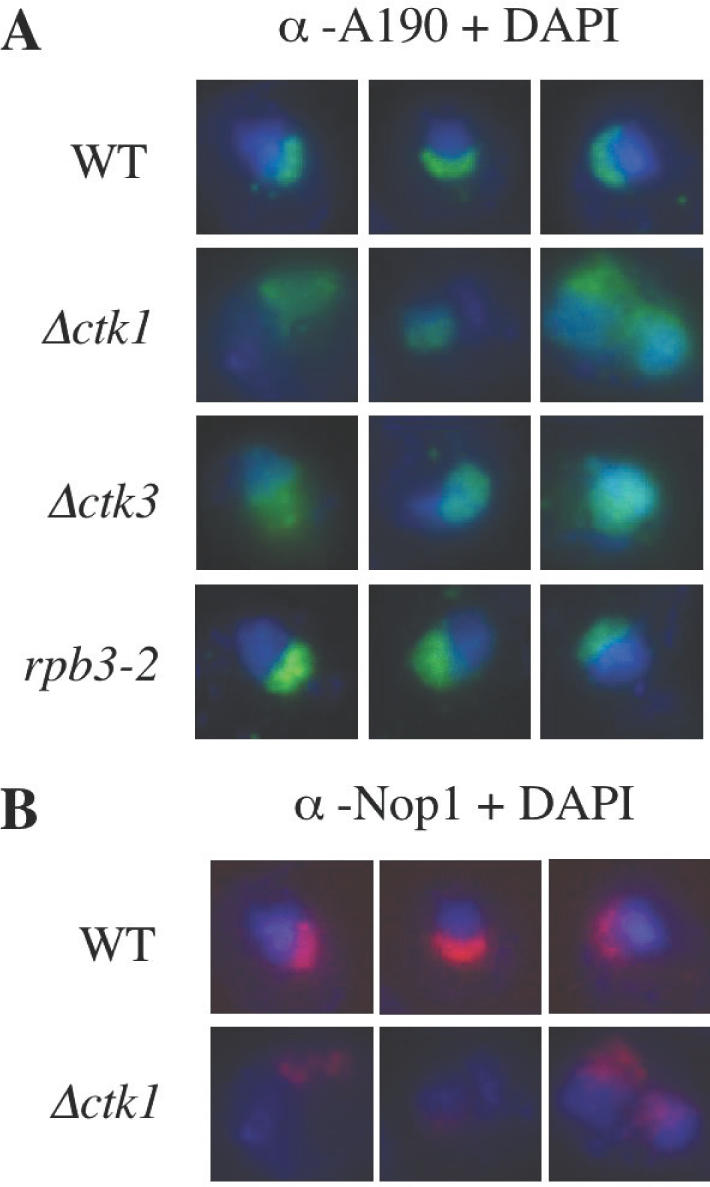
Immunofluorescence microscopy analysis of the nucleolus. Wild-type (WT), Δctk1, Δctk3 and rpb3-2 early exponential cells were analysed with either anti-A190 (A) or anti-Nop1 (B) antibodies, and DAPI staining. Three distinct representative nuclei are shown in each case.
In conclusion, ctk null mutations lead to the disruption of the nucleolar structure during early exponential growth, as evidenced by the delocalization of both Pol I and Nop1, two specific components of the nucleolus. It is interesting to note that such defects are not observed in several other mutants affected for Pol II transcription (Figure 3A, and data not shown). Because Pol I transcription plays an essential role in organizing nucleolar structure and localization (65), the results suggest that this process might be affected in Δctk cells.
Pol I transcription in Δctk1 cells
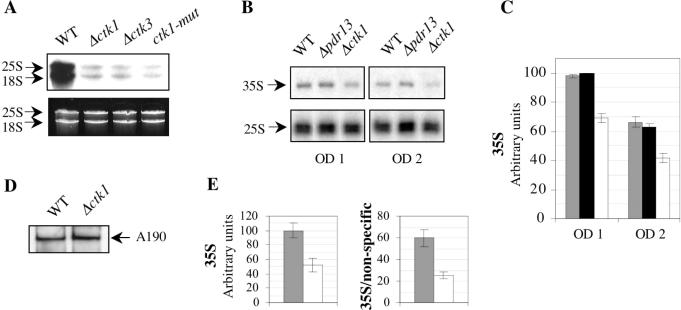
To get an insight into rRNA synthesis in ctk mutants, we realized pulse experiments in early log phase cells grown in SD-casa medium. Similar amounts of rRNA species were analysed, as determined by ethidium bromide staining (Figure 4A, lower panel). Auto-radiography revealed a decrease of 25S, 18S and 5.8S rRNA synthesis in the null Δctk1 and Δctk3 mutant strains, as compared to the isogenic wild-type strain (Figure 4A, upper panel, and data not shown). A similar result was observed with an inactive mutant of the Ctk1 kinase (G.Hautbergue, unpublished data) (Figure 4A, ctk1-mut), suggesting a substantial defect in rRNA synthesis in the absence of the Ctk1 kinase activity.
