Abstract
Transfection of cells with short double-stranded synthetic DNA molecules that contain a transcription factor binding site, known as decoy oligodeoxynucleotides (ODNs), has been proposed as a novel approach in vitro and in vivo for the study of gene regulation and for gene therapy. Once delivered into cells, decoy ODNs are predicted to bind to nuclear transcription factors, preventing their binding to consensus sequences in target genes. Using a fluorescein-labeled decoy ODN containing a consensus sequence for the AP-1 transcription factor, we show that lipid-complexed decoys were readily transfectable into cells, but were consistently detectable in the cytoplasm and not in the nucleus. The same phenomenon was observed in three different cell lines including KB-3, CHO and MDA-MB-231. The AP-1 decoy ODNs failed to inhibit the transcriptional activity of an AP-1-dependent luciferase reporter. The effect of cytoplasmic AP-1 decoy ODNs on the subcellular localization and function of c-Jun induced by the microtubule inhibitor vinblastine, which strongly induced c-Jun expression, was assessed. No difference in protein level or nuclear localization of vinblastine-induced c-Jun, or of one of its target genes, p53, was noted when cells were transfected with wild-type or mutated forms of the decoy ODNs. We suggest that subcellular localization is an unappreciated and key limiting factor for the use of transcription factor decoy ODNs that must be addressed before meaningful data interpretation can be made.
INTRODUCTION
Transfection of cells with short double-stranded synthetic DNA molecules that contain a transcription factor binding site, known as decoy oligodeoxynucleotides (ODNs), has been proposed as a novel approach in vitro and in vivo for the study of gene regulation and for gene therapy [Reviewed in (1,2)]. This approach is particularly attractive because it has the potential to specifically target transcription factors and block their ability to activate target genes. The success of this new class of compounds depends on several factors including cellular uptake efficiency, intracellular stability, cellular toxicity and non-specific effects. Several delivery vehicles have been explored to achieve efficient uptake including different lipid formulations, hemagglutinating virus of Japan (HVJ)–liposome complexes or pressure-mediated transfection (1). Since the initial report of decoy development that targeted E2F transcription factors (3), numerous transcription factor decoy ODNs have been developed including those for activator protein-1 (AP-1) (4), cyclic AMP response element (CRE) (5), nuclear factor kappa B (NFκB) (6) and CCAAT/enhancer binding protein (C/EBP) (7). Many of these agents appear to elicit cellular effects. For example, it was reported that AP-1 decoy ODNs have growth inhibitory effects on vascular smooth muscle cells in vitro (8) and neointimal formation in vivo (8,9), and similar effects were observed with E2F decoy ODNs (10). CRE decoy ODNs were shown to inhibit cancer cell growth in vitro and in vivo (5), and NFκB ODNs inhibited cytokine expression and synovial cell proliferation (11).
Demonstration of specificity is clearly of paramount importance. Specificity is often documented by preparing nuclear extracts from transfected cells and observing an inhibition of oligonucleotide probe binding in electrophoretic mobility shift assays. However, this does not constitute proof that the decoy ODN inhibited transcription factor binding in the nuclei of transfected cells, but only that the decoy ODN is present in nuclear extracts prepared from these cells. This is an important distinction because nuclear extracts made under standard conditions are relatively crude and are likely to be contaminated with other organelles. Inhibition of transcription factor activity by decoy ODNs can also be examined by reporter assays. However, in many studies, the ability of the decoy ODN to block transcriptional activity in vivo was not demonstrated (5,9,12–15). Another key requirement is to demonstrate efficient cellular uptake and nuclear localization of the decoy ODN. Cellular uptake is often monitored by fluorescence microscopy, and this can allow an evaluation of uptake efficiency. However, few if any reports on decoy ODNs have definitively proven localization to the nucleus, and in one study, NFκB ODNs were efficiently delivered to cells but were localized in the cytoplasm (16).
