Abstract
DNA polymerase μ (Pol μ) is a DNA-dependent DNA polymerase closely related to terminal deoxynucleotidyl transferase (TdT), and prone to induce template/primer misalignments and misincorporation. In addition to a proposed general role in non-homologous end joining of double-strand breaks, its mutagenic potential and preferential expression in secondary lymphoid tissues support a role in somatic hypermutation (SHM) of immunoglobulin genes. Here, we show that human Pol μ protein is expressed in the nucleus of centroblasts obtained from human tonsils, forming a characteristic foci pattern resembling that of other DNA repair proteins in response to DNA damage. Overexpression of human Pol μ in Ramos cells, in which the SHM process is constitutive, augmented the somatic mutations specifically at the variable (V) region of the immunoglobulin genes. The nature of the mutations introduced, mostly base substitutions, supports the contribution of Pol μ to mutation of G and C residues during SHM. In vitro analysis of Pol μ misincorporation on specific templates, that mimic DNA repair intermediates and correspond to mutational hotspots, indicated that many of the mutations observed in vivo can be explained by the capacity of Pol μ to induce transient template/primer misalignments.
INTRODUCTION
The primary repertoire of antibody specificities is created in the bone marrow by a DNA rearrangement process involving immunoglobulin (Ig) V (variable), D (diversity) and J (joining) gene segments (1). Following antigen encounter, germinal center (GC)-B cells (centroblasts) proliferate rapidly and undergo a further round of diversification through the somatic hypermutation (SHM) process. In SHM, a large number of mutations (10−3–10−4 mutations/bp/generation) are introduced specifically in rearranged IgV genes [reviewed in (2)]. Most mutations consist of single-nucleotide substitutions, although deletions and insertions also occur. Ig gene hypermutation exhibits a distinctive nucleotide misincorporation pattern, favoring transitions over transversions (3) and preferentially targeting G/C residues (4,5). Moreover, extensive sequence analyses have defined the consensus sequences RGYW and WA (most mutable nucleotide underlined; R is purine, Y is pyrimidine and W is A or T) as highly mutable DNA sequence motifs (6,7).
Dissection of the SHM process has yielded clear clues as to cis-acting factors [reviewed in (8)], but the molecular mechanism remains to be elucidated. At present, activation-induced cytidine deaminase (AID) is the only enzymatic activity found to be indispensable for SHM (9,10). Although initially described as an RNA editing enzyme (11), AID is also reported to deaminate dC residues directly on DNA (12–15). Recent data suggest that DNA strand breaks could trigger SHM (16–20), and it seems very likely that generation of these DNA breaks, defining the SHM hotspots, is closely related to AID activity (21–23). The ‘DNA deamination’ model proposes that AID deaminates deoxycytidine residues to uracil directly on DNA at the hypermutation domain (24); this would generate mismatches that, in the course of DNA repair, would originate the DNA breaks. Other recent work suggests the existence of AID-independent mechanisms for DNA break generation (25–28). Independently of the way in which these breaks are generated (as nicks and/or double-strand breaks, DSBs), most SHM models suggest that they trigger the mechanism(s) underlying somatic mutation. This invokes the involvement of an error-prone ‘DNA break repair’ process at the heart of the mutasome (29), which would include participation of DNA polymerase(s) and nucleolytic activities [reviewed in (30)].
The mammalian DNA polymerase family has grown considerably in the last three years, and we currently know that human cells encode at least 13 template-dependent DNA polymerases, several with clear error-prone activity (31–33). Initial analyses discarded the direct involvement of terminal deoxynucleotidyl transferase (TdT) (34) and DNA polymerase β (35) in SHM. Novel candidates for the mutagenic DNA polymerase activity (Pols ι, κ, η, ζ and μ) have thus emerged [reviewed in (30)]. Convincing data indicate that Pol η is an A/T mutator in the SHM process (7,36), and that inhibition of Pol ζ expression results in reduced SHM frequency (37,38). The participation of Pol ι in SHM is unclear since induction of SHM in BL2 cells is dependent on Pol ι (39), but Pol ι knockout mice show no significant alteration in this process (40). A putative role for Pol κ in SHM is not supported by the analysis of its KO model (41,42). In any case, these data suggest that more than one error-prone DNA polymerase is involved in process (29,30,43).
Human DNA polymerase μ (Pol μ) was the first identified novel member of the mammalian DNA Pol X family, showing both amino acid sequence and functional domain organization closely related to TdT (44). Although various biological roles were proposed for Pol μ (45), they are still matter of speculation. Based on the in vitro polymerization properties of highly purified human Pol μ, which shows unprecedented error-prone DNA synthesis on template-primer structures, and preferential mRNA expression in secondary lymphoid organs, Pol μ was suggested to be involved in SHM of Ig genes (44,45). Pol μ was recently reported to have a unique ability to promote microhomology-mediated template-primer realignments (46), to be up-regulated in response to ionizing radiation-induced DNA DSBs, and to form complexes with Ku 70/80 and XRCC4/DNA ligase IV heterodimeric complexes in the presence of DNA (47). These data support a role for Pol μ in the non-homologous end joining pathway for DSBs repair. Analysis of Pol μ-deficient mice did not support a putative role for Pol μ in SHM (48), but showed a mild impairment of Ig gene rearrangement (49), supporting a previously suggested role for Pol μ in V(D)J recombination (45).
Here, we show that human Pol μ is preferentially expressed in GC-B cells, forming discrete nuclear foci that resemble DNA damage-induced DNA repair factories. Overexpression of human Pol μ in a Burkitt's lymphoma-derived B cell line (Ramos), in which SHM is constitutive, induced an increase in somatic mutations specifically targeted to G/C residues in IgV genes. In vitro analyses using DNA substrates corresponding to hotspot sequences also support the ability of Pol μ to generate these mutations. These data support a potential role for Pol μ in the highly error-prone DNA synthesis events that appear to be associated to SHM.
MATERIALS AND METHODS
Cell lines and culture conditions
Ramos cells were kindly provided by Dr Martinez-A (CNB, Madrid) and were maintained in RPMI 1640 medium (BioWhittaker) supplemented with 10% fetal calf serum (FCS) (Gibco-BRL), 2 mM l-glutamine (BioWhittaker), HEPES buffer (10 mM, BioWhittaker) and gentamycin (50 mg/ml, BioWhittaker). The cell cultures were maintained in a humidified 37°C incubator with 5% CO2.
Rabbit antiserum preparation and testing
Recombinant purified human Pol μ was obtained as described previously (39). Outbred New Zealand rabbits received intradermal injections in multiple sites using 100 μg of purified protein emulsified with an equal volume of Freund's complete adjuvant (two times). Two intramuscular boosts of 100 μg of the same material in incomplete adjuvant were given 4 and 7 weeks later. Sera were collected 7 and 10 days after the last injection, and tested by enzyme-linked immunosorbent assay.
Western blotting
The cells (2 × 106) were lysed in 100 μl of RIPA buffer (137 mM NaCl, 20 mM Tris–HCl, pH 8, 1 mM MgCl2, 1 mM CaCl2, 10% glycerol, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 1 mg/ml leupeptin, 1 mg/ml pepstatin, 1 mM phenylmethylsulfonyl fluoride and 1 mg/ml aprotinin) for 30 min at 4°C and debris removed by centrifugation (18 000 g, 20 min). Protein concentration was determined using the Dc protein assay (Bio-Rad). Protein (20 μg) was separated in 10% SDS–PAGE and transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked for 1 h in Tris-buffered saline (25 mM Tris) with 5% non-fat dry milk (blocking buffer), followed by incubation with primary (2 h) and secondary antibody (40 min). Western blots were developed using the enhanced chemiluminescence system (Amersham Biosciences). Rabbit polyclonal anti-hPol μ antibody was used at 1:1000 dilution in blocking buffer.
Immunohistochemistry
Immunohistochemical analysis of human tonsils was performed on paraffin-embedded tissue sections. After incubation with the primary antibody (anti-human Pol μ, 1:200 dilution), immunodetection was performed with biotinylated anti-rabbit immunoglobulins, followed by peroxidase-labeled streptavidin and with diaminobenzidine chromogen as substrate. Immunostaining was performed using the TechMate 500 (DAKO) automatic immunostaining device. The labeling system and secondary antibodies were all obtained from DAKO.
