Abstract
Potent sequence selective gene inhibition by siRNA ‘targeted’ therapeutics promises the ultimate level of specificity, but siRNA therapeutics is hindered by poor intracellular uptake, limited blood stability and non-specific immune stimulation. To address these problems, ligand-targeted, sterically stabilized nanoparticles have been adapted for siRNA. Self-assembling nanoparticles with siRNA were constructed with polyethyleneimine (PEI) that is PEGylated with an Arg-Gly-Asp (RGD) peptide ligand attached at the distal end of the polyethylene glycol (PEG), as a means to target tumor neovasculature expressing integrins and used to deliver siRNA inhibiting vascular endothelial growth factor receptor-2 (VEGF R2) expression and thereby tumor angiogenesis. Cell delivery and activity of PEGylated PEI was found to be siRNA sequence specific and depend on the presence of peptide ligand and could be competed by free peptide. Intravenous administration into tumor-bearing mice gave selective tumor uptake, siRNA sequence-specific inhibition of protein expression within the tumor and inhibition of both tumor angiogenesis and growth rate. The results suggest achievement of two levels of targeting: tumor tissue selective delivery via the nanoparticle ligand and gene pathway selectivity via the siRNA oligonucleotide. This opens the door for better targeted therapeutics with both tissue and gene selectivity, also to improve targeted therapies with less than ideal therapeutic targets.
INTRODUCTION
Even the emergence of ‘targeted’ cancer therapeutics inhibiting tumor-specific proteins or pathways, such as anti-vascular endothelial growth factor (VEGF) antibody (1), has not eliminated problems of toxicity. Thus, recognition of potent, sequence-selective gene inhibition by siRNA oligonucleotides and rapid adoption as the tool of choice in cell culture has generated the expectation for their use to improve targeted therapeutics (2–5). The prospects are for siRNA to be substantially better than antibodies, in part because they are easily applicable to virtually any therapeutic target including intracellular factors and even transcription factors. Also, they promise potent gene inhibition with exquisite selectivity, even down to the level of single-nucleotide polymorphisms, and easy identification by small screens across the gene sequence of the offending protein or variant. However, this promise is tempered by their double-stranded RNA (dsRNA) oligonucleotide nature, resembling antisense, ribozymes and gene therapy (6,7). For cancer, in particular, the pharmacological hurdles are severe. Local aqueous siRNA activity has been observed for several tissues (5,8,9), but is lacking in tumors (10), and systemic exposure can induce non-specific responses, as found for CpG DNA oligonucleotides (10,11). Thus, the ‘revolutionary’ potency and selectivity of siRNA inhibitors of gene expression promises to enable improved targeted cancer therapeutics, but the means for systemic administration and targeted distribution to disseminated metastatic lesions are needed.
Since administration of aqueous siRNA, even chemically stabilized, is limited by a lack of tumor activity and non-specific responses (5,8–11), the use of gene therapy vector systems is one approach. Several efforts have evaluated cationic lipids and polymers originally developed for DNA plasmids where internalization is by non-specific electrostatic interactions (12,13). For DNA, means to improve on control of cellular interactions have focused on either addition of peptide ligands for cellular receptors or addition of steric polymer coatings to inhibit non-specific interactions, and recently their combination (14–21). The resulting systems generally are perceived of having yielded good cytoplasmic delivery, but the gene expression from the plasmid remains limited by poor trafficking into the nucleus. Recent efforts achieved some success by coupling dual antibodies to the distal end of polyethylene glycol (PEG) on a sterically stabilized liposome by a complex procedure using post modification and incurring substantial loss of antibody and siRNA (22). Nonetheless, a need exists for relatively small peptide ligands, such as a disulfide-stabilized Arg-Gly-Asp (RGD) peptide targeting integrin expression upregulated at sites of neovasculature (23–27), and self-assembling nanoparticles without the problems of liposomes (14,21). We adapted our tissue-targeted plasmid nanoparticles (14) for siRNA, with the expectation that their activity for cytoplasmic delivery would benefit siRNA since its RNA-induced silencing complex-mediated activity occurs in the cytoplasmic compartment (4). The self-assembly of cationic polymer conjugated with PEG-peptide conjugates with siRNA was found, as with plasmid DNA, to form sterically stabilized ‘layered’ nanoparticle-sized polyplexes with exposed RGD ligands, referred to here as nanoplexes. These siRNA nanoplexes were evaluated with an siRNA inhibiting the VEGF-mediated pathway of tumor angiogenesis. The results of intravenous administration into tumor-bearing animals showed evidence for sequence-specific inhibition of the target gene, reduction in angiogenesis and inhibition of tumor growth.
METHODS
Nucleic acids
Short dsRNA oligonucleotides for siRNA labeled siLuc, siLacZ, siGFP and siVEGF R2 were designed based on studies by Elbashir et al. (2), validated to lack significant interfering homology using BLAST analysis, and synthesized and purified by Dharmacon (Lafayette, CO). Two sequences were synthesized per target and combined in a 1:1 molar ratio. The target sequences used were as follows: for siLuc: aaccgctggagagcaactgca and aagctatgaaacgatatgggc, for siLacZ: aacagttgcgcagcctgaatg and aacttaatcgccttgcagcac, for siGFP: aagctgaccctgaagttcatc and aagcagcacgacttcttcaag and for siVEGF R2: aatgcggcggtggtgacagta and aagctcagcacacagaaagac [inhibition of VEGF R2 by this siRNA has been described previously (28)]. siRNA targeted against luciferase was labeled with fluorescein isothiocyanate (FITC) at the 3′ position of the sense strand with standard linkage chemical conjugation, for fluorescently activated cell sorting (FACS)-analysis and tissue distribution experiments. The luciferase-encoding pCI-Luc plasmid (pLuc) was obtained from Lofstrand Labs (Gaithersburg, MD).
Synthesis of RGD-PEG-PEI and PEG-PEI
Two PEGylated forms of branched polyethyleneimine (PEI) (P) were prepared, one with a PEG having an RGD peptide at its distal end RGD-PEG-PEI (RPP) and the other with a PEG lacking the peptide PEG-PEI (PP).
The ‘cyclic’ 10mer RGD peptide with the sequence H-ACRGDMFGCA-OH, was synthesized, oxidized to form an intramolecular disulfide bridge and purified to >95% purity by Advanced ChemTech (Louisville, KY). This sequence was derived from the integrin-binding RGD peptides identified by phage display and has been found effective for cell binding and internalization (23,29).