We have shown previously that c-Jun induction is a key cellular response to vinblastine treatment (17). Our intention was to utilize the decoy ODN approach to block vinblastine-induced c-Jun/AP-1 activity in human KB-3 carcinoma cells in order to elucidate its role. We show here that AP-1 decoy ODNs were efficiently taken up by the cells but were consistently detectable in the cytoplasm, and not in the nucleus of the transfected cells. Cytoplasmic AP-1 decoy ODNs also failed to block nuclear localization of c-Jun or affect its ability to regulate a target gene. We suggest that nuclear translocation of decoy ODNs is an important and underappreciated limiting factor that should be carefully confirmed before data derived from such experiments are interpreted.
MATERIALS AND METHODS
Cell lines
KB-3 human carcinoma, a HeLa subline, Chinese hamster ovary (CHO) and MDA-MB-231 human breast carcinoma cell lines were maintained in monolayer culture at 37°C and 5% CO2 in DMEM (KB-3) or α-MEM (CHO, MDA-MB-231) supplemented with 10% fetal bovine serum, 2 mM glutamine, 50 units/ml penicillin and 50 μg/ml streptomycin. For MDA-MB-231 cells, the medium also contained 0.01 mg/ml insulin.
AP-1 decoy oligodeoxynucleotides
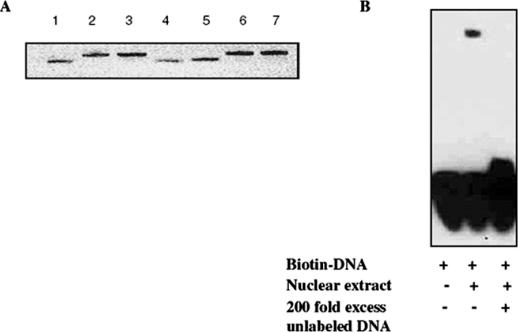
Synthetic, single-stranded, HPLC purified, phosphorothioate ODNs specific for AP-1, together with matching mutated ODN as a control, were purchased from Integrated DNA Tehnologies, Inc. (Coralville, IA). The sequences of the single-stranded ODNs were as follows: 5′-AG*CTTGTGACTCAGAAG*C*T-3′ for the AP-1 wild-type, and 5′-AG*CTTGAATCTCAGAAG*C*T-3′ for the AP-1 mutant (the consensus and the corresponding mutated sequences are underlined, and * denotes a phosphorothioate modification). Double-stranded 19mer ODNs (5 μM) were prepared in annealing buffer (10 mM Tris–HCl, pH 7.5, 50 mM NaCl, 1 mM EDTA) by melting at 95°C for 5 min followed by cooling to room temperature for 2 h. Annealing was verified by 20% polyacrylamide gel electrophoresis (Figure 1).
Figure 1.
Integrity of decoy ODN and AP-1 DNA binding. (A) Wild-type and mutated AP-1 decoy ODNs were denatured at 95°C for 5 min, cooled to room temperature for 2 h in the presence of annealing buffer, and then electrophoresed on a 20% polyacrylamide gel. DNA was visualized by ethidium bromide staining. Lanes 1, 4 and 5 are single-stranded ODNs; lanes 2 and 6 are double-stranded wild-type ODNs; and lanes 3 and 7 are double-stranded mutated ODNs. (B) AP-1-specific binding activity. AP-1 decoy ODN was biotin-labeled, 20 fmol was incubated in a binding reaction containing KB-3 cell nuclear extract and analyzed by gel electrophoresis, as described in Materials and Methods. The nuclear extract was omitted from the left lane, and the reaction in the right lane contained a 200-fold molar excess of unlabeled ODN.