Immunofluorescence microscopy
The cells were fixed in 4% paraformaldehyde (10 min, room temperature), then washed three times in PBST (PBS containing 0.1% Tween). Preparations were incubated with 20% FCS (1 h, room temperature), followed by 1 h incubation with primary antibody. After washing with PBST and staining with secondary antibody (40 min), preparations were washed and mounted in ProLong Antifade mounting medium (Molecular Probes). Rabbit anti-hPol μ antibody was used at 1:200 dilution in PBS.
Retroviral transduction and generation of Ramos cell clones overexpressing human Pol μ
Human Pol μ full-length cDNA was subcloned into the XhoI–EcoRI sites of plasmid pLZR2-IRES/gfp. This vector was derived from the retroviral transfer construct pLZR-CMV-gfp (50) by replacing the enhanced green fluorescent protein (EGFP) gene with the bicistronic cassette internal ribosomal entry site (IRES)-EGFP from pIRES2-EGFP (Clontech), preceded by a synthetic multicloning site (51). The resulting retroviral vector, confirmed by sequencing and transient expression analysis, was denominated pLZR2-hPol μ-IRES/gfp. High-titer retroviral supernatants (>5 × 106 iu/ml) were produced by transient transfection of 293T cells as described previously (50), and Ramos cells were transduced using these supernatants as described for peripheral B lymphocytes (PBLs) (50). Four days post-infection, gfp+high cells (>97% purity) were obtained by fluorescence-activated cell sorting in EPICS Elite sorter equipment. Parental Ramos cells and those transduced with either the control vector (pLZR2-IRES/gfp) or the hPol μ-encoding vector (pLZR2-hPol μ-IRES/gfp) were seeded by limiting dilution in 96 well plates. Several individual clones were maintained in culture for 3 months at a cell concentration of 106 cells/ml. Each clone was then tested for Pol μ expression by western blot analysis using rabbit anti-hPol μ antibodies. Clones showing different Pol μ expression levels, as well as control clones from both parental and empty vector-transduced cells, were selected for further molecular analysis.
Amplification of rearranged VH and Cμ regions from Ramos genomic DNA
Genomic DNA from parental and transduced Ramos cells was isolated using Tri-Reagent (Sigma) according to manufacturer's instructions. The Ramos VH region was amplified using Pfu DNA polymerase (Promega). For PCR amplification, we used an upstream primer that hybridizes at the VH4–34 gene [VH4bS, d(CAGGTGCAGCTACAGCAG)] and a downstream primer that hybridizes in the JH6 region [JhAS, d(GCTGAGG AGACGGTGACC)] (52). The reactions were performed in 10 μl vol containing 1× Pfu buffer (Promega), 200 μM each dATP, dGTP, dCTP and dTTP (GibcoBRL), 1 μM each primer and 0.6 U of Pfu DNA polymerase. The enzyme was added after a first denaturation step (5 min at 80°C) to avoid non-specific PCR products. The amplification program consisted of 30 cycles of 20 s at 95°C, 20 s at 53°C and 40 s at 72°C, followed by a final incubation step at 72°C for 10 min. The genomic Cμ 1/2 region was amplified using primers C1μS [d(GGACTTCCTTCCCGACTCCAT)] and C2μAS [d(ACGAAGACGCTCACTTTGGGA)], which hybridize at exons 1 and 2 of the Cμ heavy chain gene, respectively. Amplification was as above, except that a 60°C annealing temperature was used. The PCR products were blunt-end ligated into EcoRV-digested pZErO plasmid (Invitrogen). Following transformation in competent DH5α Escherichia coli cells and screening, positive colonies were picked and plasmid mini-preparations were made. Plasmids were sequenced in both directions using the ABIPrism BigDye Terminator cycle sequencing kit (Perkin Elmer) and automatic sequencing on an ABIPrism377 DNA sequencer XL Upgrade (Applied Biosystems/Perkin Elmer). Sequence analysis was performed using the program Multalin (http://www.toulouse.inra.fr/multalin.html) and the VBASE-database (http://www.mrc-cpe.cam.ac.uk/imt-doc/restricted/ok.html).
In vitro DNA polymerization assays on defined DNA molecules
Human Pol μ DNA polymerase activity was evaluated in vitro using 5′-labeled template-primer molecules as substrates, which were prepared by hybridization of synthetic oligonucleotides. All oligonucleotides were obtained from Invitrogen-Life Technologies and were purified by electrophoresis on 8 M urea/20% polyacrylamide gels. Oligonucleotide primers p66 [d(GTAGTAACCACTGAA)], p74 [d(GTGGGTCCTTCAGTG)], p156 [d(5′GAGGGACGGGTTGTA)], p193 [d(TCTTGGACGTGTCTA)], p210 [d(CTTCAGGGAGAGCTG)] and p218 [d(GAGCTCAAC TTCAGG)] were 5′-labeled with [γ-32P]ATP and T4 polynucleotide kinase. These 5′-32P end-labeled oligonucleotide primers were then hybridized, in the presence of 0.2 M NaCl and 60 mM Tris–HCl (pH 7.5), to the corresponding template oligonucleotides t66 [d(GGGTCCTTCAGTGGTTACTAC)], t74 [d(CCAGCTCCAGTAGTAACCACTGAAGGACCCAC)], t156 [d(ACCAACTACAACCCGTCCCTC)], t193 [d(TCGAGTCACCATATCAGTAGACACGTCCAAGA)], t210 [d(AAGAAGCAGCTCTCCCTGAAG)], and t218 [d(GCTCTCCCTGAAGTTGAGCTC)], to generate the template-primer molecules corresponding to positions 66, 74, 156, 193, 210 and 218 of the IgVH4–34 gene, respectively. DNA substrates, p74c-gap2 and p74c-gap1, were obtained by hybridization of 5′-32P end-labeled oligonucleotide primer p74 with the template oligonucleotide t74 and the downstream oligonucleotides d74g2 [d(TACTACTGGAGCTGG)] and d74g1 [d(TTACTACTGGAGCTGG)], respectively. DNA substrates, p193-gap2 and p193-gap1, were obtained by hybridization of 5′-32P end-labeled oligonucleotide primer p193 with the template oligonucleotide t193 and downstream oligonucleotides d193g2 [d(GATATGGTGACTCGA)] and d193g1 [d(TGATATGGTGACTCGA)], respectively. Polymerization reactions were carried out in a 12.5 μl vol in 50 mM Tris–HCl (pH 7.5), 2 mM MgCl2, 1 mM DTT, 4% glycerol, 0.1 mg/ml BSA, 4 nM of 5′-labeled substrate, 100 nM of purified human Pol μ and the indicated concentration of dNTPs. After incubation (30 min at 30°C), the reactions were terminated by adding gel loading buffer [95% (v/v) formamide, 10 mM EDTA, 0.1% (w/v) xylene cyanol and 0.1% (w/v) bromophenol blue]. Samples were heat-denatured (95°C for 5 min) before loading onto the gel; the products were resolved and analyzed in denaturing 8 M urea/20% PAGE and autoradiography.
RESULTS
Human Pol μ is expressed in GC-B cells (centroblasts) and forms foci-like structures
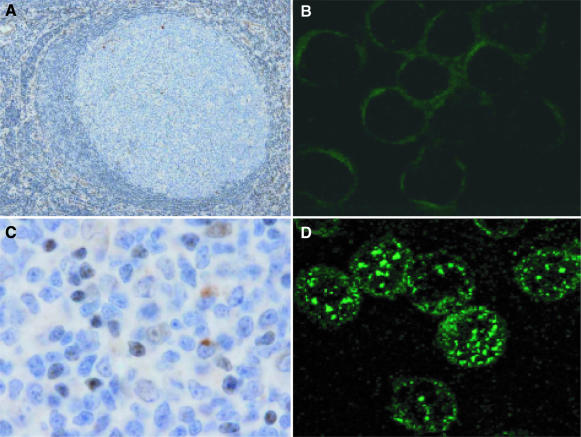
Human Pol μ mRNA is preferentially expressed in lymphoid secondary organs (44). To determine whether human Pol μ mRNA levels correlate with the amount of Pol μ protein, we generated a rabbit antibody using highly purified E.coli-expressed human Pol μ (see Materials and Methods). The IgG fraction (anti-Pol μ) was obtained and used to evaluate Pol μ levels in human GCs. Immunohistochemical analysis of human tonsil-derived secondary follicles, visualized by B220 marker staining (blue), showed a positive Pol μ signal (brown) in GCs, as shown in the magnification of an individual follicle (Figure 1A). The Pol μ-positive signal in GC-B cells was clearly intracellular (Figure 1C). Specificity was confirmed, as pre-incubation of anti-Pol μ IgG with purified recombinant human Pol μ greatly diminished staining; this pattern was not observed after incubation with anti-Pol λ antibody (data not shown).