Figure 4.
(A, B and C) In vivo transcription analyses. (A) Wild-type (WT), Δctk1, Δctk3 and ctk1-mut isogenic cells, were cultivated in SD-casa to early log phase, prior to incubation with 3H-Uracil for 30 min. After extraction, total RNA was separated by electrophoresis on urea–polyacrylamide gels prior to auto-radiography (upper panel) or ethidium bromide staining (lower panel). (B and C) Wild-type (WT), Δpdr13 and Δctk1 isogenic cells, were cultivated in SD-casa to an OD of either 1 or 2. After extraction, total RNA was analysed by primer extension with two oligonucleotides specific for the 35 and 25S rRNA species, respectively. Products were separated by electrophoresis on urea–polyacrylamide gels prior to auto-radiography (B), and 35S products analysed with a PhosphorImager (C). Grey, black and white boxes correspond to 35S amounts in WT, Δpdr13 and Δctk1 cells, respectively. Results correspond to three independent experiments. (D and E) rDNA chromatin immunoprecipitation. After cross-link, cell lysis and sonication, wild-type (WT) and Δctk1 extracts from cells grown in SD-casa to an OD of 2 were immunoprecipitated with anti-A190 antibodies. (D) Recovered proteins were analysed by western immunoblotting with anti-A190 antibodies. (E) Total and immuno-precipitated DNA were analysed by real-time PCR. Fragment corresponding to the 35S rDNA promoter (left panel), and enrichment of the 35S rDNA promoter compared to a non-specific 5S fragment (right panel). Grey and white boxes correspond to WT and Δctk1 extracts, respectively. Results correspond to five independent experiments.
Next, we performed primer extension experiments. Because mutant ctk cells exhibit a reduced growth rate (170 min) compared to wild-type cells (90 min), we also analysed Δpdr13 isogenic cells as control (66). Pdr13 encodes a member of the Hsp70 family that is implicated in pleiotropic drug resistance. PDR13 is required for normal growth, and Δpdr13 cells display a reduced growth rate similar to that of Δctk cells, both in YPD and SD-casa media (data not shown). Cells were grown to log phase in SD-casa medium (OD 1 and 2). After extraction, similar amounts of RNA were analysed by primer extension with two distinct primers allowing specific extension of the mature 25S rRNA, and the 35S rRNA precursor. Gel analysis by auto-radiography (Figure 4B) and with a PhosphorImager (data not shown), revealed similar levels of primer extension products corresponding to the mature 25S rRNA species for all strains, at OD 1 as well as 2. Levels of products corresponding to the 35S rRNA precursor were also similar in wild-type and Δpdr13 cells, whereas in contrast, they were significantly lower when RNA was extracted from Δctk1 cells (Figure 4B and C). These results indicate a substantial defect in rRNA synthesis in the absence of the Ctk1 kinase activity. This defect cannot be only the consequence of the reduced growth rate of Δctk cells as it is not observed in the slow-growing control Δpdr13 strain.
To get further insight into Pol I transcription in Δctk1 cells, we investigated Pol I recruitment at 35S rDNA promoters by chromatin immunoprecipitation experiments. After cross-link and sonication of yeast cells, Pol I was immunoprecipitated with anti-A190 antibodies, from wild-type and Δctk1 cells grown in SD-casa to mid-log phase (OD 2). After reversal of cross-links, Pol I immunoprecipitation efficiency was analysed by western blot with anti-A190 antibodies. Similar amounts of enzyme were recovered for the wild-type and Δctk1 strains (Figure 4D). Total and co-precipitated DNA were analysed by real-time PCR with two pairs of primers. The first pair allowed amplification of a specific fragment corresponding to the 35S rDNA promoter region (35S), whereas the second pair generated a fragment corresponding to the adjacent 5S region transcribed by Pol III (non-specific). While similar low levels of non-specific fragment were amplified in both extracts (data not shown), we observed a 2-fold decrease in the abundance of the 35S PCR fragment in Δctk1 cells compared to wild-type cells (Figure 4E, left panel). Thus, the enrichment ratio 35S/non-specific was lower in Δctk1 compared to wild-type extracts (Figure 4E, right panel). Similar results were observed with Δctk2 cells (data not shown), indicating that during mid-log phase, formation of the Pol I initiation complex at 35S promoters is less efficient in Δctk cells as compared to wild type. This data is consistent with the rRNA synthesis defect observed in the absence of the Ctk1 kinase.