Synthesis of RPP was carried out as follows in two steps. In the first step, to a stirring solution of RGD (60 mg) in dimethyl sulfoxide (DMSO) (600 μl) TEA [8.54 μl in 20 μl of tetrahydrofuran (THF)] was added under nitrogen. After stirring for 1 min, a solution of NHS-PEG-VS (212 mg in THF: DMSO; 300 μl:100 μl) was added in one portion. The reaction mixture was stirred at room temperature for 4 h, quenched with trifluoroacetic acid (TFA) (amount equivalent to the TEA) and the mixture was lyophilized. The intermediate RGD-PEG-VS was purified by either reverse-phase high-performance liquid chromatography (HPLC) or dialysis against water, and the compound lyophilized to give a yield of 50–90%. Conjugation was confirmed by Mass Spectral analysis (matrix-assisted laser desorption ionization).
In the second step of synthesis, 100 mg (21.7 μmol ) of the purified RGD-PEG-VS intermediate was dissolved in 1 ml of pure DMSO. To this solution, six equivalents of TEA dissolved in 0.5 ml THF was added and mixed. An aliquot of 9.4 mg (218 μmol in terms of amines) of PEI dissolved in dimethylformamide (0.5 ml) was added to the above solution and stirred at room temperature for 12 h. The completion of the conjugation was confirmed by the disappearance of RGD-PEG-VS on TLC. The reaction was terminated by the addition of an excess of TFA and lyophilized. The product was purified as the TFA salt by HPLC. Degree of conjugation of RGD-PEG to PEI was determined by proton NMR spectrometry on a 500 MHz spectrometer (Varian), from the ratio of the area under the peaks corresponding to the -CH2- protons of PEI (2.8–3.1 p.p.m.) and PEG (3.3–3.6 p.p.m.). Based on this estimate, ∼7% of the PEI amines were conjugated with RGD-PEG, or an average of about 40 RGD-PEG molecules attached to each 25 kDa PEI molecule reducing the average number of amines from 580 to 540 per PEI molecule. Percentage conjugation ranged from 7 to 9 for various syntheses.
Preparation of nanoplexes
Nanoplexes were prepared by mixing equal volumes of aqueous solutions of cationic polymer and nucleic acid to give a net molar excess of ionizable nitrogen (polymer) to phosphate (nucleic acid) over the range of 2 to 6. The electrostatic interactions between cationic polymers and nucleic acid resulted in the formation of polyplexes with average particle size distribution of about 100 nm, hence referred to here as nanoplexes.
Three forms of nanoplexes were prepared based on the three forms of PEI: P, PP and RPP. Earlier studies have reveled that conjugation of polycations used for DNA condensation with other macromolecules can lead to incomplete condensation and formation of structures with non-spherical morphology (30,31). Though we have not observed any of these problems with the conjugates used in this study, in order to avoid this potential problem, part of the polycation required for condensation was substituted with unconjugated PEI. Therefore, all RPP- and PP-nanoplexes contain PEI in molar equivalent to the conjugates, expressed in terms of amine concentration. These nanoplexes were thus prepared by first preparing an aqueous solution of cationic polymer containing RPP or PP with P in a 1:1 molar ratio in 5 mM Hepes buffer (pH 7). In a separate tube, nucleic acids (plasmid DNA and/or siRNA) were dissolved in the same buffer in the same total volume as the cationic polymer solution. The two solutions were then mixed together and vortexed for 30 s to make nanoplexes. Mean particle size distribution was determined with a Coulter N4plus particle size instrument (Beckman Coulter), and ζ-potential measurements were performed on a Coulter Delsa 440 SX instrument. Both instruments were calibrated using latex beads of defined size and mobility as standards (Beckman Coulter, Miami, FL).
Cells
Human umbilical vein endothelial cells (HUVECs) (Glycotech, Rockville, MD) were cultured in E-STIM endothelial cell culture medium (BD Biosciences, Bedford, MA). N2A murine neuroblastoma cells and SVR-bag 4 murine endothelial cells (both American Type Culture Collection, Manassas, VA) were cultured in DMEM supplemented with 10% (v/v) heat-inactivated fetal calf serum, 100 IU/ml penicillin and 100 μg/ml streptomycin (Quality Biologicals, Gaithersburg, MD). The cells were cultured at 37°C in a humidified atmosphere of 5% carbon dioxide/95% air.
siRNA serum stability
Serum stability of siRNA in aqueous solution versus in nanoplex preparations was characterized using polyacrylamide gel electrophoresis. Samples of siRNA either in aqueous solution or as a RPP-nanoplex were mixed in a 1:1 ratio with fresh serum to give 50% serum concentration and incubated at 37°C. Aliquots from different incubation times of each sample were loaded onto a gel and electrophoresis performed to visualize intact siRNA. The gel conditions were performed so that aqueous siRNA would enter the gel and run as a clearly visible band when stained with ethidium bromide. Under these electrophoresis conditions, siRNA in the nanoplex remains in the loading well and is visualized by the staining.
FACS analysis of nanoplex binding to HUVEC and N2A cells
HUVEC or N2A cells growing in a nearly confluent monolayer in 6 well plates were washed with phosphate-buffered saline (PBS) and detached using 1 mM EDTA in PBS. The cells were suspended in FACS buffer (PBS supplemented with 1% BSA, 1.26 mM CaCl2, 0.81 mM MgSO4) at 4°C, centrifuged, counted, and 1 × 105 cells were transferred to FACS tubes (BD Biosciences). The cells in 2 ml were incubated with 2 μg fluorescently labeled siRNA formulated in P- PP- or RPP-nanoplexes for 1 h at 4°C. P-, PP- and RPP-nanoplexes were made by mixing 2 μg siRNA with 0.52 μg P, (0.26 μg P + 2.4 μg PP) and (0.26 μg P + 2.9 μg RPP), respectively, to obtain complexes with N/P of 2. At the end of the incubation period, the cells were washed, fixed with 4% buffered formaldehyde and analyzed on a FACSCalibur flow cytometer (BD Biosciences). In competition experiments, the cells were pre-incubated with a 100-fold molar excess of unconjugated RGD peptide for 30 min at 4°C. Results were analyzed using WinMDI software version 2.8 (Joseph Trotter).