Figure 1.
Human Pol μ is expressed preferentially in GC-B cells forming nuclear foci. (A) Representative image of an individual GC from human tonsil sections, stained with naive B cell marker B220 (blue) and with anti-human Pol μ antibody (brown). (C) Amplified image of a GC, in which GC-B cells show intracellular human Pol μ expression (brown). (B and D) Immunofluorescence analysis of human Pol μ expression in purified peripheral B lymphocytes (B) versus human tonsil-derived centroblasts (D). Cells were incubated with rabbit anti-human Pol μ antibody (1:200 dilution), followed by fluorescein isothiocyanate-anti-rabbit secondary antibody, as described in Materials and Methods.
To further evaluate the cellular distribution of Pol μ in GC-B cells, we analyzed the expression in purified human centroblasts (B220+PNA+ tonsil B cells) using indirect immunofluorescence. Pol μ showed nuclear localization (Figure 1D), distributed in discrete spots resembling the foci in which the cellular DNA repair machinery co-localizes after the induction of DNA damage (53–55). This expression pattern was specific, as it was not found in control peripheral B cells (see Figure 1B) or when a control IgG fraction was used in the same conditions (data not shown). This protein distribution is similar to that reported for Pol μ after DSB induction in a human cell line (47). Pol μ expression data are thus consistent with a putative contribution of Pol μ in IgV gene SHM (44,45), a process that occurs specifically in the GC centroblast nucleus.
Human Pol μ overexpression in a B cell line with constitutive SHM
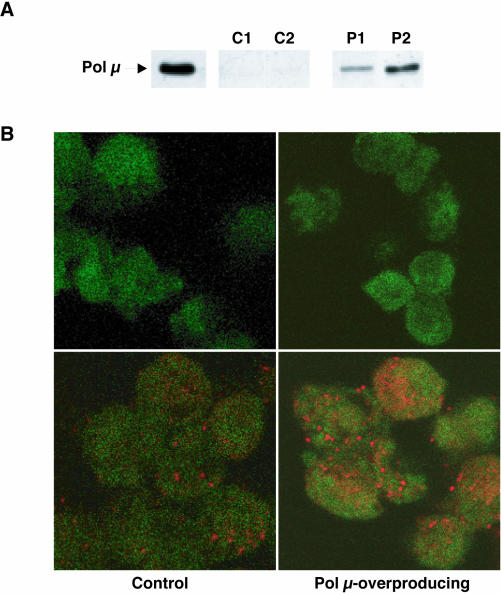
The Ramos cell line is characterized as an IgM-expressing Burkitt's lymphoma line that shows a high rate of constitutive IgV diversification during standard in vitro culture (16). Since Ramos VH diversification shows the major hallmarks of Ig gene hypermutation, this cell line is considered a good in vivo model for SHM analysis. The functional consequences of human Pol μ overexpression in Ramos cells were evaluated after retroviral transduction with human Pol μ cDNA (see Materials and Methods). Construction of empty and human Pol μ-cloned retroviral vectors, infection and clonal selection of either parental or transduced Ramos cells, were as described in Materials and Methods. As shown in Figure 2A, western blot analysis of two of the selected clones (P1 and P2) showed a marked overexpression of human Pol μ (8- and 59-fold, respectively) relative to the levels in control (empty vector-transduced) clones C1 and C2, and parental Ramos clones (data not shown). Indirect immunofluorescence analysis of clone C2 cells using anti-Pol μ (Figure 2B) showed endogenous Pol μ, levels as for Ramos cells, distributed in foci-like structures similar to those described in GC-B cells (Figure 1D). Ramos and clone C2 cells showed this pattern in equivalent proportions (∼5–10%). In contrast, a large proportion (>85%) of P2 clone cells presented a more pronounced and intense foci (Figure 2B), in good agreement with its 59-fold higher expression of Pol μ. No positive signal was detected either in Ramos cells or in clone C2 or P2 cells when pre-immune antiserum was used for analyses (Figure 2B). These results clearly indicated that Pol μ can be overexpressed in the Ramos cell line with no major alteration in viability, and that the excess protein accumulates in the natural nuclear reservoirs, forming foci-like structures.
Figure 2.
Human Pol μ overexpression in Ramos cells. (A) Western blot analysis of individual Ramos clones retrovirally transduced with the pLZR-IRES/gfp (C1, C2) and pLZR2-Pol μ-IRES/gfp (P1, P2) vectors (see Material and Methods). All cell lysates were normalized for total protein amount (20 μg loaded per lane). Recombinant human Pol μ (10 ng) was used as a positive control. (B) Indirect immunofluorescence analysis of human Pol μ overexpression in Ramos cell clones. The figure shows representative fields for control clone C2 (left panels) and Pol μ-overproducing clone P2 (right panels), which were incubated with anti-Pol μ IgGs (bottom panels) or pre-immune antiserum (upper panels). The EGFP marker used throughout the retroviral transduction process and subsequent clone selection is indicated by the green background present in all cells. The cells were incubated with rabbit anti-human Pol μ antibody (1:200 dilution), followed by rhodamine-anti-rabbit secondary antibody (red; see Materials and Methods).
Human Pol μ overexpression in Ramos cell line promotes an increase in the somatic mutation rate
To evaluate the putative implication of Pol μ in the hypermutation mechanism, we analyzed whether human Pol μ overexpression affected the final products of the SHM process. At 3.5 months post-transduction, genomic DNA was prepared from selected clones corresponding to either the parental or transduced Ramos cell line, all cultured in parallel, and Ig locus VH and Cμ regions were amplified specifically by PCR (Materials and Methods). The cloned PCR products were fully sequenced for a total of 206 clones, 184 corresponding to VH 4–34 Ig gene and 22 corresponding to the Cμ region. VH diversification in Pol μ-overexpressing Ramos cells was not influenced by a general mutator phenotype, since mutations did not occur in Cμ regions (data not shown). A database was created for specific mutational events that included mutations detected in 82 sequences from both the parental Ramos cells and from the vector-transduced clones (collectively referred to as control Ramos), and those in 102 sequences from Pol μ-overproducing clones. Mutations were defined by comparison with the canonical VH sequence from the parental Ramos cell line (GenBank sequence AY786364). The mutational database was created after the sequences (mutations) had been evaluated for phylogenetic relationships, to avoid multiple iteration of a specific mutation. The composite database consisted of 184 distinct, unselected mutated/unmutated VH sequences, covering 27.962 and 34.782 bp for control and Pol μ-overexpressing Ramos cells, respectively (Table 1). Data analysis indicated that average mutation frequency in Pol μ-overexpressing Ramos subclones increased by 290% (3.2 × 10−3 mutations/base) compared to control Ramos cells (1.1 × 10−3 mutations/base). Excess human Pol μ appeared to be integrated physiologically in the SHM machinery, as it specifically increased the mutation rate but did not induce a high frequency loss of cell surface-expressed IgM (data not shown), at difference from overexpression of TdT (16), the closest DNA polymerase relative of Pol μ (44).