Transfection of AP-1 decoy ODNs
Approximately 1.5 × 105 cells/well were plated in 12-well plates one day before transfection. When 50–60% confluent, cells were transfected with 100 nM AP-1 ODNs in 0.5 ml transfection volume and in the presence of 2 μl Oligofectamine reagent following the manufacturer's instructions (Invitrogen, Carlsbad, CA). Efficient transfection required the presence of medium containing serum and without antibiotics. After 4 h incubation at 37°C in a 5% CO2 incubator, 250 μl normal growth media was added to each well. Cellular extracts were prepared 24 h post-transfection, as described below. In some experiments, cells were treated with 30 nM vinblastine 6 h after transfection. Each experiment included transfection with wild-type AP-1 decoy ODN or mutated AP-1 decoy ODN, with untransfected cells as an additional control. The same protocol was used for all cell lines. For the other transfection reagents used, the protocols are available as Supplementary Material.
Fluorescence microscopy
ODNs were labeled with 5-carboxy fluorescein using the Label IT Nucleic Acids Labeling Kit (Mirus Corp., Madison, WI). Cells (1.5 × 105) were grown on sterile cover slips in 12-well plates and incubated with 100 nM fluorescein-labeled AP-1 ODN in the presence of 2 μl Oligofectamine reagent/0.5 ml medium/well. Fresh medium (0.25 ml) was added after 4 h incubation and the slides were prepared for examination 12, 24 and 48 h post-transfection. Briefly, cells were washed twice with phosphate-buffered saline (PBS), fixed for 20 min in 4% paraformaldehyde solution and then washed three times with PBS. In order to visualize nuclear DNA, the fixed cells were incubated for 5 min with 0.3 μM 4,6-diamidino-2-phenylindole (DAPI) (Molecular Probes, Eugene, OR) and rinsed three times for 5 min in PBS. The coverslips were mounted using Vectashield mounting medium (Vector Laboratories, Burlingame, CA). Fluorescent images were digitally photographed using an Axioskop2 wide-field deconvolution fluorescence microscope (Carl Zeiss, Thornwood, NY) with fluorescein (excitation, 492 nm; emission, 518 nm) or DAPI (excitation, 360 nm; emission, 460 nm) filter sets from Chroma Technology Corp. (Rockingham, VT). The images were acquired with Axiovision software Version 3. Uptake efficiency was estimated by comparing the number of cells exhibiting green fluorescence with the total number of cells in four independent fields, representing approximately 500 cells in total for each condition.
Electrophoretic mobility shift assay
AP-1 decoy ODNs were biotin-labeled using the Biotin 3′ End DNA Labeling Kit (Pierce Biotechnology, Rockford, IL) and labeling efficiency of 75% was verified by dot blot analysis (data not shown). For electrophoretic mobility shift assays (EMSA), a non-radioactive LightShift Chemiluminescent EMSA Kit™ (Pierce Biotechnology) was used. Binding reactions contained 20 fmol biotin-labeled ODN, 9 μg KB-3 cell nuclear extract (18), 50 μg/ml poly (dI·dC), 5% glycerol, 0.05% NP-40, 5 mM MgCl2 in the presence of binding buffer (10 mM Tris–HCl, pH 7.5, 50 mM KCl, 1 mM DTT). Competition reactions contained a 200-fold molar excess of unlabeled DNA. After 20 min incubation at room temperature, electrophoresis was performed using 6% polyacrylamide DNA retardation gels (Invitrogen, Carlsbad, CA) and then transferred to a positively charged nylon membrane (Immobilon-NY+) (Millipore Corp., Bedford, MA). After UV cross-linking, detection was performed with streptavidin–HRP conjugate antibody following the manufacturer's protocol.
AP-1-dependent luciferase assay
Approximately 1.5 × 105 cells/well were plated in 12-well plates one day before transfection. The next day, cells were cotransfected with 7 X pAP-1-Luc (Cat # 219073, Stratagene, La Jolla, CA) and pRL-TK (Cat # E2241, Promega Corp, Madison, WI) in a 10:1 ratio using Lipofectamine Plus Reagent (Invitrogen, Carlsbad, CA) and following the manufacturer's protocol. The next day, the wild-type and mutated AP-1 ODN decoys were introduced into cells using the protocol described above. To stimulate AP-1 activity, cells were treated with 12-O-tetradecanoylphorbol-13-acetate (TPA) for 16 h and luciferase activity of cell lysates was determined using the Dual-Luciferase Assay System from Promega (Madison, WI). Both luciferase activities were measured using a TD-20/20 Luminometer and AP-1 activity was normalized to Renilla activity. The results are expressed as AP-1 Firefly luciferase/Renilla luciferase activity ratios and represent the means of measurements from triplicate wells ± SD from two independent experiments.