siRNA-mediated gene-silencing effects in vitro
The SVR-bag 4 endothelial cells, constitutively expressing β-galactosidase, were seeded in a 6 well plate with 2 ml medium per well. Medium was replaced for fresh medium without serum or antibiotics, and 10 μg of siRNA against β-galactosidase or luciferase was added either in the free form or as P- (2.6 μg), PP- (1.3 μg P + 12 μg PP) or RPP- (1.3 μg P + 14.5 μg RPP) nanoplexes. In all the cases, the siRNAs complexed to the nanoparticles were prepared as an aqueous suspension described above and added to the cells once the 2 ml of medium was replaced. After 3 h, the cells were washed with PBS and serum-containing medium was added. At 48 h after transfection, the reporter enzyme activity was detected using a β-galactosidase staining kit (Active motif LLC, Carlsbad, CA). The N2A cells were seeded in a 24 well plate with 0.5 ml of medium. When nearly confluent, the medium was replaced and they were transfected with 2 μg pLuc in Lipofectamin 2000™ (Invitrogen, Carlsbad CA) according to the manufacturer's instructions, together with 1 μg of siLuc, siLacZ or siGFP in the free form or as P-, PP- or RPP-nanoplexes. Nanoparticle solutions were prepared as above and transfection was performed as described above. The concentrations were based on the plasmid or siRNA mass given above in the final medium volume of 0.5 ml. Luciferase activity was detected after 24 h, using the luciferase assay system (Promega, Madison, WI) on a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, CA). The luciferase activity of wells treated with pLuc alone was treated as 100% and used to normalize the luciferase activity of wells treated with siRNA.
Mouse tumor model
Female nude mice (6–8 weeks of age) were obtained from Taconic (Germantown, NY), kept in filter-topped cages with standard rodent chow and water available ad libitum, and a 12 h light/dark cycle. The experiments were performed according to national regulations and approved by the local animal experiments ethical committee. Subcutaneous N2A tumors were induced by inoculation of 1 × 106 N2A cells in the flank of the mice. At a tumor volume of ∼0.5–1 cm3, mice received nanoplexes or free siRNA by i.v. injection of a solution of 0.2 ml via the tail vein. The nanoplex solutions were prepared as above, at N/P ratio of 2. For tissue distribution experiments, 40 μg fluorescently labeled siRNA was injected in the free form or as P- or RPP-nanoplexes. One hour after injection, the tissues were dissected and examined with a dissection microscope fitted for fluorescence. Microscopic examination of tissues was performed with an Olympus SZX12 fluorescence microscope equipped with digital camera and connected to a PC running MagnaFire 2.0 camera software (Optronics, Goleta, CA). Pictures were taken at equal exposure times for each tissue.
In the co-delivery experiments, plasmid and siRNA were mixed in a 1:100 molar ratio, respectively (40 μg pLuc with 13 μg siRNA), and in the sequential delivery experiments 40 μg plasmid was delivered first, followed by 40 μg siRNA 2 h later (1:300 molar ratio). The tissues were dissected, weighed and put in ice-cold reporter lysis buffer (Promega) in magnetic beads containing 2 ml tubes (Q-Biogene, Carlsbad, CA), 24 h after injection of the nanoplexes. Tissues were homogenized with a Fastprep FP120 magnetic homogenizer (Q-Biogene) and samples were assayed for reporter enzyme activity using the luciferase assay system (Promega) on a Monolight 2010 luminometer (Analytical Luminescence Laboratory).
In the tumor growth inhibition studies, the experiment was started when the tumors became palpable at 7 days after inoculation of the tumor cells. Treatment consisted of 40 μg siRNA per mouse in RPP-nanoplexes every 3 days intravenously via the tail vein. Tumor growth was measured at regular intervals using a digital caliper by an observer blinded to treatment allocation. Each measurement consisted of tumor diameter in two directions of ∼90° apart. Tumor volume was calculated as, 0.52 × longest diameter × shortest diameter2 (32). At the end of the experiment, the animals were sacrificed and tumor tissue and surrounding skin was excised and put on a microscopy glass slide. Tissue examination for vascularization and angiogenesis was performed by microscopy using the Olympus microscope and camera equipment described above for fluorescent tissue measurements. Tissue was trans-illuminated to visualize blood vessels in the skin and a digital image was taken and stored as described above. Tissue was snap frozen immediately thereafter for western blotting.
Western blotting
Murine VEGF R2 expression in tumor samples was detected by western blotting. Tumor tissue was put into lysing Matrix D (Bio-Rad, Cambridge, MA) together with M-Per mammalian protein extraction reagent (Pierce). Tissue was homogenized, centrifuged and supernatants collected. Equivalent amounts of extracted protein (50 μg) were mixed with sample buffer containing 5% 2-mercaptoethanol (Bio-Rad), boiled, cooled and loaded in each lane of a 6% polyacrylamide gel. Electrophoresis was performed at 30 mA and subsequently proteins were transferred to an Immunblot polyvinylidene fluoride membrane (Bio-Rad). Membranes were blocked overnight with 3% gelatin in Tris-buffered saline (TBS). Subsequently, membranes were transferred to 1% gelatin in TTBS (10 mM Tris–HCl, 150 mM NaCl, 0.1% Tween-20) incubated with 1 μg monoclonal anti-mVEGF R2 antibody (R&D Systems) overnight. After washing twice in TTBS, goat anti-mouse immunoglobulin G peroxidase conjugate was added in 1% gelatin for 1 h, the membrane was washed twice with TTBS and subsequently once with TBS. Antibody was stained using a Bio-Rad color reagent kit for 30 min.
Statistical analyses
Luciferase expression data in vitro and tumor growth inhibition were analyzed, after log transformation to normalize the data, by ANOVA using Dunnett's post-test. As in vivo gene expression data did not follow normal distributions, data were analyzed by non-parametric Kruskal–Wallis test with Dunn's post-test to examine the significance of the differences.