Table 1. Analysis of SHM at the IgVH4–34 gene of control and Pol μ-overproducing Ramos cells.
| Control Ramos | Pol μ-overproducing Ramos | |
|---|---|---|
| Mutations | 32 | 111 |
| Total nt sequenced | 27 962 | 34 782 |
| Mutational frequency (×10−3 × Mut × base−1) | 1.1 | 3.2 |
| A/T mutated | 21.9 | 3.6 |
| G/C mutated | 78.1 | 96.4 |
| Ts/Tv | 0.46 | 0.75 |
VH mutations in Pol μ-overexpressing Ramos cells
Most independent mutations in Pol μ-overexpressing Ramos cells were single-nucleotide substitutions (109 out of 111; 98%), in accordance with reported SHM features. This is consistent with a role for Pol μ in the generation of somatic mutations in the hypermutation domain. The transition/transversion ratio (Ts/Tv) in Pol μ-overexpressing Ramos cells was 0.75 (Table 1), in contrast to the known preference for transitions over transversions in SHM. The Ts/Tv ratio in our control (parental and vector-transduced) Ramos cells was even more pronounced toward transversions, however, with a Ts/Tv ratio of 0.46 (see Table 1).
Human Pol μ overexpression influenced nucleotide target selection in Ramos cells (Table 1 and 2). The majority of mutations were found in C (59.6%) and G (36.8%) residues, rather than in A/T residues (3.6%); this nucleotide-targeting preference was similar to that showed by our control (parental and vector-transduced) Ramos cells, although more pronounced. Detailed analysis of the mutational spectra obtained following the Pol μ overexpression (Table 2) showed that C→G transversion dominated (34%), followed by C→T transition (23%); the least frequent point mutations corresponded to T→C transition and T→A transversion (1% each).
Table 2. Distribution of nucleotide substitutions in control and Pol μ-overproducing Ramos cells.
| A | C | G | T | Total | |
|---|---|---|---|---|---|
| Control Ramos (n = 32) | |||||
| A | x | 6.3 | 6.3 | ||
| C | x | 40.6 | 9.4 | 50.0 | |
| G | 9.4 | x | 18.7 | 28.1 | |
| T | 6.3 | 6.3 | 3 | x | 15.6 |
| Pol μ-overproducing Ramos (n = 109) | |||||
| A | x | 1.8 | 1.8 | ||
| C | 2.8 | x | 33.9 | 22.9 | 59.6 |
| G | 17.4 | 7.5 | x | 11.9 | 36.8 |
| T | 0.9 | 0.9 | x | 1.8 | |
n, total number of mutations.
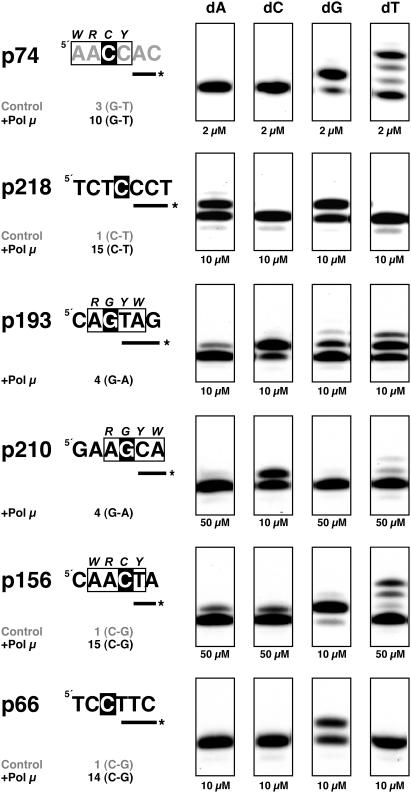
Figure 3 shows a summary of the nucleotide sequences surrounding the residues targeted by the hypermutator machinery in control and Pol μ-overexpressing Ramos cells. Data analysis indicated that the sequences most frequently mutated in Pol μ-overexpressing cells (nucleotide positions 66, 74, 156 and 218) and the mutations introduced exactly matched those in control Ramos cells. Many control cell-derived mutations were also represented in sequences from Pol μ-overexpressing Ramos cells (Figure 3). The analysis also showed good correlation between point mutations and consensus RGYW/WRCY motifs (indicated with boxes in Figure 3), predicted as mutational hotspots in the SHM process. Both control and Pol μ-overexpressing Ramos cells showed hotspot mutability, with about 45% of mutations targeted to these consensus motifs, concurring with previous reports (16).
Figure 3.
Summary of mutations and their sequence context. The nucleotide position of the mutated residues with reference to the IgVH4–34 gene is shown at left for control and Pol μ-overproducing Ramos cells. Sequences are presented in the canonical 5′ to 3′ direction, and mutated residues are indicated with white letters on a black background. RGYW/WRCY hotspot motifs are enclosed in boxes. Total number of mutations (T) and specific base substitution (Mut) at each position are also shown. Nucleotide substitutions that can be explained by Pol μ ability to promote/accept transient misalignments, either via slippage-mediated dislocation (D) or by dNTP-stabilized distortion (d) mechanisms, are indicated.
It could be inferred that Pol μ overexpression in Ramos cells reinforces the C/G mutational efficiency of the constitutive SHM machinery, regardless of whether the mutations are embedded in a RGYW/WRCY motif. This would affect the proportions of each individual mutation in the final spectrum and mutate C/G residues at the same rate.
In vitro human Pol μ activity on template-primer structures from Ramos IgVH region is largely compatible with the in vivo mutation spectrum
Purified recombinant human Pol μ behaves as an error-prone DNA-dependent DNA polymerase, acting as a strong mutator (44,46,56). To test whether the biochemical features of human Pol μ are compatible with the mutation pattern in Pol μ-overproducing Ramos cells, we analyzed in vitro misincorporation by human Pol μ on several template-primer molecules representing natural sequences. Polymerization substrates were selected according to mutational spectra from Pol μ-overexpressing Ramos cells, to generate a set of molecules corresponding to the most frequent IgVH4–34 gene mutational hotspots, irrespective of their inclusion in a RGYW/WRCY consensus motif (nucleotide positions 66, 74, 156, 193, 210 and 218; see Figures 3 and 4). Template-primer structures were obtained by hybridization of synthetic oligonucleotides corresponding to the mutational hotspots, such that the in vivo mutated nucleotide was presented either as the first templating base (positions 66, 156, 193, 210 and 218; white letters on black background in Figure 4), or as the first nucleotide to be inserted (position 74). Using a standard in vitro DNA polymerization assay and appropriate oligonucleotide primers, Pol μ misincorporation was evaluated in the presence of all complementary and non-complementary dNTPs. Nucleotide position 74 was frequently mutated from G to T (10 and 3 times in Pol μ-overexpressing and control Ramos cells, respectively; see Figure 3). Pol μ was able to introduce T in front of the template C, generating a C:T mismatch that was subsequently extended to establish the in vivo G→T transversion (Figure 4). This C:T mismatch occurs at a concentration (2 μM) similar to that of the correct nucleotide. A similar result was obtained when position 218 was tested as template (Figure 4); in this case, the efficiency of the Pol μ-generated C:A mismatch was similar to that for correct C:G base pair. After replication of the mutated strand, this misinsertion led to a C→T transition, as also observed (15 times) in Pol μ-overproducing Ramos cells. These preferred mutations could be explained by a mechanism involving misalignment of the primer terminus to be paired to the adjacent templating base, through a dislocation mechanism originally described for Pol β [reviewed in (57)], and Pol μ (46). In addition to the changes introduced at positions 74 and 219, those at positions 22, 167 and 338 can be also explained by such a mistemplating mechanism (see Figure 3).
Figure 4.
Human Pol μ misincorporation analysis on template-primer structures derived from IgV mutational hotspots. Sequences containing various nucleotide positions mutated in the IgVH4–34 gene of Ramos cells are presented in the canonical 5′ to 3′ direction, with the exception of position 74, whose sequence (in gray) corresponds to the complementary strand. The number of mutations and base substitution for each position is indicated below each sequence. Mutated residues are indicated with white letters on a black background. Labeled oligonucleotide primers used are depicted as a line marked with asterisk at the 5′ end. RGYW/WRCY hotspot motifs are indicated with boxes. Polymerization assays were carried out as described in Materials and Methods, in the presence of the indicated concentrations of each dNTP. After 30 min at 30°C, primer extension was analyzed by 8 M urea/PAGE and autoradiography.