Cell extraction and immunoblotting
Whole-cell extracts were prepared as described previously (17) and nuclear and cytoplasmic subcellular fractions were prepared using extraction kits from Active Motif (Carlsbad, CA). Protein concentration was measured and normalized using the Bio-Rad protein assay. Immunoblotting was performed as described previously using 60 μg/lane (18). Antibodies to c-Jun (H-79), ERα (MC-20), actin (I-19) and caspase-3 (sc7148) were from Santa Cruz Biotechnology (Santa Cruz, CA); antibody to p53 was from NeoMarkers (Fremont, CA).
RESULTS AND DISCUSSION
Double-stranded AP-1 decoy ODNs were prepared as described in Materials and Methods and analyzed by gel electrophoresis for purity and integrity (Figure 1A). To confirm the binding ability, electrophoretic mobility shift assays were performed. As shown in Figure 1B, a retarded band was observed when the AP-1 decoy ODN was incubated with nuclear proteins from KB-3 cells, which have been shown previously to exhibit AP-1 binding activity (18). A 200-fold molar excess of wild-type ODN out-competed AP-1 binding to the biotinylated probe, demonstrating specificity.
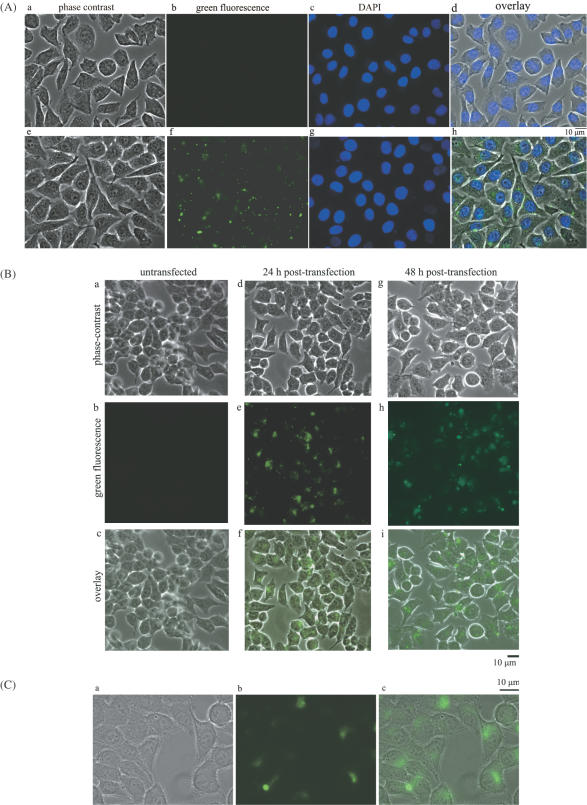
KB-3 cells were transfected with fluorescein-labeled AP-1 decoy ODNs. Uptake efficiency, determined as described in Materials and Methods, was routinely 80% under standard transfection conditions and was similar for both wild-type and mutated ODNs (Figure 2A and B). Closer inspection of the cells revealed however that the fluorescent signal was extranuclear, and most often appeared in a distinctive perinuclear pattern perhaps associated with the endoplasmic reticulum or Golgi apparatus (Figure 2C). This extranuclear localization was evident at both early (12 h) and late (48 h) time-points of transfection, and thus did not represent an eventual fate of the ODN.
Figure 2.