Pol I specific in vitro transcription is impaired in the absence of Ctk1
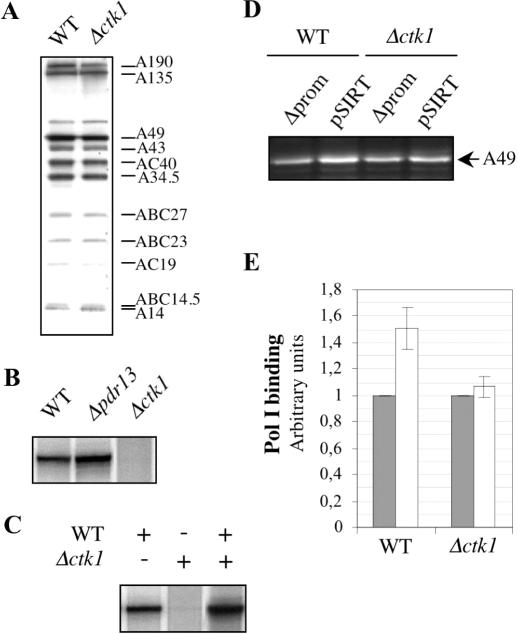
To confirm that the defect in rRNA synthesis observed in vivo was at the level of Pol I transcription machinery, we next investigated Pol I specific transcription in extracts from Δctk1 cells. PA600 fraction containing the complete Pol I transcription apparatus, i.e. the Pol I and the general transcription factors required for Pol I specific transcription (61), was purified from wild-type, Δpdr13, and Δctk1 cells. Western blot analysis did not reveal any Pol II or Pol III specific subunit in the PA600 fraction, showing that the major polymerase form present in this fraction is the Pol I (data not shown). We first assessed Pol I catalytic activity by performing non-specific transcription assays with poly (dA-dT) as template. With such a template, RNA synthesis does not rely upon general transcription factors. We observed similar results with wild-type, Δpdr13 and Δctk1 extracts (data not shown). Pol I levels and subunit composition, as analysed by western immunoblotting, were similar in all PA600 extracts (Figure 5A, and data not shown). Altogether, the data show that the catalytic activity of the Pol I enzyme is not affected in either Δpdr13 or Δctk1 cells. For specific in vitro transcription assays, we used the YepSIRT plasmid that carries an rDNA unit containing a large deletion of the sequence transcribed by Pol I (62). Transcription of this mini-gene in wild-type, Δpdr13, and Δctk1 PA600 fractions, was analysed by electrophoresis on a urea–polyacrylamide gel. Autoradiography revealed a specific RNA for both the wild-type and Δpdr13 extracts, whereas in contrast, no specific transcript was detected for the Δctk1 extract (Figure 5B), unless blots were overexposed (data not shown). The same result was observed with a linearized template (data not shown), suggesting that the Δctk1 PA600 defect does not take place at the level of transcription termination. To determine whether specific transcription impairment was due to the presence of an inhibitor, wild-type and Δctk1 mutant, extracts were mixed prior to incubation with YepSIRT. We observed an efficient specific transcription (Figure 5C), showing that the defective Δctk1 extract does not contain any inhibitor of transcription. Despite the fact that the Pol I enzyme from Δctk1 cell extract is catalytically active, in vitro Pol I specific transcription is dramatically deficient.
Figure 5.
In vitro analyses. (A, B and C) Transcription assay. (A) Pol I from wild-type (WT) and Δctk1 PA600 fractions were analysed by western blot with anti-Pol I antibodies. (B) Specific in vitro transcription of a mini rDNA transcription unit was carried out with PA600 fractions purified from wild-type (WT), Δpdr13 and Δctk1 strains. (C) Specific in vitro transcription with different combinations of wild-type (WT) and Δctk1 PA600 fractions. (D and E) In vitro binding of Pol I to the 35S rDNA promoter. (D) After incubation with immobilized templates containing (pSIRT), or not (Δprom), the 35S rDNA promoter, bound proteins were eluted from released templates and analysed by western immunoblot with anti-Pol I antibodies. (E) Quantification of three independent experiments. Grey boxes, Δprom template; white boxes, pSIRT template.