RESULTS
siRNA nanoplex colloidal properties
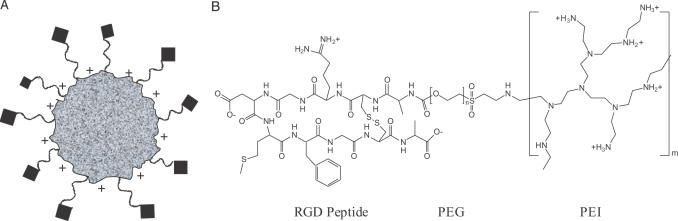
The siRNA nanoplex was developed with a modular approach to design molecular conjugates that incorporate the three functional requirements: self-assembly, formation of a steric polymer protective surface layer and exposed ligands. To design an siRNA nanoplex, we revisited materials used originally for plasmid DNA including PEI for the polycation complexing agent, PEG for steric stabilization and peptide ligands containing an RGD motif to provide tumor selectivity due to their ability to target integrins expressed on activated endothelial cells in tumor vasculature. While peptides containing an RGD motif can bind several integrins, their specificity is determined by the flanking amino acid sequence as well as the conformation of the binding domain (33). In this study, we used a ‘cyclic’ RGD peptide whose integrin-binding domain is conformationally constrained by a disulfide bond. It has an identical amino acid sequence within the cyclic region of a peptide that was shown to cause cell binding and internalization by receptor-mediated pathway when expressed on filamentous phage (23,29). This peptide was shown to inhibit cell attachment to fibronectin- and vitronectin-coated plates in a sequence-specific manner. Furthermore, when coupled to an oligolysine this peptide showed receptor-mediated DNA delivery in a variety of cells, including endothelial cells (34). In order to facilitate chemical conjugation to PEG, for these studies, an alanine residue was added to each end of this peptide outside the cyclic region (23,29). The targeted siRNA nanoplexes (shown schematically in Figure 1A) were prepared by chemical synthesis of tripartite polymer conjugates (Figure 1B) with a cationic polymer, a steric polymer and a peptide ligand (RPP), followed by nanoparticle self-assembly by mixing with nucleic acid in aqueous solution (14). This RPP conjugate allows individualized optimization or chemical replacement for each functional domain.
Figure 1.
Design of targeted self-assembling siRNA nanoplex and activity in vitro. (A) Schematic structure of the targeted self-assembling nanoplex. Electrostatic interactions between negatively charged siRNA and cationic polymer result in the formation of a complex (speckled particle) that is protected by exposed steric polymer strands (wavy lines), and which gains target selectivity through a targeting ligand moiety coupled to the distal end of the steric polymer (diamonds). (B) Schematic structure of the RPP-polymer. The polymer is designed with the three functional domains needed to obtain the targeted self-assembling nanoplex as described in (A). Branched PEI with a molecular weight of ∼25 kDa (∼581 monomer units) is used as the cationic complexing polymer and contains on average 25% primary, 50% secondary and 25% tertiary amines. Approximately 7% of the amines are modified with PEG with an average molecular weight of 3.4 kDa (80 monomer units). The PEG is conjugated to a folded RGD peptide containing a disulfide bridge between two cysteine residues, a ligand for integrins.
Upon mixing purified RPP with an aqueous nucleic acid solution, the cationic domain of the conjugate binds to negatively charged nucleic acid driving self-assembly to form a nanoparticle dispersion. Studies found that stable nanoplexes could be formed at an amine (PEI) to phosphate (nucleic acid) ratio (N/P) of 2:1. Particle size and ζ-potential results at this ratio are given in Table 1. The mean size of either RPP- or PP-nanoplexes was small, between 0.07 and 0.10 μm. Particle size remained largely unchanged for 9 days, a period for which particle size was monitored. In contrast, the mean particle size of P-nanoplexes was larger, between 0.12 and 0.17 μm, and aggregated within 24 h. The ζ-potential of P-nanoplexes with siRNA was found to be highly positive, 35 ± 4 mV, as typically found with plasmid DNA, but incorporation of the PEG conjugate of PEI resulted in a reduction in the ζ-potential to 5 ± 6 mV and 6 ± 1 mV for PP-nanoplexes and RPP-nanoplexes, respectively. Surface charge polarity and amplitude depended on the ratio of the two components, but at the same ratio amplitude decreased when PEG was present (data not shown), indicating that a steric polymer layer is formed on the nanoplex surface (14,20,21).
Table 1. Particle characterization of siRNA nanoplexesa.
| siRNA nanoplex | Mean particle size (μm) | ζ-potential (mV) |
|---|---|---|
| PEI | 0.12 ± .04 | 35 ± 4 |
| PP | 0.09 ± .01 | 5 ± 6 |
| RPP | 0.09 ± .01 | 6 ± 1 |
aamine:phosphate ratio of 2:1, average values of three preparations.
These measurements of particle size and ζ-potential indicate that the RPP-nanoplexes formed with siRNA exhibit colloidal surface properties indicative of an outer steric polymer layer and potentially exposed RGD ligand to mediate cell-binding selectivity. This nanoplex self-assembly occurs by simple mixing of aqueous solutions of RPP conjugates with siRNA (RPP-nanoplexes). Their colloidal and biological properties were compared either to preparations with precursor conjugates lacking the RGD peptide (PP-nanoplexes) or to unconjugated PEI (P-nanoplexes).
In Vitro ligand-mediated uptake and gene inhibition
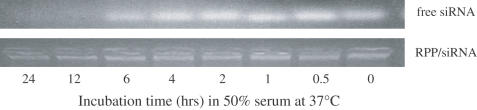
In vitro studies were performed to characterize the ligand-mediated uptake and intracellular activity of the entrapped siRNA. For use of microscopic imaging and FACS analysis, these studies relied on standard, commercially available FITC conjugates of siRNA. Initially, some stability studies were performed to characterize lability of the siRNA oligonucleotides in 50% serum in aqueous solution and in the nanoplex. The results, shown in Figure 2, demonstrate that serum incubation of siRNA oligonucleotides in aqueous solution resulted in degradation of the oligonucleotide (loss of band in the gel) after several hours, but siRNA in the nanoplex was protected for the entire length of the studies, 12 h. Additionally, microscopy studies of tumor cell transfection in culture using FITC-siRNA showed that as found by the siRNA supplier, this standard FITC conjugate did not interfere with siRNA gene inhibition activity and FITC fluorescence in cells correlated with loss of reporter gene expression (data not shown). These results support cell culture and animal administration where the siRNA is exposed to serum nucleases, at least for periods up to a few hours and for longer periods in the case of siRNA nanoplexes.