Cellular uptake and subcellular localization of AP-1 decoy ODNs in KB-3 cells. Representative micrographs of KB-3 cells transfected with fluorescent-labeled AP-1 decoy ODNs (represented as a green signal). (A) Panels (a–d) represent untransfected cells; panels (e–h) represent 12 h post-transfection. (a and e) Phase-contrast images; (b and f) green fluorescence images; (c and g) DAPI nuclear staining; (d and h) merged images. (B) Panels (a–c) represent untransfected cells; panels (d–f) represent 24 h post-transfection; panels (g–i) represent 48 h post-transfection. (a, d and g) Phase-contrast images; (b, e and h) are fluorescent images; (c, f and i) merged images. (C) Images were taken using the 100× objective 48 h post-transfection. (a) Phase-contrast; (b) green fluorescence; (c) merged images. Extranuclear fluorescence is evident.
To further assess whether the AP-1 decoy ODNs were able to modulate AP-1 transcriptional activity, we used an AP-1-dependent luciferase reporter assay, as described under Materials and Methods. TPA-stimulated AP-1 luciferase activity was readily observed in cells, and was unaffected by the AP-1 decoy ODNs, using either the wild-type or mutated AP-1 decoys. The results are available as supplementary data (Figure S1) and are fully consistent with our observation of extranuclear decoy localization.
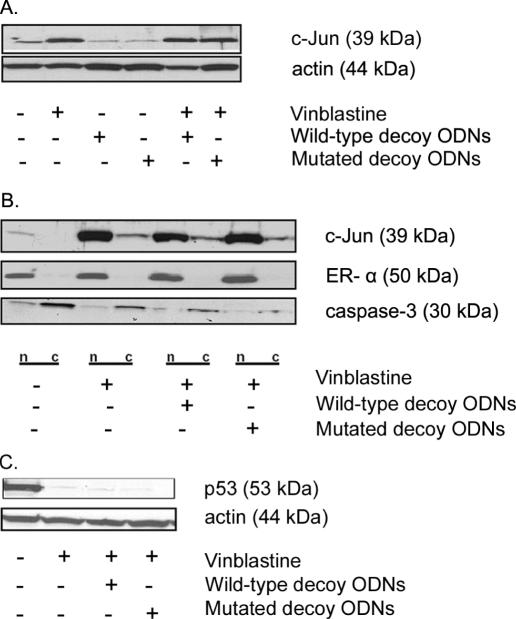
Because the main aim was to inhibit vinblastine-induced c-Jun function, we reasoned that the decoy ODNs, although not localized to the nucleus, might still be effective if they were able to inhibit c-Jun synthesis or to sequester newly synthesized c-Jun. Treatment of KB-3 cells with vinblastine resulted in increased protein expression of c-Jun, as previously described (17), and this was unaffected by prior transfection with AP-1 decoy ODNs (Figure 3A). In order to determine whether the AP-1 decoy ODNs affected nuclear localization of c-Jun, KB-3 cells were treated with decoy ODNs and then treated with vinblastine, and nuclear and cytoplasmic fractions were prepared. Immunoblotting for estrogen receptor-α and procaspase-3 was performed as nuclear and cytoplasmic markers, respectively (Figure 3B). It is evident that the induced c-Jun was mainly located in the nuclear fraction and that the decoy ODNs had no effect on nuclear localization of the induced c-Jun following vinblastine treatment (Figure 3B). One of the main roles of c-Jun is to transcriptionally downregulate p53 expression (19), and we have shown that p53 downregulation is one of the key functions of vinblastine-induced c-Jun in KB-3 cells (17). As shown in Figure 3C, vinblastine caused downregulation of p53 expression as expected, and this was unaffected by the AP-1 decoy ODNs. These results demonstrated that the AP-1 decoy ODNs were ineffective and were unable to inhibit the transcriptional activity of c-Jun, in keeping with their non-nuclear location.
Figure 3.