We next analysed in vitro Pol I binding to the DNA template. Mini-genes from the YepSIRT and Δprom (identical to the YepSIRT construct except that the 35S promoter has been deleted; O. Gadal, unpublished data) plasmids were amplified by PCR with a downstream oligonucleotide that was biotinylated. After purification, PCR products were bound to magnetic beads coupled with streptavidin. PA600 fractions derived from wild-type and Δctk1 cells were incubated with the immobilized templates in the same conditions as specific transcription assays, except that UTP, GTP and CTP were omitted (see Materials and Methods). Bound proteins were eluted after release of the templates by enzymatic digestion, and analysed by western blot with anti-Pol I antibodies (Figure 5D, and data not shown). Quantification was performed from A49, A43, AC40 and A34.5 subunits. We observed that Pol I from wild-type and Δctk1 fractions was similarly bound to the mini-gene in the absence of promoter sequences (Figure 5D and E, Δprom). This is due to the high affinity of RNA polymerases for non-specific DNA (67). However, in the presence of promoter sequences, we observed an increase in Pol I binding only in the case of wild-type extract (Figure 5D and E, pSIRT). Although Pol I from Δctk1 extract exhibits the same non-specific DNA binding properties than wild-type Pol I, no promoter-dependent binding is observed. This result is consistent with specific in vitro transcription data.
We conclude that the reduction in rRNA synthesis observed in vivo is due to an impairment of the Pol I transcription machinery.
DISCUSSION
Altogether, our results reveal that CTDK-I, previously described as a class II factor, is also involved in the Pol I transcription process: (i) Δctk cells exhibit defects in nucleolar structure. Such defects are not observed with several other Pol II transcription mutants. (ii) In vivo and in vitro defects in Pol I transcription are observed in the absence of Ctk1, but not in the case of a slow-growth mutant control, and (iii) Ctk1 is present in the nucleolus and interacts directly and specifically with Pol I.
In yeast, substrates other than the CTD have been described for the CTD kinases Kin28 (43,44) and Srb10 (45–48,43). All these substrates are class II transcription factors, in agreement with the fact that so far, the function of CTD kinases has been restricted to mRNA synthesis. CTDK-I being involved in rRNA synthesis raises the question of whether the Pol I might be a new substrate of the Ctk1 kinase. Although we do not rule out this possibility, several lines of evidence suggest that it might not be the case: (i) most cyclin-dependent kinases can associate with several cyclins either sequentially, or in diverse cellular compartments, and cyclins often dictate their kinase specificity (68). Indeed, physical interaction between a cyclin subunit and its kinase substrate has been reported in both yeast and mammals (69–71), whereas neither the Ctk2 cyclin nor the Ctk3 co-cyclin subunit display a detectable interaction with Pol I. (ii) A stable interaction is generally not observed between a kinase and its substrate. Interestingly, during the Ctk1 purification process, we never evidenced any interaction between Ctk1 and its known substrate, namely the CTD, consistent with the fact that Ctk1 has never been identified as a component of either the Pol II holoenzyme or the Pol II elongator complex. In contrast, co-immunoprecipitation experiments demonstrate physical interaction between Ctk1 and Pol I.
It has been suggested that some nucleolar proteins are not specifically targeted to the nucleolus, but rather are retained in this compartment as a consequence of their functional interactions with other components (72). We suggest that Pol I might be the docking site of the Ctk1 kinase, thus targeting CTDK-I to a nucleolar substrate. Such hypothesis is reminiscent of the recruitment of the CTD phosphatase Fcp1 that has a docking site distinct from its CTD substrate (73).
We have tried to rescue in vitro Pol I transcription from Δctk mutant extract by the addition of purified CTDK-I. Although CTDK-I could efficiently phosphorylate a recombinant CTD, its addition did not rescue specific transcription in Δctk extract (data not shown). Several reasons can account for this negative result (e.g. it is possible that a cofactor required for Ctk1 function in the Pol I transcription process is not co-purified with Ctk1) that is in agreement with previous data showing that only CF, UAF–TBP, and Pol I–Rrn3 are required for efficient in vitro transcription (57). It is thus possible that Ctk1 is not active in vitro for Pol I transcription. Although Ctk1 is not essential in vivo for Pol I transcription, prior phosphorylation of its substrate in vivo would be required to observe efficient transcription in vitro.
Altogether our data suggest that Ctk1 might be involved in Pol I transcription during the initiation step, the major step of Pol I transcription regulation (74). In vitro experiments indicate that Pol I binding to the 35S promoter is defective in the absence of Ctk1, and Pol I occupancy at the 35S promoter is decreased in Δctk cells as observed by chromatin immunoprecipitation experiments. Furthermore, in yeast exponential cells most Pol I is not associated with Rrn3, and similarly, most Pol I is not associated with the Ctk1 kinase. Because the fraction of cellular Ctk1 that co-purifies with Pol I also co-purifies with Rrn3, it is possible that in vivo, Ctk1 is associated with Pol I–Rrn3 the only form of enzyme competent for transcription initiation.