Figure 2.
Degradation of siRNA in serum. Degradation of siRNA oligonucleotides when exposed to serum was measured for RPP-nanoplexes and compared with aqueous siRNA. The siRNA nanoplex, or aqueous siRNA, was incubated in 50% serum from 0 to 24 h and then the remaining intact siRNA determined by gel electrophoresis. Staining of the siRNA bound in nanoplexes shows it remains in the loading well without evidence for degradation, while the aqueous siRNA runs as a brightly stained band that diminishes with incubation time.
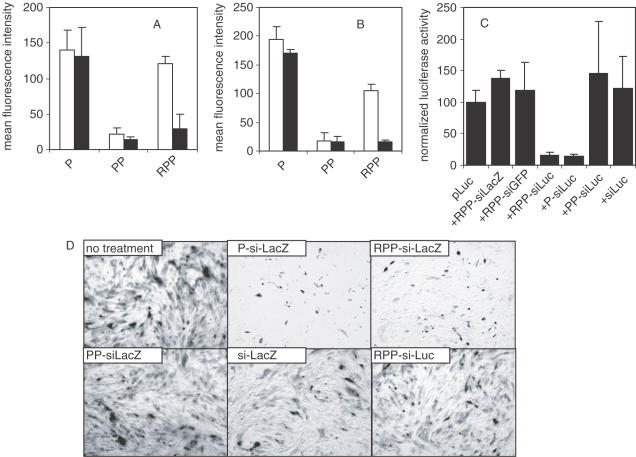
FACS analysis of endothelial (Figure 3A) and tumor cell (Figure 3B) binding studies using FITC-labeled siRNA shows that in the absence of ligand, the steric polymer surface layer reduces non-specific cell binding by 6- to 8-fold (P-nanoplexes versus PP-nanoplexes in Figure 3A and B), correlating with a reduction in ζ-potential. The presence of the peptide restores binding in a ligand-specific manner (RPP-nanoplexes in Figure 3A and B), despite maintenance of a near neutral surface charge. This binding is inhibited by the addition of unconjugated RGD peptide via competitive binding (closed bars in Figure 3A and B). These observations show that the RPP-nanoplex has both a PEG surface layer that provides steric inhibition of non-specific electrostatic cell binding and, more importantly, an RGD ligand-mediated cell binding, presumably through integrins expressed on both cell types studied (25,35).
Figure 3.
In vitro activity of siRNA nanoplex. (A and B) siRNA nanoplex binding to cells. 1 × 105 HUVEC (A, open bars) or N2A cells (B, open bars) were incubated with 2 μg fluorescently labeled siRNA formulated in P-, PP- or RPP-nanoplexes for 1 h at 4°C. Both cell types express integrins. After the incubation period, the cells were washed, fixed with 4% buffered formaldehyde and cell-bound fluorescence analyzed by FACS analysis. Unshielded positively charged P-nanoplexes showed relatively high cell binding to both cell types. Shielding of the charged nanoplex with PEG (PP-nanoplexes) reduced cell interaction, which was restored by coupling of the RGD peptide to the distal end of the PEG-shield (RPP-nanoplexes). Pre-incubation of HUVEC (A, closed bars) or N2A (B, closed bars) with a 100-fold molar excess of RGD peptide reduced binding of RPP-nanoplexes while leaving binding of P-or PP-nanoplexes unaffected, indicating that the binding of RPP-nanoplexes to cells is mediated by the RGD peptide targeting ligand. (C) Luciferase silencing in vitro. N2A cells were transfected with 2 μg luciferase plasmid, using cationic lipids without siRNA or cotransfected with 1 μg siRNA formulated as RPP-siLacZ, RPP-siGFP, RPP-siLuc, P-siLuc, PP-siLuc or free siLuc. Sequence-specific silencing of luciferase expression with siLuc is observed for RPP- and P-nanoplexes. Luciferase activity of cells treated with various agents were normalized assuming the activity of cells transfected with luciferase plasmid to be 100%. (D) β-Galactosidase silencing in vitro. SVR-bag 4 endothelial cells, constitutively expressing β-galactosidase were left untreated or incubated with 10 μg of siRNA as follows: P-siLacZ, RPP-siLacZ, PP- siLacZ, RPP-siLuc or free siLacZ. After 3 h incubation, the cells were washed and 48 h after transfection, the cells were stained for β-galactosidase activity. Only panels P-siLacZ and RPP-siLacZ show clear inhibition of β-galactosidase expression.
Gene-silencing activity of the preparations was determined with cell culture assays of reporter gene-targeted siRNA using both transient and endogenous gene expression assays. Both P- and RPP-nanoplexes inhibit transient luciferase (pLuc) expression (Figure 3C) or endogenous β-galactosidase expression (Figure 3D) in a sequence-specific manner. Thus, both non-specific and ligand-mediated cellular uptake can enable sequence-specific siRNA activity in these cells. A lack of inhibition with aqueous siRNA or PP-nanoplexes reflects poor intracellular availability but for different reasons: siRNA in aqueous solution has a strong anionic character reducing intracellular uptake while in the case of PP-nanoplexes cellular uptake is diminished by the surface PEG steric layer. Thus, in vitro gene-silencing activity of the different siRNA nanoplex preparations is paralleled by their cell interaction and intracellular uptake.