Effect of AP-1 decoy ODNs on vinblastine-mediated c-Jun induction, nuclear expression and function with respect to p53 downregulation. KB-3 cells were either untreated or treated with vinblastine (30 nM, 16 h) in the presence or in the absence of the wild-type or mutated AP-1 decoy ODNs (100 nM, 6 h prior to vinblastine) as indicated. (A) Whole-cell extracts were prepared, subjected to SDS–PAGE and immunoblotted with antibodies to c-Jun or actin as a loading control. (B) Nuclear (n) and cytoplasmic (c) extracts were prepared and subjected to SDS–PAGE and immunoblotting with c-Jun-specific antibody. Estrogen receptor-α and procaspase-3 expression were monitored as nuclear and cytoplasmic markers, respectively. Reduced levels of procaspase 3 were observed with vinblastine treatment due to processing of the cleaved form not evident on the gel. (C) Whole-cell extracts were prepared, subjected to SDS–PAGE and immunoblotted with p53-specific antibody. Actin was used as a loading control.
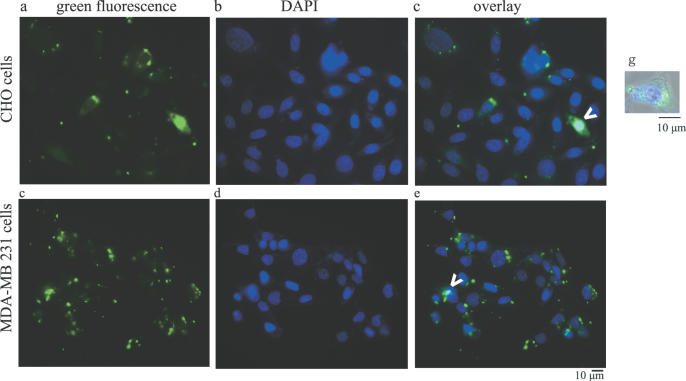
In order to demonstrate whether these results were more generally applicable, two other cell lines were examined. Upon transfection of CHO cells, green fluorescent AP-1 decoy ODN was observed intracellularly, but was clearly distinct in location from DAPI-stained nuclei (Figure 4, panels a, b and c). Similarly, in MDA-MB-231 cells, punctuate green fluorescence was distinct from nuclear DNA staining (Figure 4, panels d, e and f). In both cases, an occasional cell with coincident green and blue fluorescence, representing nuclear localized decoy ODN, was observed. A representative CHO cell, with merged color and phase-contrast imaging, is shown in Figure 4, panel g, and serves to emphasize the non-nuclear localization of the AP-1 decoy ODNs.
Figure 4.
Subcellular localization of AP-1 decoy ODNs in CHO and in MDA-MB-231 cells. Representative micrographs of cells transfected with fluorescent-labeled AP-1 decoy ODNs (represented as a green signal). Panels (a–c) represent CHO cells; panels (d–f) represent MDA-MB-231 cells. (a and d) Green fluorescence; (b and e) DAPI staining; (c and f) merged images. The arrows indicate rare cells with nuclear decoy ODNs. Panel g represents an image of a representative CHO cell with phase contrast, green fluorescence and blue fluorescence merged that was acquired using the 100× objective. Extranuclear decoy ODN localization is evident.
Several other DNA delivery reagents were utilized to achieve nuclear uptake of the decoy ODNs. Thus, we used Polyfect Transfection Reagent (Qiagen) that is an activated-dendrimer; Transfectin Lipid Reagent (Biorad) that is a mixture of cationic compound and a co-lipid (1,2-dioleoyl-sn-glycero-3-phoshoethanolamine); and Fugene 6 Transfection Reagent (Roche Applied Science) that has a non-liposomal formulation. For each formulation, we observed intracellular but extranuclear decoy localization in KB-3 cells, although uptake efficiency was lower when compared with our standard Oligofectamine method. No green fluorescence was observed in cells transfected with Fugene 6 reagent.
