Abstract
Dictyostelium discoideum has proven an exceptionally powerful system for studying numerous aspects of cellular and developmental functions. The relatively small (∼34 Mb) chromosomal genome of Dictyostelium and high efficiency of targeted gene disruption have enabled researchers to characterize many specific gene functions. However, the number of selectable markers in Dictyostelium is restricted, as is the ability to perform effective genetic crosses between strains. Thus, it has been difficult to create multiple mutations within an individual cell to study epistatic relationships among genes or potential redundancies between various pathways. We now describe a robust system for the production of multiple gene mutations in Dictyostelium by recycling a single selectable marker, Blasticidin S resistance, using the Cre-loxP system. We confirm the effectiveness of the system by generating a single cell carrying four separate gene disruptions. Furthermore, the cells remain sensitive to transformation for additional targeted or random mutagenesis requiring Blasticidin selection and for functional expression studies of mutated or tagged proteins using other selectable markers.
INTRODUCTION
The eukaryote Dictyostelium discoideum has many advantages that make it amenable to molecular manipulation for the analyses of gene function (1,2). The chromosomal genome of Dictyostelium is haploid and relatively small (∼34 Mb), although its transcriptome contains ∼11 000 genes (2). Targeting mutations in non-essential genes by homologous recombination is often at a relatively high frequency (>20%) and methods for generating genome-wide mutations by insertional (REMI) mutagenesis and for recovering the targeted genes are also well developed (3). In addition, many transformation vectors exist for the temporal- or spatial-specific expression of mutated or tagged proteins. Dictyostelium grow as individual cells, but initiate a multicellular developmental program upon starvation (1,2). Thus, growth and development are distinct and separable events, facilitating the isolation and maintenance of strains carrying gene mutations that are required for the development but not for the growth.
Dictyostelium transformants can be selected using auxotrophy (uracil, thymidine) or antibiotic [neomycin (Neo, G418), Blasticidin S and hygromycin] resistance (4). Auxotrophic selection requires cells with specific mutations and growth in specialized media. The G418 selection often yields cells carrying concatamerized, transforming DNA fragments and sometimes multiple insertions (5). In contrast, Blasticidin-resistance (Bsr) selection is very efficient, tolerates no background growth in standard medium and generally results in cells carrying only single-copy integrants (5,6). Consequently, most targeted and REMI mutageneses in Dictyostelium now utilize Bsr. However, since genetic crossing in Dictyostelium is inefficient, it is not a simple procedure to generate strains carrying multiple mutations (7). Yet, such multiple mutated strains are critical for studying the interaction among various genes or for examining functional genetic redundancies. Furthermore, strains carrying multiple mutations must still be sensitive to further transformation for expression studies of tagged (e.g. GFP and RFP) or mutated proteins for the analyses of complex and dynamic processes. Finally, in the absence of additional selection procedures, it is not possible to utilize a REMI screen for genetic suppression of Bsr-generated gene disruptants.
To overcome these genetic limitations, we significantly simplified strategies developed for directed recombination/deletion between specific DNA sequences to facilitate the targeted disruption of multiple genes in a single cell. We have adapted the Cre-loxP recombination system (8,9) for use in Dictyostelium to permit the recycling of Bsr and the continuous generation of new targeted mutations. Briefly, a gene targeting construct was created that contains a Bsr-cassette flanked (floxed) by loxP recombination sites; translational stop codons in all six reading frames were also added outside the loxP sites. Transient expression of the Cre recombinase removes the Bsr expression cassette through intramolecular recombination but does not remove the translational stops. This event creates a nonsense mutation within the targeted gene in a Blasticidin-sensitive cell line. We have confirmed the molecular recombination events by sequence analyses and proved the robustness of the system by creating cell lines with multiple targeted mutations.