When nucleotide position 193 was tested as template, the preferred Pol μ-generated mismatch was G:T. After one replication round, it originated the same single base substitution (G→A transition; see Figure 3) detected in vivo (four times) in Pol μ-overexpressing Ramos cells. The preference for the G:T mismatch can be explained if Pol μ is also able to use the adjacent template nucleotide (dA) without realignment of the primer, favoring insertion of a T residue. A mechanism in which template misalignment is stabilized by an incoming dNTP was originally described for Pol β (58). A similar ‘misincorporation’ preference was observed when position 210 was tested as template (Figure 4); in this case, the G→A transition observed in vivo (four times) was also compatible with a dNTP-stabilized transient misalignment event promoted by Pol μ in vitro. In addition to the changes introduced at positions 193 and 210, those at positions 52, 81, 86, 134, 143, 189, 216, 231, 237, 240, 254, 256, 288, 290, 293, 301, 311 and 324 can be also explained by such a mistemplating mechanism (Figure 3).
When position 156 was tested as template, Pol μ generated the C:C mismatch (Figure 4), which resulted in a C→G transversion like that obtained (15 times) in vivo in Pol μ-overexpressing Ramos cells after one replication round. In this case, however, a higher concentration of non-complementary dNTPs (50 μM) was necessary, and other mismatches (C:T, C:A) were also generated. In contrast, when position 66 was tested as template (Figure 4), no misincorporation was observed in vitro, and the in vivo mutations found at this position (14 C→G transversions; Figure 3) thus could not be explained directly by the sole action of Pol μ.
Pol μ mutator ability is stimulated on DNA strand break repair intermediates
Since single or double-strand breaks could be one of the initiator events for the SHM mechanism, and assuming that Pol μ may have a role in DNA DSB repair (45–47), we examined whether different substrates representing DNA repair intermediates affected Pol μ mutator ability in vitro. It is worth noting that human Pol μ, like other X family DNA polymerases (59–61), shows a marked preference for binding DNA substrates that contain short gaps (J.F. and L.B., unpublished results). We thus examined the mutational ability of Pol μ on nicked versus gapped (1 or 2 nt) DNA substrates compared to DNA template-primer substrate.
Pol μ used a nicked DNA substrate, preferentially inserting the nucleotide complementary to the template base immediately adjacent to the nick (data not shown), which suggests Pol μ capacity for limited strand-displacement. Pol μ inserted the other 3 nt very poorly, indicating that nicked DNA is a poor substrate for Pol μ mutagenesis compared to the template/primer, in which several templating bases are available (unpaired).
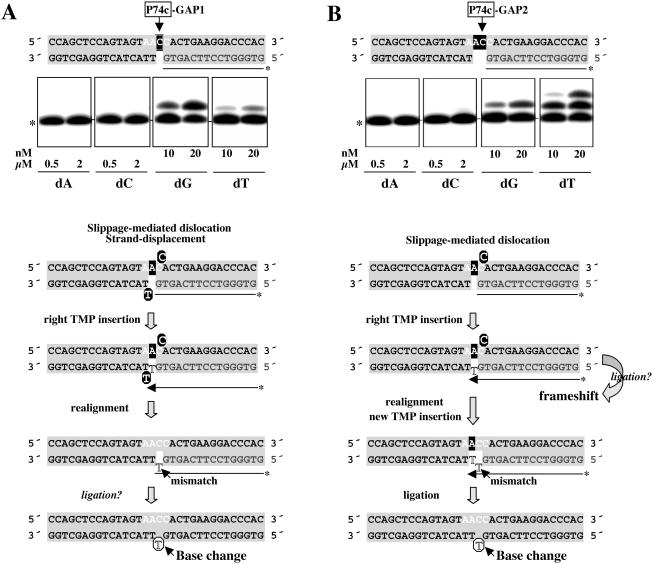
When position 74, previously described as a mutational hotspot in Pol μ-overexpressing Ramos cells, was evaluated in a 1 nt gapped DNA substrate (Figure 5A), Pol μ preferentially inserted the nucleotide (dG) complementary to the only available templating base (C), although it was also able to insert dT at the same nucleotide concentration (10–20 nM). As for template/primer sequences, this ‘mismatch specificity’ can be explained by a slippage-mediated dislocation mechanism, in which the next adjacent base (dA) can be used for templating dT insertion (see the scheme at Figure 5A). In this case, availability of the adjacent base as template implies minimal displacement of the downstream DNA strand. After realignment and potential ligation of the mismatched terminus to the downstream strand, the G→T base substitution would be established at position 74.
Figure 5.
Pol μ misincorporation analysis at gapped-DNA intermediates containing position 74. DNA substrates in which position 74 is embedded in gaps of 1 nt (A) or 2 nt (B) were obtained as described in Materials and Methods. Labeled oligonucleotide primers are shown in gray letters. The WRCY hotspot motif sequence containing position 74 (arrow) is indicated with white letters. Templating residues available at each step are indicated with white letters on black background. Polymerization assays were carried out as described in Materials and Methods, in the presence of the indicated concentrations of each individual dNTP. After 30 min at 30°C, primer extension was analyzed by 8 M urea/PAGE and autoradiography. Mobility of the unextended primer is indicated with an asterisk at the left of the autoradiograph. A scheme detailing the different stages of the slippage-mediated dislocation model that explains the main misincorporation is shown (see text for further details). Transiently misaligned nucleotides generated by dislocation or strand displacement are indicated with white letters on black circles. Mismatched nucleotides are indicated with unfilled letters.
When position 74 was tested in a 2 nt gapped substrate (Figure 5B), two bases were available for templating the first insertion, with no strand displacement required. The template misalignment potential of Pol μ was thus enhanced on this substrate, since incorporation of the non-complementary dTTP was even more efficient than that of the complementary nucleotide. As depicted in the scheme at Figure 5B, immediate ligation of this dislocation intermediate after TMP insertion would lead to a frameshift mutation, concurring with previous speculations (46). A second insertion event leading to a +2 product was readily obtained with dTTP alone, however, suggesting that the dislocation event is transient. After primer realignment, formation of a new A:T pair would establish the C:T mismatch as a single base substitution, precluding formation of a −1 frameshift product. It is also possible that, after template realignment and mismatch formation, a mismatch-extender DNA poymerase such as Pol ζ, (62) could enhance the reaction. A final ligation step, implying a matched primer terminus, would generate the base substitution seen in in vivo experiments.
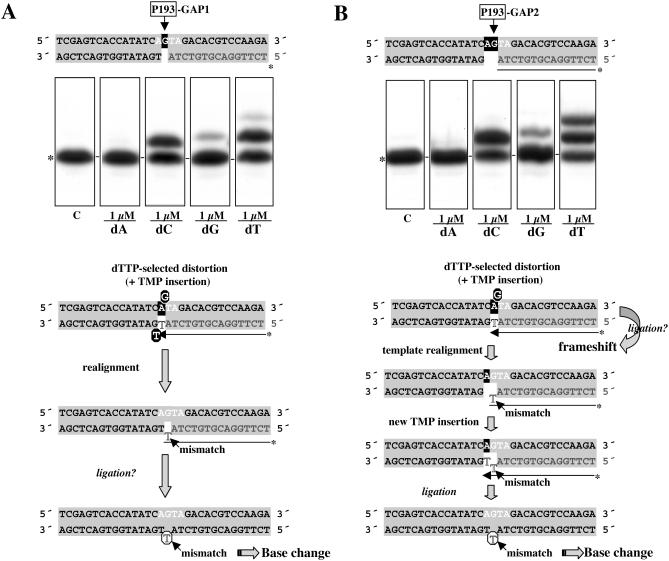
When position 193 was tested as template in gapped-DNA substrates (see Figure 6A and B), the preferred Pol μ-catalyzed misincorporation observed (G:T) was compatible with the G→A transition found in Pol μ-overproducing Ramos cells. This specificity again supports a mistemplating mutation mechanism that does not require template iteration, since the distortion in the template strand is favored/maintained by an incoming nucleotide complementary to an adjacent templating base (see schemes at Figure 6), and may (1nt-gap; Figure 6A) or may not (2 nt gap; Figure 6B) require strand displacement. Template distortion is also sufficiently transient in this case to allow creation of a G:T mismatch that can be further elongated (in the case of the 2 nt gap) to avoid the formation of a −1 frameshift product.
Figure 6.