In conclusion, we have shown that AP-1 decoy ODNs are readily transfectable into cells, but fail to localize to the nucleus in the majority of transfected cells using several transfection reagents commonly utilized for this purpose. Similar results were found for three different cell lines, suggesting that this is not a cell-specific phenomenon. Cytoplasmic localization of NFκB decoy ODNs has also been reported (16), suggesting that this finding is not limited to AP-1 decoy ODNs. In KB-3 cells, the AP-1 decoy ODNs were ineffective at blocking AP-1-dependent transactivation and c-Jun function induced by vinblastine. These results suggest that nuclear translocation of decoy ODNs is a further limiting factor in their use. Many studies of decoy ODNs have not reported subcellular localization and have not demonstrated specific inhibitory effects on transcription. Therefore, the reported biological effects may be non-specific in nature. We suggest that nuclear localization should be confirmed before data derived from such experiments are interpreted, and that this represents an important complement to experiments demonstrating inhibition of transcriptional activity in vivo.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health Grant CA75577 from the National Cancer Institute. The use of the facilities in the University of Arkansas of Medical Sciences Digital and Confocal Microscopy Laboratory supported by NIH Grants 1 P20 RR 16460 and PAR-98-092, 1-R24 CA82899 is acknowledged.
REFERENCES
- 1.Dzau V. (2002) Transcription factor decoy. Circ. Res., 90, 1234–1236. [DOI] [PubMed] [Google Scholar]
- 2.Morishita R., Higaki,J., Tomita,N. and Ogihara,T. (1998) Application of transcription factor ‘decoy’ strategy as means of gene therapy and study of gene expression in cardiovascular disease. Circ. Res., 82, 1023–1028. [DOI] [PubMed] [Google Scholar]
- 3.Morishita R., Gibbons,G.H., Horiuchi,M., Ellison,K.E., Nakama,M., Zhang,L., Kaneda,Y., Ogihara,T. and Dzau,V.J. (1995) A gene therapy strategy using a transcription factor decoy of the E2F binding site inhibits smooth muscle proliferation in vivo. Proc. Natl Acad. Sci. USA, 92, 5855–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morishita R., Gibbons,G.H., Horiuchi,M., Kaneda,Y., Ogihara,T. and Dzau,V.J. (1998) Role of AP-1 complex in angiotensin II-mediated transforming growth factor-beta expression and growth of smooth muscle cells: using decoy approach against AP-1 binding site. Biochem. Biophys. Res. Commun., 243, 361–367. [DOI] [PubMed] [Google Scholar]
- 5.Park Y.G., Nesterova,M., Agrawal,S. and Cho-Chung,Y.S. (1999) Dual blockade of cyclic AMP Response Element- (CRE) and AP-1-directed transcription by CRE-transcription factor decoy oligonucleotide. J. Biol. Chem., 274, 1573–1580. [DOI] [PubMed] [Google Scholar]
- 6.Morishita R., Sugimoto,T., Aoki,M., Kida,I., Tomita,N., Moriguchi A., Maeda,K., Sawa,Y., Kaneda,Y., Higaki,J. and Ogihara,T. (1997) In vivo transfection of cis element ‘decoy’ against nuclear factor-kappaB binding site prevents myocardial infarction. Nature Med., 3, 894–899. [DOI] [PubMed] [Google Scholar]
- 7.Wagner A.H., Krzesz,R., Gao,D., Schroeder,C., Cattaruzza,M. and Hecker,M. (2000) Decoy oligodeoxynucleotide characterization of transcription factors controlling endothelin-B receptor expression in vascular smooth muscle cells. Mol. Pharmacol., 58, 1333–1340. [DOI] [PubMed] [Google Scholar]