MATERIALS AND METHODS
The floxed-Bsr targeting vector pLRBLP
The loxP recombination site (Figure 1A) consists of two highly conserved inverted repeats separated by a variable spacer region (10). The spacer region determines the specificity of loxP recombination (11). Only loxP sites separated by identical spacer sequences can recombine. The relative orientation of the spacer between two loxP sites determines the type of recombination event (11). Intramolecular recombination between two loxP sites in the same orientation will circularize and delete the intervening sequences, leaving a single loxP site (8,9,11).
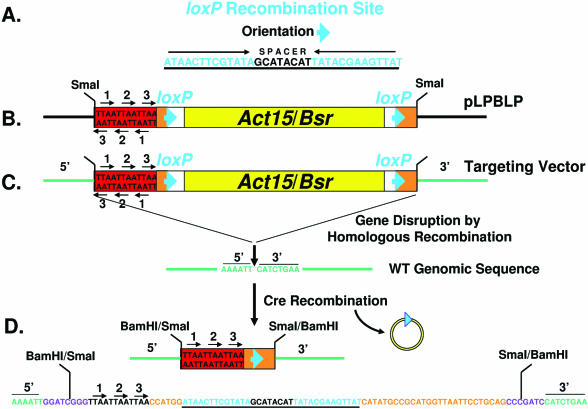
Figure 1.
A strategy for Cre-loxP recycling of the Bsr selectable marker. (A) The loxP recombination site includes an inverted repeat separated by a spacer sequence. (B) The gene targeting vector pLPBLP was constructed with loxP sites in the same orientation flanking both sides of the Bsr expression cassette act15/Bsr. An oligonucleotide cassette was also added that had translational stop codons in all six reading frames. The restriction enzyme sites outside of the floxed-Bsr cassette, such as SmaI (see Materials and Methods), permits the cloning of 5′ and 3′ gene sequences for targeted disruption. (C) The floxed-Bsr cassette of pLPBLP was inserted into the gene DDB0183838 sequence as indicated (see also Figure 2A). A BamHI site was engineered into the targeted gene sequence. This was filled and blunt-end ligated to the SmaI sites flanking the floxed-Bsr cassette to generate the gene-targeting vector. WT cells were transformed for gene disruption by homologous recombination and selected for resistance to Blasticidin S. (D) Transient expression of Cre promotes recombination and deletion of sequences between the two loxP sites in the disrupted gene DDB0183838. A 73 nt sequence remains that includes the translational stop cassette and a single loxP site. The sequence presented was determined directly from a PCR-amplified fragment of the region (see Figure 2B) and matches the predicted recombination event. The disrupted target gene sequences are in green, the loxP site is underlined and the stop codons are indicated by arrows. The site of the BamHI/SmaI fusion is also indicated.
Two loxP sites were engineered in the same orientation within the pBluescriptKS plasmid and six stop codons in all six reading frames were placed outside of the loxP sites (Figure 1B). Finally, the Dictyostelium Bsr expression cassette (act15/Bsr) was inserted between the two loxP sites to create the floxed-Bsr gene disruption vector pLPBLP (Figure 1B). Transcriptional regulation of Bsr is under control of the Dictyostelium actin 15 promoter and the Dictyostelium actin 8 terminator (6). The entire fragment is 1.5 kb and is flanked by SmaI sites, permitting the simple cloning into any gene of interest (Figure 1B). Unique cloning sites are also present: KpnI-SalI-AccI-ClaI-HindIII-SmaI—act15/Bsr—PstI-SmaI-BamHI-SpeI-NotI
The pLPBLP vector has been deposited with the Dicty Stock Center for distribution (see http://dictybase.org/StockCenter/StockCenter.html).
Transformation for homologous recombination
The floxed-Bsr cassette of pLPBLP was used to engineer various gene-targeting disruption constructs by standard cloning procedures. The DNA-mediated transformation was with linear vectors (4) and clonal selection (10–14 days) used 10 μg/ml Blasticidin S (MP Biomedicals) (6). Genomic DNA was purified (High Pure PCR Template Kit; Roche), and targeted gene disruptions were preliminarily identified by PCR. Several PCRs were performed. One PCR design used a primer from within the Bsr cassette and another primer outside of the targeting fragment. Another PCR examined the presence of the wild-type or disrupted gene using primers that flanked the floxed-Bsr insertion site. Southern blot hybridizations were used for absolute confirmation of gene disruption.