Pol μ misincorporation analysis at gapped-DNA intermediates containing position 193. DNA substrates in which position 193 is embedded in gaps of 1 nt (A) or 2 nt (B) were obtained as described in Materials and Methods. Labeled oligonucleotide primers are shown in gray letters. The RGYW hotspot motif sequence containing position 193 (arrow), is indicated in white letters. Templating residues available at each step are indicated in white letters on black background. Polymerization assays and primer extension analysis were as in Figure 5. Mobility of the unextended primer is indicated with an asterisk at the left of the autoradiograph. A scheme is shown detailing the different stages of the dNTP-stabilization-mediated model that explains the main misincorporation observed is shown (see text for further details). Transiently misaligned nucleotides generated by dNTP-stabilized distortion or strand displacement are indicated with white letters on black circles. Mismatched nucleotides are indicated with unfilled letters.
DISCUSSION
Human DNA Pol μ is a recently identified DNA polymerase of the mammalian Pol X family, closely related to TdT (44). Recombinant purified Pol μ nonetheless has intrinsic biochemical properties differentiable from those of TdT; thus, although Pol μ shows mild terminal transferase activity, its catalytic polymerization efficiency is strongly enhanced by the presence of a template strand (44). Pol μ and TdT also differ in their expression patterns: while TdT expression is specifically restricted to primary lymphoid tissues, Pol μ mRNA expression is preferentially associated to secondary lymphoid tissues (44). Here, we confirmed this preferential expression at the protein level, showing association of human Pol μ to tonsil-derived GC-B cells. Moreover, Pol μ is localized entirely in the nuclear compartment of purified human centroblasts, forming foci-like structures very similar to those formed by several DNA repair proteins in response to DNA damage (53–55). Pol μ was proposed to be part of the cellular response to DNA DSB, because it directly interacts with protein factors involved in the end joining pathway, and it forms nuclear foci which coincide with those formed by γH2AX histone (47). However, the Pol μ foci observed in Ramos, as those observed in sorted centroblasts (this manuscript) do not require exogenous damage suggesting a more specific role of Pol μ in processes, as SHM, that could correlate with the appearance of specific (targeted) DSBs. In agreement with Pol μ participation in SHM, murine Pol μ expression is up-regulated in response to immunization, and Pol μ protein is found in GC that developed in immunized spleen (D. Lucas, M. T. Lain, M. A. Gonzalez, O. Dominguez, J. F. Ruiz, J. Chamorro, C. Martinez-A, L. Blanco and A. Bernad, manuscript submitted).
Pol μ activity could contribute to the SHM process
As shown in this paper, human Pol μ overexpression in an in vivo system that recreates the SHM process, such as the Burkitt's lymphoma-derived Ramos cell line, did not alter cell viability and physiology parameters, but increased the mutational rate at IgVH regions of Ramos cells (2.8-fold compared to control cells). This effect was specific of IgVH genes, as no mutations were detected in the of IgM locus constant region (Cμ), and IgM surface expression was unaffected. Mutations in Pol μ-overexpressing cells were mainly base substitutions, targeted preferentially to C and G residues (94% of all mutations detected). This indicates that human Pol μ overexpression enhances the previously reported natural bias (16), increasing mutation at G/C residues. In contrast to our data, TdT overexpression in Ramos cells did not increase base substitutions, but significantly affected the membrane IgM expression as a consequence of untemplated nucleotide insertions at DNA breaks scattered throughout the IgV domain (16).
In accordance with the mutation spectra from the Ramos in vivo model, analysis of Pol μ polymerization activity on hotspot-containing DNA substrates supported the Pol μ ability to generate the same specific mutations in vitro. Biochemical analysis showed that Pol μ mutational specificity is dictated largely by the sequence context; whereas a single-nucleotide repeat is sufficient to induce slippage-mediated template misalignment (dislocation), as reported by Zhang et al. (46), other sequence contexts are less predictable, but effectively trigger misalignments. As shown here for positions 193 and 210, and for other non-specific sequence contexts (data not shown), Pol μ can promote mutations based on a dNTP stabilization-mediated mechanism of template misalignment (58). Based on its dislocation capacity, a simplistic model was proposed, in which Pol μ would promote mainly frameshift mutations rather than base substitutions, as an argument against a role of Pol μ in SHM (46). Our data indicate that in vivo or in vitro Pol μ-dependent mutations led preferentially to single-nucleotide substitutions. This suggests that Pol μ-induced template distortions are transient, leading to the mismatched pairs responsible for the base substitutions. Moreover, Pol μ mutational abilities are greatly enhanced on gapped DNA substrates, concurring with a ‘DNA repair’ model that fixes certain mutations throughout the SHM process.
These findings support a potential role for Pol μ in the physiological SHM process acting preferentially by promoting mutation on G/C base pairs in the course of error-prone DNA breaks-mediated repair.
How many polymerases contribute to SHM?
Considering all mechanistic evidences, a plausible model for SHM envisions formation of programmed DNA breaks (single- or double-strand breaks) in the hypermutation domain of the Ig locus, to recruit several unique DNA polymerases (Polι, Pol μ, Polη, mutator polymerases, and Pol ζ, mismatch-extender polymerase) that could act in concert for the error-prone repair of initial or further-processed lesions (after exonucleolytic action). The notion that multiple DNA polymerases are involved in SHM has been previously proposed by several groups (29,30,43). However, the preferential participation of any of the DNA polymerases involved would be determined by the nature of the initiating lesion, or controlled by other proteins responsible for their specific recruitment. The final output of the reaction would be conditioned by the biochemical properties of each enzyme and the sequence context in which it can act. This concurs with evidence for the existence of two distinct mutators or mutational phases, one directed to A/T residues and other to G/C residues (63). Indeed, it was suggested that Pol η promotes A/T mutations during SHM (7,36,64), and here we show that Pol μ would act preferentially on G/C residues, as also proposed for Pol ι (39). Finally, after resolution and ligation, several cellular repair systems could allow fixation of some of the mutations introduced, erasing others. Mouse knockout models analyses were expected to provide a definitive answer to the putative individual roles of the various DNA polymerases involved in SHM. Nonetheless, homozygous knockout mice for the Pol ζ gene suffer early embryonic lethality (65–67), and direct analysis of its role in Ig SHM remains unevaluated. Moreover, analyses of Pol μ [(48); D. Lucas, M. T. Lain, M. A. Gonzalez, O. Dominguez, J. F. Ruiz, J. Chamorro, C. Martinez-A, L. Blanco and A. Bernad, manuscript submitted] and Pol ι (40) homozygous knockout mice show no gross defect in the hypermutation mechanism. It is thus possible that the lack of an SHM-deficient phenotype is due to redundancy among the specialized DNA polymerases for the short gap-filling highly error-prone DNA synthesis that seems to be linked to SHM process. Further analyses, including generation of double or multiple knockout mice, are needed to define the contribution of Pol μ and the other error-prone DNA polymerases to the SHM process.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Catherine Mark for editorial support. This work was supported by Ministerio de Ciencia y Tecnología Grants BMC 2003-00186 (to L.B.) and BMC 2003-00186 (to A.B.) and by an institutional grant to Centro de Biología Molecular ‘Severo Ochoa’ from Fundación Ramón Areces. The Department of Immunology and Oncology is supported by the Spanish Council for Scientific Research (CSIC) and by Pfizer. J.F.R. was recipient of a fellowship from the Ministerio de Educación y Ciencia. D.L. and E.G.P. were fellows of the Comunidad Autónoma de Madrid.