Tumor uptake, targeted gene inhibition and phenotypic effect
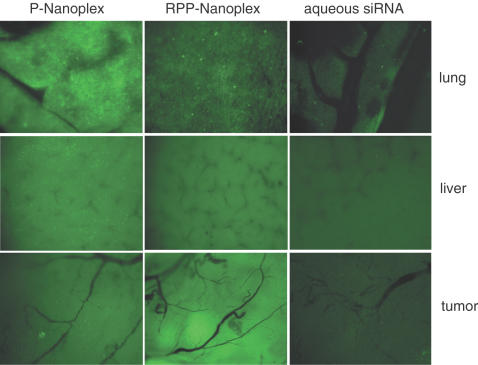
Since both RPP-nanoplexes and P-nanoplexes display activity in cell culture, they were selected for in vivo studies in tumor-bearing mice. Studies were performed to determine whether increased tumor levels of siRNA can be achieved by siRNA nanoplexes administered by i.v. injection to tumor-bearing animals. Imaging of FITC-labeled siRNA uptake into established neuroblastoma N2A tumors in nude mice was used to observe tumor accumulation. Fluorescence microscopy to detect FITC fluorescence in tumor, lung and liver are shown in Figure 4 for animals administered aqueous siRNA, P-nanoplexes and RPP-nanoplexes. Intravenous administration of aqueous siRNA did not produce appreciable FITC-siRNA fluorescence in the tumor. Also, for this sample very little FITC fluorescence can be observed in the liver and even less in the lung. These results are most likely a reflection of rapid clearance of the FITC-siRNA into the urine, poor tissue accumulation except liver and potentially metabolic instability resulting in rapid excretion or liver metabolism of the FITC. The lack of tumor fluorescence from the aqueous FITC-siRNA indicates that any instability of the FITC linkage that would result in loss of FITC from nanoplex preparations will not yield tumor fluorescence (since it was not yielded by aqueous FITC-siRNA, which is more labile in serum than that in the nanoplex as shown in Figure 2 above). This conclusion is confirmed by a lack of tumor FITC fluorescence when P-nanoplexes were administered (Figure 4). This lack of tumor FITC fluorescence demonstrates that neither the P-nanoplexes accumulate in the tumor to any detectable extent nor does any FITC linkage stability in the siRNA nanoplex result in artifactual tumor fluorescence. On the other hand, the FITC-siRNA in P-nanoplexes does produce appreciable FITC-siRNA fluorescence in liver and especially in lung with a punctate profile (Figure 4). Again, this finding further supports the utility of the FITC-siRNA labeling since many plasmid studies have revealed the clearance of P-nanoparticles to distribute into these two tissues and with the punctate pattern observed here with siRNA. In contrast, RPP-nanoplexes produced appreciable FITC-siRNA fluorescence in the tumor, but poor liver and lung accumulation and a reduced punctate fluorescence pattern. This provides strong evidence that the RPP nanoplex potentially exhibits reduced non-specific tissue interactions reducing the liver and lung uptake and accumulation in tumor due to ligand binding. Also, P-nanoplexes but not RPP-nanoplexes produced occasional minor adverse effects such as piloerection, also reported to occur with plasmid DNA and attributed to non-specific charge interactions with blood components, aggregation and trapping of aggregates in tissue capillaries (21,36). Since the FITC fluorescent label is covalently attached to the siRNA with a linkage routinely used for oligonucleotides with known in vivo stability, the fluorescence distribution observed in these tissues likely corresponds to distribution of siRNA and not to FITC linkage instability. Importantly, the studies of siRNA serum stability in Figure 2 clearly demonstrate that the siRNA in the RPP-nanoplex is protected from degradation for several hours and is more stable than aqueous siRNA. Moreover, the fluorescent microscopy tissue distribution studies were performed after a very short time frame, 1 h, which was much shorter than the 6 h serum stability observed for aqueous siRNA and 12 h stability observed for the RPP-nanoplex. Furthermore, even administration of the FITC-conjugated siRNA in aqueous form, which is more sensitive to degradation in serum, did not result in any significant accumulation in any of the tissues measured. These results indicate that the RPP siRNA nanoplex gives increased tumor levels of siRNA molecules and led to studies of siRNA biological activity in the tumor described below.
Figure 4.
Tissue distribution of siRNA nanoplexes in N2A tumor-bearing mice. Mice received 40 μg fluorescently labeled siRNA by intravenous injection as P-nanoplexes (left column), or RPP-nanoplexes (middle column) or aqueous solution (right column). One hour after injection, tissues were dissected and examined on a fluorescence microscope. Pictures were taken at equal exposure times for each tissue. P-nanoplexes show punctate fluorescence in all organs, especially high in lung and liver. RPP-nanoplexes show lower fluorescence levels in lung tissue with a punctate distribution and lower non-punctate fluorescence in liver. Higher levels of fluorescence were observed in the tumor as compared with P-nanoplexes. Fluorescence levels after administration of free siRNA were much lower in all organs, as compared with either of the two nanoplex formulations.
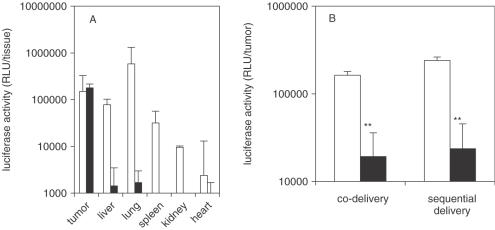
The tumor accumulation observed with RPP-nanoplexes may occur through RGD peptide specificity for tumor endothelial cells (23–27) or by passive tumor accumulation through ‘enhanced permeability and retention’ effect (14,19,21,37), or both. While there are differences in these two mechanisms, the key issue is whether distribution into the tumor achieves cellular uptake and siRNA activity. This was evaluated using transient expression of luciferase in tumor and its inhibition mediated by an siRNA targeted against luciferase mRNA. Since RPP-nanoplexes can be used for tumor-targeted delivery of siRNA as well as plasmid DNA, the experiment was carried out in two different ways, co-delivery of plasmid with siRNA or sequential delivery of plasmid followed by siRNA 2 h later, all using RPP-nanoplex. Tumor-targeted luciferase activity, achieved by the delivery of luciferase plasmid using RPP-nanoplexes to tumor-bearing mice, is shown in Figure 5A. The plasmid expression profile showing highest expression in tumor tissue parallels the tissue distribution observed with fluorescent siRNA (Figure 5A). Note that RPP-nanoplexes with plasmids also represent an attractive tissue selective gene delivery system in its own right, including constructs to express siRNA in vivo (R. Schiffelers, A. Ansari, C.J. Snel, M.H.A.M. Fens, G. Molema, G. Zhou, G. Strahan, G. Storm, M.C. Woodle and P.V. Scaria, manuscript in preparation). RPP-nanoplexes with siRNA inhibited plasmid expression by ∼90% (P < 0.01) regardless of whether administered simultaneously (co-delivery) or 2 h later (sequential delivery) and was highly sequence specific (Figure 5B). These experiments clearly demonstrate that the RPP-nanoplex can be used for the delivery of siRNA to tumor tissue through i.v. administration and can provide sequence-specific inhibition of gene expression in tumor. Next, whether the siRNA delivery achieved through RPP-mediated delivery into tumor would be sufficient to provide therapeutic efficacy was tested using an siRNA targeted to an endogenous therapeutic gene. For these studies, siRNA targeting murine VEGF R2 was selected and used with RPP-nanoplexes since it is a pivotal factor in tumor angiogenesis (1,13,38). For therapeutic effects on this gene, however, the siRNA requires delivery into host (murine) endothelial cells within the tumor to elicit a phenotypic effect on tumor growth. Efficacy studies were performed with siRNA inhibiting expression of murine VEGF R2, characterized in cell culture (data not shown). Studies were performed with this therapeutic gene siRNA administered intravenously as RPP-nanoplexes every 3 days. The results, shown in Figure 6, show strong inhibition of tumor growth rate (Figure 6A) and this was sequence specific. This result suggests that the RPP siRNA nanoplex acts through an endothelial cell uptake mechanism.