The Dictyostelium Cre expression vector pDEX-NLS-cre
We used a Cre protein (NLS-cre) engineered with a nuclear localization signal to promote high efficiency in vivo recombination between loxP sites. The coding region of NLS-cre was amplified from plasmid pTZ-Cre (12) with a 5′-flanking sequence that had been optimized for Dictyostelium A+T bias. The NLS-cre fragment was cloned into the EcoRI site that lies downstream of the Dictyostelium actin 15 (act15) promoter in the Dictyostelium expression vector pDEXRH (13), which also carries a G418-resistance cassette. The resulting Cre-expressing plasmid was termed pDEX-NLS-cre.
The pDEX-NLS-cre vector has been deposited with the Dicty Stock Center for distribution (see http://dictybase.org/StockCenter/StockCenter.html).
Cre transformation/expression and selection
Bsr knockout mutants were electroporated (4) with pDEX-NLS-cre. Approximately 107 growth-phase cells were washed at 0°C, once in 17 mM K-phosphate buffer, pH 6.0 and once in electroporation buffer (EP; 50 mM sucrose, 10 mM Na-K-phosphate buffer, pH 6.1). The cell pellet was then resuspended in 700 μl EP and the suspension gently mixed with ∼35 μg pDEX-NLS-cre DNA at 0°C in a 4 mm electroporation cuvette. Electroporation was for 1 ms, at 1 kV and 10 uF with the Gene Pulser Xcell (Biorad); two pulses were applied at 5 s intervals. After electroporation, the cells were transferred to a 9 cm plastic petri dish and shaken at ∼40 r.p.m. for 15 min at 20°C. The suspension was adjusted to 2 mM CaCl2 and 2 mM MgCl2 and shaking was continued for another 15 min. Finally, 12 ml axenic growth medium (14) was added, and the cells were allowed to recover for 24 h.
After the 24 h recovery period, the cells were placed in axenic medium (14) containing 10–20 μg/ml of G418 (Invitrogen Life Technologies) to select for pDEX-NLS-cre transformants. Selection in G418 continued for 3–10 days. The cells were then plated for clonal isolation by standard dilution. The clonal cell lines were subsequently picked in replica onto two different plates. The first was a standard SM agar plate (14) using growing Klebsiella aerogenes (K.a.) as the nutrient source. The second was a non-nutrient phosphate (30 ml) agar plate (17 mM Na/K-phosphate, pH 6.0 in 1.5% agar) layered with 500 μl of a concentrated K.a. suspension in Na/K-phosphate buffer and 120 μl of Blasticidin S at 10 mg/ml; the final concentration of Blasticidin S in the agar plate was 40 μg/ml. All Dictyostelium cells grow on SM K.a. plates, but only Dictyostelium that have retained the Bsr cassette grow rapidly in the presence of Blasticidin S. A WT parental cell was always used for growth control. This selection procedure allows for the rapid screening of hundreds of Cre transformants.
Cells not growing in Blasticidin S were replica plated into 24 well microtitre plates containing either axenic media, axenic media with Blasticidin S or axenic media with G418. Each transformation gave different frequencies for selective and optimized growth. Genomic DNA from Blasticidin- and G418-sensitive cell lines was analyzed by PCR, Southern blot hybridization and/or sequencing of PCR-amplified fragments to confirm Cre recombination and also the absence of both the Bsr and Neo expression cassettes. Usually, >95% of cells selected by growth criteria showed appropriate Cre recombination. This method was used to multiply target four diaphanous-related formin (drf) genes: drf1 on chromosome 5, accession no. J812237; drf2 on chromosome 4, accession no. AJ748258; drf3 on chromosome 1, accession no. AJ812236; and drf4 on chromosome 3, accession no. AJ812734.