REFERENCES
- 1.Tonegawa S. (1983) Somatic generation of antibody diversity. Nature, 302, 575–581. [DOI] [PubMed] [Google Scholar]
- 2.Papavasiliou F.N. and Schatz,D.G. (2002) Somatic hypermutation of immunoglobulin genes: merging mechanisms for genetic diversity. Cell, 109, S53–S44. [DOI] [PubMed] [Google Scholar]
- 3.Rajewsky K. (1996) Clonal selection and learning in the antibody system. Nature, 381, 751–758. [DOI] [PubMed] [Google Scholar]
- 4.Betz A.G., Neuberger,M.S. and Milstein,C. (1993) Discriminating intrinsic and antigen-selected mutational hotspots in immunoglobulin V genes. Immunol. Today, 14, 405–411. [DOI] [PubMed] [Google Scholar]
- 5.Bachl J. and Wabl,M. (1996) An immunoglobulin mutator that targets G:C base pairs. Proc. Natl Acad. Sci. USA, 93, 851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogozin I.B. and Kolchanov,N.A. (1992) Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim. Biophys. Acta, 1171, 11–18. [DOI] [PubMed] [Google Scholar]
- 7.Rogozin I.B., Pavlov,Y.I., Bebenek,K., Matsuda,T. and Kunkel,T.A. (2001) Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nature Immunol., 2, 530–536. [DOI] [PubMed] [Google Scholar]
- 8.Storb U., Peters,A., Klotz,E., Kim,N., Shen,H.M., Hackett,J., Rogerson,B. and Martin,T.E. (1998) Cis-acting sequences that affect somatic hypermutation of Ig genes. Immunol. Rev., 162, 153–160. [DOI] [PubMed] [Google Scholar]
- 9.Muramatsu M., Kinoshita,K., Fagarasan,S., Yamada,S., Shinkai,Y. and Honjo,T. (2000) Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell, 102, 553–563. [DOI] [PubMed] [Google Scholar]
- 10.Revy P., Muto,T., Levy,Y., Geissmann,F., Plebani,A., Sanal,O., Catalan,N., Forveille,M., Dufourcq-Labelouse,R., Gennery,A., Tezcan,I., Ersoy,F., Kayserili,H., Ugazio,A.G., Brousse,N., Muramatsu,M., Notarangelo,L.D., Kinoshita,K., Honjo,T., Fischer,A. and Durandy,A. (2000) Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell, 102, 565–575. [DOI] [PubMed] [Google Scholar]
- 11.Muramatsu M., Sankaranand,V.S., Anant,S., Sugai,M., Kinoshita,K., Davidson,N.O. and Honjo,T. (1999) Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem., 274, 18470–18476. [DOI] [PubMed] [Google Scholar]
- 12.Petersen-Mahrt S.K., Harris,R.S. and Neuberger,M.S. (2002) AID mutates E.coli suggesting a DNA deamination mechanism for antibody diversification. Nature, 418, 99–103. [DOI] [PubMed] [Google Scholar]
- 13.Dickerson S.K., Market,E., Besmer,E. and Papavasiliou,F.N. (2003) AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med., 197, 1291–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bransteitter R., Pham,P., Scharff,M.D. and Goodman,M.F. (2003) Activation-induced cytidine deaminase deaminates deoxycitidine on single-stranded DNA but requires the action of RNase. Proc. Natl Acad. Sci. USA, 100, 4102–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaudhuri J., Tian,M., Khuong,C., Chua,K., Pinaud,E. and Alt,F.W. (2003) Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature, 422, 726–730. [DOI] [PubMed] [Google Scholar]
- 16.Sale J.E. and Neuberger,M.S. (1998) TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity, 9, 859–869. [DOI] [PubMed] [Google Scholar]
- 17.Bross L., Fukita,Y., McBlane,F., Demolliere,C., Rajewsky,K. and Jacobs,H. (2000) DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity, 13, 589–597. [DOI] [PubMed] [Google Scholar]
- 18.Papavasiliou F.N. and Schatz,D.G. (2000) Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature, 408, 216–221. [DOI] [PubMed] [Google Scholar]
- 19.Kong Q. and Maizels,N. (2001) DNA breaks in hypermutating immunoglobulin genes: evidence for a break-and-repair pathway of somatic hypermutation. Genetics, 158, 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zan H., Wu,X., Komori,A., Holloman,W.K. and Casali,P. (2003) AID-dependent generation of resected double-strand DNA breaks and recruitment of Rad52/Rad51 in somatic hypermutation. Immunity, 18, 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pham P., Bransteitter,R., Petruska,J. and Goodman,M.F. (2003) Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature, 424, 103–107. [DOI] [PubMed] [Google Scholar]
- 22.Beale R.C., Petersen-Mahrt,S.K., Watt,I.N., Harris,R.S., Rada,C. and Neuberger,M.S. (2004) Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol., 337, 585–596. [DOI] [PubMed] [Google Scholar]
- 23.Yu K., Huang,F.T. and Lieber,M.R. (2004) DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J. Biol. Chem., 279, 6496–6500. [DOI] [PubMed] [Google Scholar]
- 24.Rada C., Williams,G.T., Nilsen,H., Barnes,D.E., Lindahl,T. and Neuberger,M.S. (2002) Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol., 12, 1748–1755. [DOI] [PubMed] [Google Scholar]
- 25.Papavasiliou F.N. and Schatz,D.G. (2002) The activation-induced deaminase functions in a postcleavage step of the somatic hypermutation process. J. Exp. Med., 195, 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bross L., Muramatsu,M., Kinoshita,K., Honjo,T. and Jacobs,H. (2002) DNA double-strand breaks: prior to but not sufficient in targeting hypermutation. J. Exp. Med., 195, 1187–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bross L. and Jacobs,H. (2003) DNA double strand breaks occur independent of AID in hypermutating Ig genes. Clin. Dev. Immunol., 10, 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zan H., Wu,X., Komori,A., Holloman,W.K. and Casali,P. (2003) AID-dependent generation of resected double-strand DNA breaks and recruitment of Rad52/Rad51 in somatic hypermutation. Immunity, 18, 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faili A., Aoufouchi,S., Gueranguer,Q., Zober,C., Leon,A., Bertocci,B., Weill,J.C. and Reynaud,C.A. (2002) AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nature Immunol., 3, 815–821. [DOI] [PubMed] [Google Scholar]
- 30.Gearhart P.J. and Wood,R.D. (2001) Emerging links between hypermutation of antibody genes and DNA polymerases. Nature Rev. Immunol., 1, 187–192. [DOI] [PubMed] [Google Scholar]
- 31.Hübscher U., Maga,G. and Spadari,S. (2002) Eukaryotic DNA polymerases. Annu. Rev. Biochem., 71, 133–163. [DOI] [PubMed] [Google Scholar]
- 32.Goodman M.F. (2002) Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem., 71, 17–50. [DOI] [PubMed] [Google Scholar]
- 33.Friedberg E.C., Wagner,R. and Radman,M. (2002) Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science, 296, 1627–1630. [DOI] [PubMed] [Google Scholar]
- 34.Texido G., Jacobs,H., Meiering,M., Kuhn,R., Roes,J., Muller,W., Gilfillan,S., Fujiwara,H., Kikutani,H., Yoshida,N., Amakawa,R., Benoist,C., Mathis,D., Kishimoto,T., Mak,T.W. and Rajewsky,K. (1996) Somatic hypermutation occurs in B cells of terminal deoxynucleotidyl transferase-, CD23-, interleukin-4-, IgD- and CD30-deficient mouse mutants. Eur. J. Immunol., 26, 1966–1969. [DOI] [PubMed] [Google Scholar]
- 35.Esposito G., Texido,G., Betz,U.A., Gu,H., Muller,W., Klein,U. and Rajewsky,K. (2000) Mice reconstituted with DNA polymerase beta-deficient fetal liver cells are able to mount a T cell-dependent immune response and mutate their Ig genes normally. Proc. Natl Acad. Sci. USA, 97, 1166–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng X., Winter,D.B., Kasmer,C., Kraemer,K.H., Lehmann,A.R. and Gearhart,P.J. (2001) DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nature Immunol., 2, 537–541. [DOI] [PubMed] [Google Scholar]