Figure 5.
Inhibition of plasmid-mediated reporter gene expression in vivo by siRNA-nanoplexes. (A) RPP-plasmid mediated luciferase expression in tumor. N2A-tumor-bearing mice received a single intravenous injection of 40 μg pLuc in either P-nanoplexes (open bars) or RPP-nanoplexes (closed bars). After 24 h following administration, tissues were dissected, homogenized and assayed for luciferase activity. The tissues obtained from mice treated with plasmid in P-nanoplexes show no tumor-specific expression and injection was associated with side effects. Tissues obtained from mice treated with plasmid in RPP-nanoplexes displayed appreciable levels of luciferase expression only in the tumor (n = 5). (B) Plasmid delivery combined with siRNA delivery. N2A-tumor-bearing mice were treated (i.v. injection) with pLuc plasmid as in 5A but received either 13 μg siRNA simultaneously (co-delivery) or 40 μg siRNA 2 h later (sequential delivery) in RPP-nanoplexes. In both the cases, expression of luciferase in the tumor treated with irrelevant siRNA (open bars) was similar to the expression obtained after injection of plasmid alone (A) and treatment with Luc siRNA in RPP-nanoplexes results in ∼90% gene silencing (closed bars) (n = 5).
Figure 6.
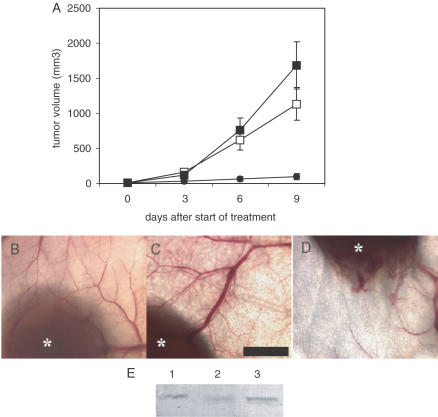
Tumor growth inhibition by VEGF R2 siRNA nanoplexes. (A) Tumor growth inhibition by siRNA RPP-nanoplexes. Mice were inoculated with N2A tumor cells and left untreated (open squares) or treated every 3 days by tail vein injection with RPP-nanoplexes with siLacZ (filled squares) or siVEGF R2 (filled circles) at a dose of 40 μg per mouse. Treatment was started at the time-point that the tumors became palpable (∼20 mm3). Only VEGF R2-sequence-specific siRNA inhibited tumor growth, whereas treatment with LacZ siRNA did not affect tumor growth rate as compared with untreated controls (n = 5). (B–D) Neovascularization in tumors treated with siRNA RPP-nanoplexes. Representative tumors excised at the end of the tumor growth inhibition experiment (A) were examined using low magnification light microscopy. Trans-illumination of tumor and surrounding skin tissue shows strong neovascularization in mice left untreated (B) and mice treated with RPP-nanoplexes with siRNA-LacZ (C). In contrast, mice treated with RPP-nanoplexes with VEGF R2 siRNA showed low neovascularization and erratic branching of blood vessels (D). Asterisks indicate tumor tissue. Bar = 2 mm. (E) VEGF R2 expression in tumor tissue after treatment with siRNA RPP-nanoplexes. Representative tumors removed at the end of the tumor growth inhibition experiment (A) were homogenized and VEGF R2-expression levels measured by western blotting. The largest tumors in the group treated with mVEGF R2 siRNA in RPP-nanoplexes and the smallest tumors in the other two groups were removed at the end of the tumor growth study 3 days following the final treatment. Lane 1 is untreated tumor, Lane 2 is VEGF R2 siRNA treated tumor and Lane 3 is LacZ siRNA treated tumor.
Further studies were performed to obtain molecular and mechanistic evidence for mVEGF R2 siRNA gene inhibition within the tumor. Tumor angiogenesis was characterized along with VEGF R2 expression levels. Reduced tumor growth rate was paralleled by reduction in blood vessels in the vicinity immediately surrounding the tumor (Figure 6B–D). Additionally, the few blood vessels visible in the tumor treated with mVEGF R2 siRNA (Figure 6D) exhibit evidence of erratic branching expected from silencing VEGF R2 expression (1,38). Expression of VEGF R2 in treated tumors was also reduced in a sequence-specific manner (Figure 6E). Taken together, these results support the interpretation that the tumor inhibition by siVEGF R2 in RPP-nanoplexes occurs as a result of effective delivery of the siRNA into tumor vasculature producing a sequence-specific inhibition of VEGF R2 expression, tumor angiogenesis and growth.
DISCUSSION
The layered, self-assembled siRNA nanoplex shows both tissue-specific delivery and siRNA gene-specific targeting with intravenous systemic administration. Thus, this system combines tissue-targeted selectivity with the gene sequence selectivity of siRNA. The RGD peptides used here to target integrin expression on neovasculature proved effective for targeting tumor neovasculature. Nonetheless, the modular chemical design allows substitution with other ligands, or combinations of ligands, to selectively target other tissues. The modular design of this layered nanoplex provides a versatile system that can be tailored to the requirements of the physiology and to the requirements of the nucleic acid. The use of synthetic siRNA oligonucleotides compacted and protected within a nanoparticle has several potential advantages over a gene therapy approach for the expression of dsRNA RNAi agents, including adaptability to alternative chemical forms of nucleic acids and avoidance of virus safety and immunogenicity problems. Nonetheless, new chemical polymer conjugates and the nanoparticles they form face their own hurdles, but offer a much wider range of materials limited only by the scope of polymer chemistry.