As an alternative, one can bypass the entire G418 and Blasticidin S selections, and directly analyze clonal populations of cells after transient Cre expression. Following electroporation, the cells were clonally diluted into microtitre dishes at 0.5 cells/well and grown in the absence of Blasticidin S or G418 selection. While this strategy requires less time, it is also less efficient; in general, <5% of the colonies isolated may be pure Cre recombinants. This approach was used to multiply target DDB0183838 and DDB0184318 (see http://dictybase.org/), separated by 1.2 Mb on chromosome 6.
Finally, it should be noted that we were unable to promote in vivo recombination between loxP sites by direct electroporation of Cre protein. Our approach was modeled after the success observed with restriction enzymes during REMI mutagenesis (3), but we did not exploit all possible experimental variations.
RESULTS
Validation of Cre-mediated recombination
We created a disrupting gene construct of DDB0183838 (see Figure 1), transformed it into Dictyostelium, selected cells for Bsr growth and isolated individual colonies. Genomic DNA was prepared from the clonal isolates and homologous recombination monitored by PCR using primers within the gene sequence that lay outside of the floxed-Bsr insertion (Figure 1C). The endogenous, WT control band was ∼450 bp, while the gene containing the floxed-Bsr insert was ∼2000 bp (Figure 2A). Southern blot hybridization was used to confirm gene disruptions and the absence of secondary sites of insertion. The frequency for this targeted disruption was ∼80%.
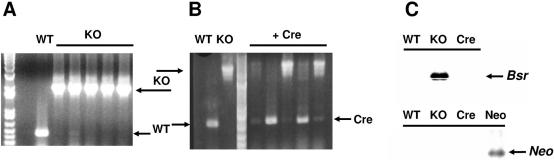
Figure 2.
Cre recombination deletes the Bsr selectable marker. (A) Genomic sequences flanking the insertion site of gene DDB0183838 depicted in Figure 1C were amplified by PCR and analyzed by agarose gel electrophoresis. WT is the endogenous wild-type gene sequence of ∼450 bp. Knockout mutants (KO) are clonal isolates of cells transformed and selected for resistance to Blasticidin S; insertion of the floxed-Bsr cassette yields a fragment of ∼2 kb. (B) A KO isolate from Figure 2A was transformed for transient expression of Cre, and several clonal isolates were randomly screened for functional recombination by PCR as in Figure 1A. Wild-type is the endogenous WT gene sequence of ∼450 bp. KO is the gene sequence carrying the insertion of the floxed-Bsr cassette (∼2 kb). Recombination between loxP sites generates a fragment that is ∼520 bp. The fragment was sequenced for confirmation (see Figure 1D). (C) Genomic DNA from WT, KO and Cre cells were probed by southern blot hybridization for the presence of Bsr and Neo gene sequences. DNA from a Neo-expressing transgenic cell line was included as a control.
A Bsr knockout cell line was chosen and transformed transiently to express NLS-cre. The cells were then clonally diluted into microtitre dishes at 0.5 cells/well and grown in the absence of Blasticidin S or G418 selection. PCR was used to assess the loss of Bsr sequences via recombination between loxP sites (Figure 1D). The Cre-mediated recombinant fragment is ∼520 bp, 73 bp larger than the endogenous, WT 450 bp band (Figures 1D and 2B). To confirm the specificity of the recombination event, we sub-cloned and sequenced the WT and Cre-generated fragments. As seen in Figures 1C and 1D, there was an exact recombination between the loxP sites. The Bsr cassette was removed but the in-frame stop codons and a single loxP site remained. This created a nonsense mutation in the gene. The resulting cell line was unable to grow in either Blasticidin S or G418, indicating that neither Bsr of pLPBLP nor Neo of the Cre plasmids were integrated into the genome. This conclusion was confirmed by Southern blot hybridization (Figure 2C). This simple Cre recombination and selection approach had an efficiency of ∼3% for the isolation of pure populations of Cre-recombined cells. An alternative approach using G418 selection (see Materials and Methods) and described below for drf gene targeting can increase this frequency to >95%.