- 37.Zan H., Komori,A., Li,Z., Cerutti,A., Schaffer,A., Flajnik,M.F., Diaz,M. and Casali,P. (2001) The translesion DNA polymerase zeta plays a major role in Ig and bcl-6 somatic hypermutation. Immunity, 14, 643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diaz M., Verkoczy,L.K., Flajnik,M.F. and Klinman,N.R. (2001) Decreased frequency of somatic hypermutation and impaired affinity maturation but intact germinal center formation in mice expressing antisense RNA to DNA polymerase zeta. J. Immunol., 167, 327–335. [DOI] [PubMed] [Google Scholar]
- 39.Faili A., Aoufouchi,S., Flatter,E., Gueranger,Q., Reynaud,C.A. and Weill,J.C. (2002) Induction of somatic hypermutation in immunoglobulin genes is dependent on DNA polymerase iota. Nature, 419, 944–947. [DOI] [PubMed] [Google Scholar]
- 40.McDonald J.P., Frank,E.G., Plosky,B.S., Rogozin,I.B., Masutani,C., Hanaoka,F., Woodgate,R. and Gearhart,P.J. (2003). 129-derived strains of mice are deficient in DNA polymerase ι and have normal immunoglobulin hypermutation. J. Exp. Med., 198, 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schenten D., Gerlach,V.L., Guo,C., Velasco-Miguel,S., Hladik,C.L., White,C.L., Friedberg,E.C., Rajewsky,K. and Esposito,G. (2002) DNA polymerase kappa deficiency does not affect somatic hypermutation in mice. Eur. J. Immunol., 32, 3152–3160. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu T., Shinkai,Y., Ogi,T., Ohmori,H. and Azuma,T. (2002) The absence of DNA polymerase kappa does not affect somatic hypermutation of the mouse immunoglobulin heavy chain gene. Immunol. Lett., 86, 265–270. [DOI] [PubMed] [Google Scholar]
- 43.Diaz M. and Casali,P. (2002) Somatic immunoglobulin hypermutation. Curr. Opin. Immunol., 14, 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dominguez O., Ruiz,J.F., Lain de Lera,T., Garcia-Diaz,M., Gonzalez,M.A., Kirchhoff,T., Martinez-A,C., Bernad,A. and Blanco,L. (2000) DNA polymerase mu, (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J., 19, 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruiz J.F., Dominguez,O., Lain de Lera,T., Garcia-Diaz,M., Bernad,A. and Blanco,L. (2001) DNA polymerase mu, a candidate hypermutase? Philos. Trans. R. Soc. Lond., B, Biol. Sci., 356, 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y., Wu,X., Yuan,F., Xie,Z. and Wang,Z. (2001) Highly frequent frameshift DNA synthesis by human DNA polymerase mu. Mol. Cell. Biol., 21, 7995–8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahajan K.N., McElhinny,A.N., Mitchell,B.S. and Ramsden,D.A. (2002) Association of DNA polymerase μ (pol μ) with ku and ligase IV: role for pol μ in end-joining double-strand break repair. Mol. Cell. Biol., 22, 5194–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bertocci B., De Smet,A., Flatter,E., Dahan,A., Bories,J.C., Landreau,C., Weill,J.C. and Reynaud,C.A. (2002) Cutting edge: DNA polymerases mu and lambda are dispensable for Ig gene hypermutation. J. Immunol., 168, 3702–3706. [DOI] [PubMed] [Google Scholar]
- 49.Bertocci B., De Smet,A., Berek,C., Weill,J.C. and Reynaud,C.A. (2003) Immunoglobulin kappa light chain gene rearrangement is impaired in mice deficient for DNA polymerase mu. Immunity, 19, 203–211. [DOI] [PubMed] [Google Scholar]
- 50.Yang S., Delgado,R., King,S.R., Woffendin,C., Barker,C.S., Yang,Z.Y., Xu,L., Nolan,G.P. and Nabel,G.J. (1999) Generation of retroviral vector for clinical studies using transient transfection. Hum. Gene Ther., 10, 123–132. [DOI] [PubMed] [Google Scholar]
- 51.Abad J.L., Serrano,F., San Roman,A., Delgado,R.Bernad,A. and Gonzalez M.A. (2002) Single-step multiple retroviral transduction of human T cells. J. Gene Med., 4, 27–37. [PubMed] [Google Scholar]
- 52.Tomlinson I.M. (1997) V Base database of human antibody genes. Medical Research Council, Centre for Protein Engineering, UK.
- 53.Raderschall E., Bazarov,A., Cao,J., Lurz,R., Smith,A., Mann,W., Ropers,H.H., Sedivy,J.M., Golub,E.I., Fritz,E. and Haaf,T. (2002) Formation of higher-order nuclear Rad51 structures is functionally linked to p21 expression and protection from DNA damage-induced apoptosis. J. Cell. Sci., 115, 153–164. [DOI] [PubMed] [Google Scholar]
- 54.Stewart G.S., Wang,B., Bignell,C.R., Taylor,A.M and Elledge,S.J. (2003) MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature, 421, 961–966. [DOI] [PubMed] [Google Scholar]
- 55.Robinson J.G., Elliot,J., Dixon,K. and Oakley,G.G. (2004) Replication protein A and the Mre11/Rad50/Nbs1 complex co-localize and interact at sites of stalled replication forks. J. Biol. Chem., 279, 34802–34810. [DOI] [PubMed] [Google Scholar]
- 56.Covo S., Blanco,L. and Livneh,Z. (2004) Lession bypass by human DNA polymerase mu reveals a template-dependent, sequence-independent nucleotidyl transferase activity. J. Biol. Chem., 279, 859–865. [DOI] [PubMed] [Google Scholar]
- 57.Bebenek K. and Kunkel,T.A. (2000) Streisinger revisited: DNA synthesis errors mediated by substrate misalignments. Cold Spring Harb. Symp. Quant. Biol., 65, 81–91. [DOI] [PubMed] [Google Scholar]
- 58.Efrati E., Tocco,G., Eritka,R., Wilson,S.H. and Goodman,M.F. (1997) Abasic translesion synthesis by DNA polymerase beta violates the “A-rule”. Novel types of nucleotide incorporation by human DNA polymerase beta at an abasic lesion in different sequence contexts. J. Biol. Chem., 272, 2559–2569. [DOI] [PubMed] [Google Scholar]
- 59.Chagovetz A.M., Sweasy,J.B. and Preston,B.D. (1997) Increased activity and fidelity of DNA polymerase beta on single-nucleotide gapped DNA. J. Biol. Chem., 272, 27501–27504. [DOI] [PubMed] [Google Scholar]
- 60.Garcia-Diaz M., Bebenek,K., Sabariegos,R., Dominguez,O., Rodriguez,J., Kirchhoff,T., Garcia-Palomero,E., Picher,A.J., Juarez,R., Ruiz,J.F., Kunkel,T.A. and Blanco,L (2002) DNA polymerase lambda, a novel DNA repair enzyme in human cells. J. Biol. Chem., 277, 13184–13191. [DOI] [PubMed] [Google Scholar]
- 61.Garcia-Diaz M., Bebenek,K., Krahn,J.M., Blanco,L., Kunkel,T.A. and Pedersen,L.C. (2004) A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology. Mol. Cell, 13, 561–572. [DOI] [PubMed] [Google Scholar]
- 62.Johnson R.E., Washington,M.T., Haracska,L., Prakash,S. and Prakash,L. (2000) Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature, 406, 1015–1019. [DOI] [PubMed] [Google Scholar]
- 63.Rada C., Ehrenstein,M.R., Neuberger,M.S. and Milstein,C. (1998) Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity, 9, 135–141. [DOI] [PubMed] [Google Scholar]
- 64.Pavlov Y.I., Rogozin,I.B., Galkin,A.P., Aksenova,A.Y., Hanaoka.,F., Rada,C. and Kunkel,T.A. (2002) Correlation of somatic hypermutation specificity and A-T base pair substitution errors by DNA polymerase eta during copying of a mouse immunoglobulin kappa light chain transgene. Proc. Natl Acad. Sci. USA, 99, 9954–9959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bemark M., Khamlichi,A.A., Davies,S.L., Neuberger,M.S. (2000) Disruption of mouse polymerase zeta (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr. Biol., 10, 1213–1216. [DOI] [PubMed] [Google Scholar]
- 66.Esposito G., Godindagger,I., Klein,U., Yaspo,M.L., Cumano,A. and Rajewsky,K. (2000) Disruption of the Rev31-encoded catalytic subunit of polymerase zeta in mice results in early embryonic lethality. Curr. Biol., 10, 1221–1224. [DOI] [PubMed] [Google Scholar]
- 67.Wittschieben J., Shivji,M.K., Lalani,E., Jacobs,M.A., Marini,F., Gearhart,P.J., Rosewell,I., Stamp,G. and Wood,R.D. (2000) Disruption of the developmentally regulated Rev31 gene causes embryonic lethality. Curr. Biol., 10, 1217–1220. [DOI] [PubMed] [Google Scholar]