Several lines of evidence indicate that at the level of Pol II transcription, CTDK-I function is growth-related. (i) The stability of the activating subunit, the Ctk2 cyclin, is correlated to cell density (23), (ii) CTK1 deletion affects the pattern of CTD phosphorylation, a pattern shown to be regulated in response to environmental signals (14,25), and (iii) ctk mutants exhibit defects in the transcriptional induction of various classes of genes in response to growth conditions [(25) V.G., unpublished data], indicating that CTDK-I functions in a gene-specific fashion in response to growth conditions. It is possible that CTDK-I may play a regulatory role related to environmental signals in both Pol II and Pol I transcription processes, consistent with the fact that this kinase is not essential to cell viability.
Interestingly, Fath et al. (75) recently reported that dephosphorylation of Pol I by the CTD phosphatase Fcp1 is required for efficient rRNA synthesis. It thus appears that enzymes involved in CTD phosphorylation/dephosphorylation are also implicated in Pol I transcription in yeast. In higher eukaryotes, several components previously described as class II specific factors, have also revealed to be involved in rDNA transcription (58,76,77). For example, in mouse, the class II general transcription initiation factor TFIIH has been shown to be an integral component of the Pol I holoenzyme required for in vitro rRNA synthesis, providing a link between Pol I transcription and transcription-coupled repair of active ribosomal genes (58,78). Given the strong functional similarities between gene expression in yeast and mammals, it is likely that similar to CTDK-I, CTD kinases will be implicated in mammals in the control of other transcription systems, a means for eukaryotic cells to coordinate the control of mRNA and rRNA synthesis in response to changes in the environment or in the cell growth rate (2,79). An attractive possibility is that CTDK-I links distinct nuclear machineries, thus participating in the global transcriptional response to environmental signals.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to S. Chédin, G. Peyroche and A. Sentenac for critical reading of the manuscript. C.B. was supported by the Fondation pour la Recherche Medicale (FDT20030926549). G.H. was supported by the Association pour la Recherche contre le Cancer. V.G. was supported by the Centre National de la Recherche Scientifique.
REFERENCES
- 1.Sentenac A. (1985) Eukaryotic RNA polymerases. CRC Crit. Rev. Biochem., 18, 31–90. [DOI] [PubMed] [Google Scholar]
- 2.Warner J.R. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci., 24, 437–440. [DOI] [PubMed] [Google Scholar]
- 3.Nomura M. (1998) Transcription factors used by Saccharomyces cerevisiae RNA polymerase I and the mechanism of initiation. In Paule,M.R. (ed.), Transcription of Ribosomal RNA Genes by Eukaryotic RNA Polymerase I. Springer Verlag, Berlin, Germany, pp. 155–172. [Google Scholar]
- 4.Keys D.A., Lee,B.S., Dodd,J.A., Nguyen,T.T., Vu,L., Fantino,E., Burson,L.M., Nogi,Y. and Nomura,M. (1996) Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev., 10, 887–903. [DOI] [PubMed] [Google Scholar]
- 5.Aprikian P., Moorefield,B. and Reeder,R.H. (2001) New model for the yeast RNA polymerase I transcription cycle. Mol. Cell. Biol., 21, 4847–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bordi L., Cioci,F. and Camilloni,G. (2001) In vivo binding and hierarchy of assembly of the yeast RNA polymerase I transcription factors. Mol. Biol. Cell, 12, 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fath S., Milkereit,P., Peyroche,G., Riva,M., Carles,C. and Tschochner,H. (2001) Differential roles of phosphorylation in the formation of transcriptional active RNA polymerase I. Proc. Natl Acad. Sci. USA, 98, 14334–14339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riggs D.L., Peterson,C.L., Wickham,J.Q., Miller,L.M., Clarke,E.M., Crowell,J.A. and Sergere,J.C. (1995) Characterization of the components of reconstituted Saccharomyces cerevisiae RNA polymerase I transcription complexes. J. Biol. Chem., 270, 6205–6210. [DOI] [PubMed] [Google Scholar]
- 9.Clarke E.M., Peterson,C.L., Brainard,A.V. and Riggs,D.L. (1996) Regulation of the RNA polymerase I and III transcription systems in response to growth conditions. J. Biol. Chem., 271, 22189–22195. [DOI] [PubMed] [Google Scholar]
- 10.Stefanovsky V.Y., Pelletier,G., Hannan,R., Gagnon-Kugler,T., Rothblum,L.I. and Moss,T. (2001) An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell, 8, 1063–1073. [DOI] [PubMed] [Google Scholar]