Validation of Bsr recycling
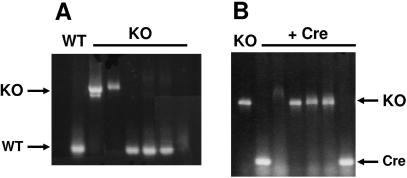
We next tested the efficacy of Cre-loxP for recycling of Blasticidin S selection. A second gene, DDB0184318, disruption fragment was prepared using pLPBLP. This was used to transform the Cre recombination, Blasticidin-sensitive cell line of Figure 2B. The frequency for the second site disruption was ∼20% (Figure 3A). The same frequency was obtained using a WT, parental cell line. Secondary, Cre transformation of the double knockout cell line was able to drive recombination within the new floxed-Bsr insertion (Figure 3B).
Figure 3.
Cre-recombined cells are susceptible to additional Bsr recycling. (A) A Cre-recombinant isolate from Figure 2B was transformed for a secondary gene disruption within DDB0184318 and selected for resistance to Blasticidin S. Genomic sequences flanking the insertion site of gene DDB0184318 were examined by PCR amplification. WT is the endogenous wild-type gene sequence; KO mutants are selected clonal isolates with the insertion of the floxed-Bsr cassette. (B) A KO isolate from Figure 3A was transformed for transient expression of Cre, and several clonal isolates were randomly screened for functional recombination by PCR as in Figure 2A. KO is the gene sequence carrying the insertion of the floxed-Bsr cassette. Recombination between loxP sites generates a fragment that is only ∼70 bp larger than the WT sequence.
Highly efficient generation of multiple gene disruptions in Dictyostelium
The Dictyostelium genome contains six drf genes (2,15). To test the robustness of the Cre-loxP system, we chose four drf genes (drf1, drf2, drf3 and drf4) for targeted disruption through four alternating rounds of gene inactivation followed by Bsr deletion through transient expression of NLS-cre. As depicted in Figure 4A and exemplified for drf1, each targeting vector contained the floxed-Bsr cassette inserted between 5′ and 3′ sequences that were specific for each drf gene. After transformation with linearized constructs, Blasticidin-resistant, drf knockout mutants were identified by PCR using sequence-specific primers and genomic DNA of clonal isolates (see Figure 4B). The floxed-Bsr cassette of drf-null cells was deleted by transient transfection using pDEX-NLS-cre, initial growth in the presence of G418, and final screening on selective K.a. plates (see Materials and Methods). Genomic DNA from transformants that were sensitive to both Blasticidin S and G418 was assayed by PCR in the absence of the Bsr (Figure 4C and D) and Neo cassettes (see also Figure 2C).
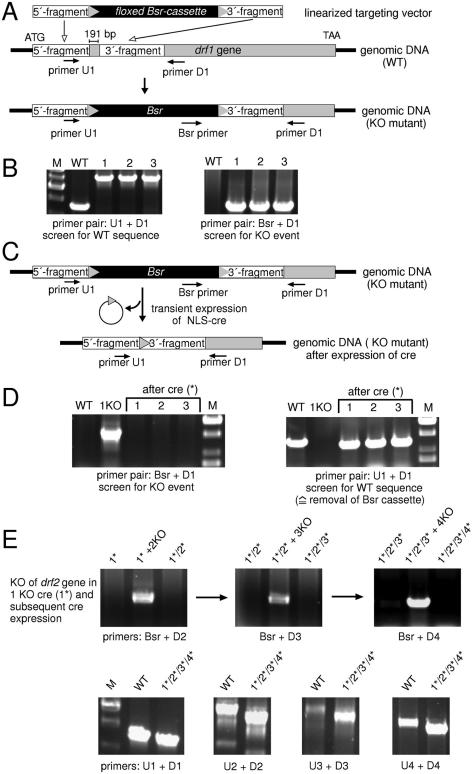
Figure 4.