- 8.Ahn J.D., Morishita,R., Kaneda,Y., Lee,S.J., Kwon,K.Y., Choi,S.Y., Lee,K.U., Park,J.Y., Moon,I.J., Park,J.G., Yoshizumi,M., Ouchi,Y. and Lee,I.K. (2002) Inhibitory effects of novel AP-1 decoy oligodeoxynucleotides on vascular smooth muscle cell proliferation in vitro and neointimal formation in vivo. Circ. Res., 90, 1325–1332. [DOI] [PubMed] [Google Scholar]
- 9.Kume M., Komori,K., Matsumoto,T., Onohara,T., Takeuchi,K., Yonemitsu,Y. and Sugimachi,K. (2002) Administration of a decoy against the activator protein-1 binding site suppresses neointimal thickening in rabbit balloon-injured arteries. Circulation, 105, 1226–1232. [DOI] [PubMed] [Google Scholar]
- 10.Ahn J.D., Morishita,R., Kaneda,Y., Kim,H.S., Chang,Y.C., Lee,K.U., Park,J.Y., Lee,H.W., Kim,Y.H. and Lee,I.K. (2002) Novel E2F decoy oligodeoxynucleotides inhibit in vitro vascular smooth muscle cell proliferation and in vivo neointimal hyperplasia. Gene Ther., 9, 1682–1692. [DOI] [PubMed] [Google Scholar]
- 11.Tomita N., Morishita,R., Tomita,S., Gibbons,G.H., Zhang,L., Horiuchi,M., Kaneda,Y., Higaki,J., Ogihara,T. and Dzau,V.J. (2000) Transcription factor decoy for NFkappaB inhibits TNF-alpha-induced cytokine and adhesion molecule expression in vivo. Gene Ther., 7, 1326–1332. [DOI] [PubMed] [Google Scholar]
- 12.Tomita T., Takano,H., Tomita,N., Morishita,R., Kaneko,M., Shi,K., Takahi,K., Nakase,T., Kaneda,Y., Yoshikawa,H. and Ochi,T. (2000) Transcription factor decoy for NFkappaB inhibits cytokine and adhesion molecule expressions in synovial cells derived from rheumatoid arthritis. Rheumatology, 39, 749–757. [DOI] [PubMed] [Google Scholar]
- 13.Viedt C., Dechend,R., Fei,J., Hansch,G.M., Kreuzer,J. and Orth,S.R. (2002) MCP-1 induces inflammatory activation of human tubular epithelial cells: involvement of the transcription factors, nuclear factor-kappaB and activating protein-1. J. Am. Soc. Nephrol., 13, 1534–1547. [DOI] [PubMed] [Google Scholar]
- 14.Penolazzi L., Lambertini,E., Borgatti,M., Piva,R., Cozzani,M., Giovannini,I., Naccari,R., Siciliani,G. and Gambari,R. (2003) Decoy oligodeoxynucleotides targeting NF-kappaB transcription factors: induction of apoptosis in human primary osteoclasts. Biochem. Pharmacol. 66, 1189–1198. [DOI] [PubMed] [Google Scholar]
- 15.Maeshima Y., Kashihara,N., Yasuda,T., Sugiyama,H., Sekikawa,T., Okamoto,K., Kanao,K., Watanabe,Y., Kanwar,Y.S. and Makino,H. (1998) Inhibition of mesangial cell proliferation by E2F decoy oligodeoxynucleotide in vitro and in vivo. J. Clin. Invest. 101, 2589–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griesenbach U., Cassady,R.L., Cain,R.J., du Bois,R.M., Geddes,D.M. and Alton,E.W. (2002) Cytoplasmic deposition of NF-kB decoy oligonucleotides is insufficient to inhibit bleomycin-induced pulmonary inflammation. Gene Ther., 9, 1109–1115. [DOI] [PubMed] [Google Scholar]
- 17.Fan M., Goodwin,M. and Chambers,T.C. (2001) The c-Jun NH2-terminal protein kinase/AP-1 pathway is required for efficient apoptosis induced by vinblastine. Cancer Res., 61, 4450–4458. [PubMed] [Google Scholar]
- 18.Berry A., Goodwin,M., Moran,C. and Chambers,T.C. (2001) AP-1 activation and altered AP-1 composition in association with increased phosphorylation and expression of specific Jun and Fos family proteins induced by vinblastine in KB-3 cells. Biochem. Pharmacol., 62, 581–591. [DOI] [PubMed] [Google Scholar]
- 19.Schreiber M., Kolbus,A., Piu,F., Szabowski,A., Mohle-Steinlein,U., Tian,J., Karin M., Angel,P. and Wagner,E.F. (1999) Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev., 13, 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.