- 11.Corden J.L. (1990) Tails of RNA polymerase II. Trends Biochem. Sci., 15, 383–387. [DOI] [PubMed] [Google Scholar]
- 12.Dahmus M.E. (1996) Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem., 271, 19009–19012. [DOI] [PubMed] [Google Scholar]
- 13.Komarnitsky P., Cho,E.J. and Buratowski,S. (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev., 14, 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patturajan M., Schulte,R.J., Sefton,B.M., Berezney,R., Vincent,M., Bensaude,O., Warren,S.L. and Corden,J.L. (1998) Growth-related changes in phosphorylation of yeast RNA polymerase II. J. Biol. Chem., 273, 4689–4694. [DOI] [PubMed] [Google Scholar]
- 15.Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- 16.Hengartner C.J., Myer,V.E., Liao,S., Wilson,C.J., Koh,S.S. and Young,R.A. (1998) Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell, 2, 43–53. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez C.R., Cho,E.J., Keogh,M.C., Moore,C.L., Greenleaf,A.L. and Buratowski,S. (2000) Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol., 20, 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao S.-M., Zhang,J., Jeffery,D.A., Koleske,A.J., Thompson,C.M., Chao,D.M., Viljoen,M., van Vuuren,H.J.J. and Young,R.A. (1995) A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature, 374, 193–196. [DOI] [PubMed] [Google Scholar]
- 19.Murray S., Udupa,R., Yao,S., Hartzog,G. and Prelich,G. (2001) Phosphorylation of the RNA polymerase II carboxy-terminal domain by the Bur1 cyclin-dependent kinase. Mol. Cell. Biol., 21, 4089–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keogh M.C., Podolny,V. and Buratowski,S. (2003) Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol. Cell. Biol., 23, 7005–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sterner D.E., Moon Lee,J., Hardin,S.E. and Greenleaf,A.L. (1995) The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol. Cell. Biol., 15, 5716–5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J.M. and Greenleaf,A.L. (1991) CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr., 1, 149–167. [PMC free article] [PubMed] [Google Scholar]
- 23.Hautbergue G. and Goguel,V. (1999) The yeast C-type cyclin Ctk2p is phosphorylated and rapidly degraded by the ubiquitin-proteasome pathway. Mol. Cell. Biol., 19, 2527–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hautbergue G. and Goguel,V. (2001) Activation of the cyclin-dependent kinase CTDK-I requires the heterodimerization of two unstable subunits. J. Biol. Chem., 15, 8005–8013. [DOI] [PubMed] [Google Scholar]
- 25.Patturajan M., Conrad,N.K., Bregman,D.B. and Corden,J.L. (1999) Yeast carboxyl-terminal domain kinase I positively and negatively regulates RNA polymerase II carboxyl-terminal domain phosphorylation. J. Biol. Chem., 274, 27823–27828. [DOI] [PubMed] [Google Scholar]
- 26.Lee J.M. and Greenleaf,A.L. (1997) Modulation of RNA polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J. Biol. Chem., 272, 10990–10993. [DOI] [PubMed] [Google Scholar]
- 27.Jona G., Wittschieben,B.O., Svejstrup,J.Q. and Gileadi,O. (2001) Involvement of yeast carboxy-terminal domain kinase I (CTDK-I) in transcription elongation in vivo. Gene, 267, 31–36. [DOI] [PubMed] [Google Scholar]
- 28.Lindstrom D.L. and Hartzog,G.A. (2001) Genetic interactions of Spt4-Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics, 159, 487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho E.J., Kobor,M.S., Kim,M., Greenblatt,J. and Buratowski,S. (2001) Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev., 15, 3319–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skaar D.A. and Greenleaf,A.L. (2002) The RNA polymerase II CTD kinase CTDK-I affects pre-mRNA 3′ cleavage/polyadenylation through the processing component Pti1p. Mol. Cell, 10, 1429–1439. [DOI] [PubMed] [Google Scholar]
- 31.Ahn S.H., Kim,M. and Buratowski,S. (2004) Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell. Biol., 13, 67–76. [DOI] [PubMed] [Google Scholar]
- 32.Morris D.P. and Greenleaf,A.L. (2000) The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem., 275, 39935–39943. [DOI] [PubMed] [Google Scholar]