Strategy for the generation of a quadruple drf-null cell line. (A) Generation of a drf1-null cell line. The 5′ and 3′ specific sequences of the drf1 gene were cloned into pLPBLP. The linear targeting vector was then used to disrupt the drf1 gene by homologous recombination. Targeted integration also caused a small deletion in the gene. A similar approach was used to construct targeting vectors for drf genes 2, 3 and 4. (B) WT cells and three independent Bsr mutants were examined by PCR amplification employing two different sets of drf1 primers. Left panel: the primer combination of U1 and D1 (see A) identifies WT and homologously recombined drf1 sequences. Upon homologous recombination, the drf1 PCR product is ∼1.3 kb larger than that of WT for all three cell lines shown. Right panel: the primer combination of Bsr and D1 (see A) specifically identifies only the homologous recombination event. (C) Strategy for deletion of Bsr by transient expression of NLS-cre. Deletion of the floxed-Bsr cassette leaves a sequence of ∼70 nt. (D) PCR analysis of drf1-null derived cells following transient expression of NLS-cre (*). All three clonal cell lines that were sensitive to Blasticidin and G418 for growth and were also deleted of the floxed-Bsr cassette. (E) Elimination of three additional drf genes. Top panel: three additional alternating rounds of gene inactivation followed by the subsequent elimination of the floxed/Bsr-cassette through transient expression of NLS-cre were performed to obtain a single mutant cell line lacking four drf genes. Each PCR screen used a common Bsr upstream primer and a downstream primer that was specific to drf genes 2, 3 or 4 (see A). Bottom panel: DNA from WT cells and the quadruple mutant 1*/2*/3*/4* was amplified by PCR using U and D primers specific to each drf gene to illustrate the stability of the four single loxP sites in the genome of the quadruple mutant. Each of the original homologous recombination events was designed to delete sequences from the endogenous gene (see A). Following the Cre-mediated recombination, an additional ∼70 bp remains that includes the translational stop cassette and a single loxP site (see Figures 1 and 4C), but still, the PCR products amplified from each gene of the 1*/2*/3*/4* mutant are always smaller than that of the WT gene fragment.
This strategy proved to be successful for the elimination of three additional drf genes in the same cell line (Figure 4E). As a final step of confirmation, we analyzed the rearrangements of the four drf genes in the quadruple mutant. Each locus was amplified with U and D specific primers (see Figure 4A), and their PCR products were compared with those obtained from WT genomic DNA. Since drf gene fragments were deleted during each of the targeted homologous recombination events (see Figure 4A), the PCR products amplified from the mutant DNA were always smaller than those from WT.
DISCUSSION
We have demonstrated a highly effective system to recycle a single selectable marker for the generation of multiple gene disruptions within a single cell. The technique described greatly surpasses the efficiencies of approaches that rely on parasexual genetics or fortuitous molecular events dictated by co-transformation for isolating Dictyostelium with multiple mutations (7,16).
The difficulty to generate Dictyostelium cell lines with multiple gene mutations has restricted the analyses of complex biochemical and genetic pathways and the identification of processes controlled by multigene families or functional redundancy (17). We believe that the described Bsr recycling approach largely overcomes this limitation. The cells deleted of Bsr are sensitive to additional rounds of gene disruption or to REMI mutagenic screening. In addition, the cells can be engineered for the regulated expression of specific protein variants, for global screening, for complementation expression or for gene interference by RNAi or antisense methodologies (3,18–20).
There are some potential engineering confines that should be considered when designing targeting constructs using the floxed-Bsr system. Gene disruptions that carry the entire Bsr cassette usually do not produce a stable transcript or protein fragment. However, we frequently detect normal levels of mRNA expression after the floxed-Bsr is removed via Cre recombination. Although the mRNA now encodes an in-frame nonsense codon that prevents the synthesis of a full-length protein, there is high probability for the production of truncated protein variants. The position of the engineered nonsense codon within an individual gene dictates the protein variant produced, and different protein truncations can cause distinct phenotypic effects. This feature can be used to an advantage if one wishes to examine the function of proteins with specific C-terminal truncations; otherwise floxed-Bsr insertions should be designed near the 5′ end of the gene. Parental and Cre-recombined cells must always be compared to ensure that Bsr deletion (or Cre-transformation) has not created a dominant (or secondary) phenotype.
After each successful Cre recombination event, one loxP site will remain in the genome. If two loxP-targeted genes were located on the same chromosome, there is a theoretical potential for Cre-mediated recombination between the loxP sites of the two genes and thus, for deletion of the entire genomic region between them. However, since Dictyostelium has a haploid genome, large deletions will be lethal, making such recombinations unlikely to be a common occurrence. Indeed, we were able to successfully disrupt genes DDB0183838 and DDB0184318 that are located on the same chromosome, but separated by ∼1.2 Mb. While small deletions are possible, they will be readily identified during genomic mapping by the PCR or southern blot hybridizations that are required to confirm the desired, site-specific recombination event. Nonetheless, there are simple modifications that can be used to prevent recombination between genes. The spacer sequence of the loxP site conveys recombination specificity (11). Thus, different floxed-Bsr cassettes will not recombine. Alternatively, the Cre-loxP system can be coupled with other recombination systems, such as Flp-FRT and φC31-att (9), for increased specificity and flexibility.
The Cre-loxP approach should also be easily adapted for many of the applications proven so effective for mouse genetics (8,9). Gene constructs can be engineered that become disrupted only upon the conditional expression of Cre. Many Dictyostelium transformation vectors already exist that direct gene expression with specific developmental patterns. In addition, the expression can also be regulated by a tetracycline-responsive system (21). Thus, it should be possible to study the effects of gene loss at specific developmental stages or in specific cell types. We do not believe that high-level expression of Cre will cause deleterious biological effects in Dictyostelium by inducing recombination within WT chromosomes. We have not observed any abnormal phenotypes upon the expression of Cre from a constitutive promoter (data not shown). Furthermore, sequence analysis of the entire Dictyostelium genome does not reveal any endogenous cryptic loxP sites, as are suggested to occur in genomes of higher complexity (22).
Conditional Cre expression can also be used to provide an additional layer of transcriptional regulation during development. Recombination between loxP sites can be used to induce gene expression by directly fusing a promoter and gene target separated by a floxed inactivating sequence, or conversely, to promote gene repression by deleting an element essential for transcription.
The utility of Cre-loxP recombination is also not limited to Bsr recycling for gene inactivation. Once loxP sites have been inserted into a gene of interest, it now becomes a site for recombination with exogenous transforming DNA elements for gene replacement studies (8,9,11). The loxP constructs are thus facile targets for the creation of gene knockins for the expression of mutant or tagged protein variants or novel promoter/reporter fusions.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs J. Brzostowski, F. Comer, L. Hennighausen, T. Khurana, V. McMains, C. Parent and D. Rosel for continued advice and critical comments. We also thank R. Fässler for plasmid pTZ-Cre, and Dr K. Kibler and Ms C. Su for technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 413).