- 33.Xiao T., Hall,H., Kizer,K.O., Shibata,Y., Hall,M.C., Borchers,C.H. and Strahl,B.D. (2003) Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev., 17, 654–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ostapenko D. and Solomon,M.J. (2003) Budding yeast CTDK-I is required for DNA damage-induced transcription. Eukaryot. Cell, 2, 274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hurt E., Luo,M.J., Rother,S., Reed,R. and Strasser,K. (2004) Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc. Natl Acad. Sci. USA, 101, 1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall N.F. and Price,D.H. (1995) Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem., 270, 12335–12338. [DOI] [PubMed] [Google Scholar]
- 37.Price D.H. (2000) P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol., 20, 2629–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serizawa H., Makela,T.P., Conaway,J.W., Conaway,R.C., Weinberg,R.A. and Young,R.A. (1995) Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature, 374, 280–282. [DOI] [PubMed] [Google Scholar]
- 39.Shiekhattar R., Mermelstein,F., Fisher,R.P., Drapkin,R., Dynlacht,B., Wessling,H.C., Morgan,D.O. and Reinberg,D. (1995) Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature, 374, 283–287. [DOI] [PubMed] [Google Scholar]
- 40.Lu H., Fisher,R.P., Bailey,P. and Levine,A.J. (1997) The CDK7-cycH-p36 complex of transcription factor IIH phosphorylates p53, enhancing its sequence-specific DNA binding activity in vitro. Mol. Cell. Biol., 17, 5923–5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akoulitchev S., Chuikov,S. and Reinberg,D. (2000) TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature, 407, 102–106. [DOI] [PubMed] [Google Scholar]
- 42.Kim J.B. and Sharp,P.A. (2001) Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem., 276, 12317–12323. [DOI] [PubMed] [Google Scholar]
- 43.Guidi B.W., Bjornsdottir,G., Hopkins,D.C., Lacomis,L., Erdjument-Bromage,H., Tempst,P. and Myers,L.C. (2004) Mutual targeting of mediator and the TFIIH kinase Kin28. J. Biol. Chem., 279, 29114–29120. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y., Kung,C., Fishburn,J., Ansari,A.Z., Shokat,K.M. and Hahn,S. (2004) Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol. Cell. Biol., 24, 1721–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirst M., Kobor,M.S., Kuriakose,N., Greenblatt,J. and Sadowski,I. (1999) GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol. Cell, 3, 673–678. [DOI] [PubMed] [Google Scholar]
- 46.Chi Y., Huddleston,M.J., Zhang,X., Young,R.A., Annan,R.S., Carr,S.A. and Deshaies,R.J. (2001) Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev., 15, 1078–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vincent O., Kuchin,S., Hong,S.P., Townley,R., Vyas,V.K. and Carlson,M. (2001) Interaction of the Srb10 kinase with Sip4, a transcriptional activator of gluconeogenic genes in Saccharomyces cerevisiae. Mol. Cell. Biol., 21, 5790–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nelson C., Goto,S., Lund,K., Hung,W. and Sadowski,I. (2003) Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature, 421, 187–190. [DOI] [PubMed] [Google Scholar]
- 49.Longtine M.S., McKenzie,A., Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- 50.Milkereit P. and Tschochner,H. (1998) A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J., 17, 3692–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buhler J.M., Sentenac,A. and Fromageot,P. (1974) Isolation, structure, and general properties of yeast ribonucleic acid polymerase A (or I). J. Biol. Chem., 249, 5963–5970. [PubMed] [Google Scholar]
- 52.Huet J., Buhler,J.M., Sentenac,A. and Fromageot,P. (1975) Dissociation of two polypeptide chains from yeast RNA polymerase A. Proc. Natl Acad. Sci. USA, 72, 3034–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smid A., Riva,M., Bouet,F., Sentenac,A. and Carles,C. (1995) The association of three subunits with yeast RNA polymerase is stabilized by A14. J. Biol. Chem., 270, 13534–13540. [DOI] [PubMed] [Google Scholar]
- 54.Buhler J.M., Huet,J., Davies,K.E., Sentenac,A. and Fromageot,P. (1980) Immunological studies of yeast nuclear RNA polymerases at the subunit level. J. Biol. Chem., 255, 9949–9954. [PubMed] [Google Scholar]
- 55.Aris J.P. and Blobel,G. (1988) Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell. Biol., 107, 17–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmitt M.E., Brown,T.A. and Trumpower,B.L. (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res., 18, 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keener J., Josaitis,C.A., Dodd,J.A. and Nomura,M. (1998) Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J. Biol. Chem., 273, 33795–33802. [DOI] [PubMed] [Google Scholar]