REFERENCES
- 1.Aubry L. and Firtel,R. (1999) Integration of signaling networks that regulate Dictyostelium differentiation. Annu. Rev. Cell Dev. Biol., 15, 469–517. [DOI] [PubMed] [Google Scholar]
- 2.Kreppel L., Fey,P., Gaudet,P., Just,E., Kibbe,W.A., Chisholm,R.L. and Kimmel,A.R. (2004) dictyBase: a new Dictyostelium discoideum genome database. Nucleic Acids Res., 32(Database issue), D332–D333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuspa A. and Loomis,W.F. (1992) Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc. Natl Acad. Sci. USA, 89, 8803–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pang K.M., Lynes,M.A. and Knecht,D.A. (1999) Variables controlling the expression level of exogenous genes in Dictyostelium. Plasmid, 41, 187–197. [DOI] [PubMed] [Google Scholar]
- 5.Barth C., Fraser,D.J. and Fisher,P.R. (1998) Co-insertional replication is responsible for tandem multimer formation during plasmid integration into the Dictyostelium genome. Plasmid, 39, 141–153. [DOI] [PubMed] [Google Scholar]
- 6.Sutoh K. (1993) A transformation vector for Dictyostelium discoideum with a new selectable marker bsr. Plasmid, 30, 150–154. [DOI] [PubMed] [Google Scholar]
- 7.King J. and Insall,R.H. (2003) Parasexual genetics of Dictyostelium gene disruptions: identification of a ras pathway using diploids. BMC Genet., 4, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sauer B. (2002) Cre/lox: one more step in the taming of the genome. Endocrine, 19, 221–228. [DOI] [PubMed] [Google Scholar]
- 9.Branda C.S. and Dymecki,S.M. (2004) Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev. Cell, 6, 7–28. [DOI] [PubMed] [Google Scholar]
- 10.Hoess R.H. and Abremski,K. (1984) Interaction of the bacteriophage P1 recombinase Cre with the recombining site loxP. Proc. Natl Acad. Sci. USA, 81, 1026–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoess R.H., Wierzbicki,A. and Abremski,K. (1986) The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res., 14, 2287–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potocnik A.J., Brakebusch,C. and Fässler,R. (2000) Fetal and adult hematopoietic stem cells require beta1 integrin function for colonizing fetal liver, spleen, and bone marrow. Immunity, 12, 653–663. [DOI] [PubMed] [Google Scholar]
- 13.Faix J., Gerisch,G. and Noegel,A.A. (1992) Overexpression of the csA cell adhesion molecule under its own cAMP-regulated promoter impairs morphogenesis in Dictyostelium. J. Cell. Sci., 102, 203–214. [DOI] [PubMed] [Google Scholar]
- 14.Sussman M. (1987) Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell. Biol., 28, 9–29. [DOI] [PubMed] [Google Scholar]
- 15.Glöckner G., Eichinger,L., Szafranski,K., Pachebat,J.A., Bankier,A.T., Dear,P.H., Lehmann,R., Baumgart,C., Parra,G., Abril,J.F. et al. (2002) Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature, 418, 79–85. [DOI] [PubMed] [Google Scholar]
- 16.Betapudi V., Shoebotham,K. and Egelhoff,T.T. (2004) Generation of double gene disruptions in Dictyostelium discoideum using a single antibiotic marker selection. Biotechniques, 36, 106–112. [DOI] [PubMed] [Google Scholar]
- 17.Witke W., Schleicher,M. and Noegel,A.A. (1992) Redundancy in the microfilament system: abnormal development of Dictyostelium cells lacking two F-actin cross-linking proteins. Cell, 68, 53–62. [DOI] [PubMed] [Google Scholar]
- 18.Martens H., Novotny,J., Oberstrass,J., Steck,T.L., Postlethwait,P. and Nellen,W. (2002) RNAi in Dictyostelium: the role of RNA-directed RNA polymerases and double-stranded RNase. Mol. Biol. Cell, 13, 445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson D.N. and Spudich,J.A. (2000) Dynacortin, a genetic link between equatorial contractility and global shape control discovered by library complementation of a Dictyostelium discoideum cytokinesis mutant. J. Cell. Biol., 150, 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spann T.P., Brock,D.A., Lindsey,D.F., Wood,S.A. and Gomer,R.H. (1996) Mutagenesis and gene identification in Dictyostelium by shotgun antisense. Proc. Natl Acad. Sci. USA, 93, 5003–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blaauw M., Linskens,M.H. and van Haastert,P.J. (2000) Efficient control of gene expression by a tetracycline-dependent transactivator in single Dictyostelium discoideum cells. Gene, 252, 71–82. [DOI] [PubMed] [Google Scholar]
- 22.Sauer B. (1996) Multiplex Cre/lox recombination permits selective site-specific DNA targeting to both a natural and an engineered site in the yeast genome. Nucleic Acids Res., 24, 4608–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]