- 58.Iben S., Tschochner,H., Bier,M., Hoogstraten,D., Hozak,P., Egly,J.M. and Grummt,I. (2002) TFIIH plays an essential role in RNA polymerase I transcription. Cell, 109, 297–306. [DOI] [PubMed] [Google Scholar]
- 59.Kuras L. and Struhl,K. (1999) Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature, 399, 609–613. [DOI] [PubMed] [Google Scholar]
- 60.Kuras L., Borggrefe,T. and Kornberg,R.D. (2003) Association of the Mediator complex with enhancers of active genes. Proc. Natl Acad. Sci. USA, 100, 13887–13891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Milkereit P., Schultz,P. and Tschochner,H. (1997) Resolution of RNA polymerase I into dimers and monomers and their function in transcription. Biol. Chem., 378, 1433–1443. [DOI] [PubMed] [Google Scholar]
- 62.Musters W., Knol,J., Maas,P., Dekker,A.F., van Heerikhuizen,H. and Planta,R.J. (1989) Linker scanning of the yeast RNA polymerase I promoter. Nucleic Acids Res., 17, 9661–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tollervey D., Lehtonen,H., Carmo-Fonseca,M. and Hurt,E.C. (1991) The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J., 10, 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan Q., Linask,K.L., Ebright,R.H. and Woychik,N.A. (2000) Activation mutants in yeast RNA polymerase II subunit RPB3 provide evidence for a structurally conserved surface required for activation in eukaryotes and bacteria. Genes Dev., 14, 339–348. [PMC free article] [PubMed] [Google Scholar]
- 65.Oakes M., Nogi,Y., Clark,M.W. and Nomura,M. (1993) Structural alterations of the nucleolus in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Mol. Cell. Biol., 13, 2441–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hallstrom T.C., Katzmann,D.J., Torres,R.J., Sharp,W.J. and Moye-Rowley,W.S. (1998) Regulation of transcription factor Pdr1p function by an Hsp70 protein in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cho E.J., Takagi,T., Moore,C.R. and Buratowski,S. (1997) mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev., 11, 3319–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller M.E. and Cross,F.R. (2001) Cyclin specificity: how many wheels do you need on a unicycle? J. Cell Sci., 114, 1811–1820. [DOI] [PubMed] [Google Scholar]
- 69.Ewen M.E., Sluss,H.K., Sherr,C.J., Matsushime,H., Kato,J. and Livingston,D.M. (1993) Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell, 73, 487–497. [DOI] [PubMed] [Google Scholar]
- 70.Schulman B.A., Lindstrom,D.L. and Harlow,E. (1998) Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl Acad. Sci. USA, 95, 10453–10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilson W.A., Mahrenholz,A.M. and Roach,P.J. (1999) Substrate targeting of the yeast cyclin-dependent kinase Pho85p by the cyclin Pcl10p. Mol. Cell. Biol., 19, 7020–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dundr M. and Misteli,T. (2002) Nucleolomics: an inventory of the nucleolus. Mol. Cell, 9, 5–7. [DOI] [PubMed] [Google Scholar]
- 73.Chambers R.S., Wang,B.Q., Burton,Z.F. and Dahmus,M.E. (1995) The activity of COOH-terminal domain phosphatase is regulated by a docking site on RNA polymerase II and by the general transcription factors IIF and IIB. J. Biol. Chem., 270, 14962–14969. [DOI] [PubMed] [Google Scholar]
- 74.Reeder R.H. (1999) Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog. Nucleic Acid Res. Mol. Biol., 62, 293–327. [DOI] [PubMed] [Google Scholar]
- 75.Fath S., Kobor,M.S., Philippi,A., Greenblatt,J. and Tschochner,H. (2004) Dephosphorylation of RNA polymerase I by Fcp1p is required for efficient rRNA synthesis. J. Biol. Chem., 279, 25251–25259. [DOI] [PubMed] [Google Scholar]
- 76.Bradsher J., Auriol,J., Proietti de Santis,L., Iben,S., Vonesch,J.L., Grummt,I. and Egly,J.M. (2002) CSB is a component of RNA Pol I transcription. Mol. Cell, 10, 819–829. [DOI] [PubMed] [Google Scholar]
- 77.Lin C.Y., Tuan,J., Scalia,P., Bui,T. and Comai,L. (2002) The cell cycle regulatory factor TAF1 stimulates ribosomal DNA transcription by binding to the activator UBF. Curr. Biol., 12, 2142–2146. [DOI] [PubMed] [Google Scholar]
- 78.Hoogstraten D., Nigg,A.L., Heath,H., Mullenders,L.H., van Driel,R., Hoeijmakers,J.H., Vermeulen,W. and Houtsmuller,A.B. (2002) Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell, 10, 1163–1174. [DOI] [PubMed] [Google Scholar]
- 79.Nomura M. (1999) Regulation of ribosome biosynthesis in Escherichia coli and Saccharomyces cerevisiae: diversity and common principles. J. Bacteriol., 181, 6857–6864. [DOI] [PMC free article] [PubMed] [Google Scholar]