The results obtained here open up new perspectives for the development of targeted cancer therapeutics with two levels of selectivity using intravenously targeted siRNA within layered nanoparticles. To utilize siRNA gene silencing of endogenous genes in a therapeutic setting, however, identification of target cell types and target genes is required, in addition to an understanding of the temporal and spatial gene-expression effect required for efficacy. Exquisite pathological tissue selectivity using a single gene, single-nucleotide polymorphism or splice variant may be possible with siRNA, but identification of such unique therapeutic targets is proving difficult. This hurdle becomes much less of a requirement for targeted therapeutics when tissue selective delivery is combined with an inhibitor of a less than ideal therapeutic target. Even more importantly, however, long before such therapeutic benefits are realized, tissue-targeted siRNA permits vast improvements in characterizing gene and RNA function in animal models of human disease that can help to identify the most selective targets for targeted therapeutics, not just siRNA. For targeted cancer therapeutics, tumor targeting of siRNA of different sequences but virtually identical physical chemistry promises to enable inhibition of multiple tumor selective pathways in one therapeutic formulation.
Acknowledgments
ACKNOWLEDGEMENTS
Many discussions have been very helpful including with Alberto Gabizon, Hamid Ghandahari, Wim Henning, Dexi Liu, James Mixson, Misha Papisov and Samuel Zalipsky. This study was partly funded by the Dutch Cancer Society (Grant No. UU 2001-2185).
REFERENCES
- 1.Ferrara N., Hillan,K.J., Gerber,H.-P. and Novotny,W. (2004) Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nature Rev. Drug Discov., 3, 391–400. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir S.M., Harborth,J., Weber,K. and Tuschl,T. (2002) Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods, 26, 199–213. [DOI] [PubMed] [Google Scholar]
- 3.Paddison P.J. and Hannon,G.J. (2003) siRNAs and shRNAs: skeleton keys to the human genome. Curr. Opin. Mol. Ther., 5, 217–224. [PubMed] [Google Scholar]
- 4.Carpenter A.E. and Sabatini,D.M. (2004) Systematic genome-wide screens of gene function. Nature Rev. Genet., 5, 11–22. [DOI] [PubMed] [Google Scholar]
- 5.Ganju P. and Hall,J. (2004) Potential applications of siRNA for pain therapy. Expert Opin. Biol. Ther., 4, 531–542. [DOI] [PubMed] [Google Scholar]
- 6.Song E., Lee,S.K., Wang,J., Ince,N., Ouyang,N., Min,J., Chen,J., Shankar,P. and Lieberman,J. (2003) RNA interference targeting Fas protects mice from fulminant hepatitis. Nature Med., 9, 347–351. [DOI] [PubMed] [Google Scholar]
- 7.Davidson B.L. and Paulson,H.L. (2004) Molecular medicine for the brain: silencing of disease genes with RNA interference. Lancet Neurol., 3, 145–149. [DOI] [PubMed] [Google Scholar]
- 8.Tolentino M.J., Brucker,A.J., Fosnot,J., Ying,G.S., Wu,I.H., Malik,G., Wan,S. and Reich,S.J. (2004) Intravitreal injection of vascular endothelial growth factor small interfering RNA inhibits growth and leakage in a nonhuman primate, laser-induced model of choroidal neovascularization. Retina, 24, 132–138. [DOI] [PubMed] [Google Scholar]
- 9.Layzer J.M., McCaffrey,A.P., Tanner,A.K., Huang,Z., Kay,M.A. and Sullenger,B.A. (2004) In vivo activity of nuclease-resistant siRNAs. RNA, 10, 766–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filleur S., Courtin,A., Ait-Si-Ali,S., Guglielmi,J., Merle,C., Harel-Bellan,A., Clezardin,P. and Cabon,F. (2003) siRNA-mediated inhibition of vascular endothelial growth factor severely limits tumor resistance to antiangiogenic thrombospondin-1. Cancer Res., 63, 3919–3922. [PubMed] [Google Scholar]
- 11.Krieg A.M. (2002) CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol., 20, 709–760. [DOI] [PubMed] [Google Scholar]
- 12.Sorensen D.R., Leirdal,M. and Sioud,M. (2003) Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J. Mol. Biol., 327, 761–766. [DOI] [PubMed] [Google Scholar]
- 13.Takei Y., Kadomatsu,K., Yuzawa,Y., Matsuo,S. and Muramatsu,T.A (2004) Small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer Res., 64, 3365–3370. [DOI] [PubMed] [Google Scholar]
- 14.Woodle M.C., Scaria,P., Ganesh,S., Subramanian,K., Titmas,R., Cheng,C., Yang,J., Pan,Y., Weng,K., Gu,C. and Torkelson,S. (2001) Sterically stabilized polyplex: ligand-mediated activity. J. Control Release, 74, 309–311. [DOI] [PubMed] [Google Scholar]
- 15.Langer R. (2001) Drug delivery: drugs on target. Science, 293, 58–59. [DOI] [PubMed] [Google Scholar]
- 16.Suh W., Han,S.-O.,Yu,L. and Kim,S.W. (2002) An angiogenic, endothelial-cell-targeted polymeric gene carrier. Mol. Ther., 6, 664–672. [PubMed] [Google Scholar]
- 17.Hood J.D., Bednarski,M., Frausto,R., Guccione,S., Reisfeld,R.A., Xiang,R. and Cheresh,D.A. (2002) Tumor regression by targeted gene delivery to the neovasculature. Science, 296, 2404–2407. [DOI] [PubMed] [Google Scholar]
- 18.Yamada T., Iwasaki,Y., Tada,H., Iwabuki,H., Chuah,M.K., VandenDriessche,T., Fukuda,H., Kondo,A., Ueda,M., Seno,M., Tanizawa,K. and Kuroda,S. (2003) Nanoparticles for the delivery of genes and drugs to human hepatocytes. Nat. Biotechnol., 21, 885–890. [DOI] [PubMed] [Google Scholar]
- 19.Verbaan F.J., Oussoren,C., van Dam,I.M., Takakura,Y., Hashida,M., Crommelin,D.J., Hennink,W.E. and Storm,G. (2004) Steric stabilization of poly(2-(dimethylamino)ethyl methacrylate)-based polyplexes mediates prolonged circulation and tumor targeting in mice. J. Gene Med., 6, 64–75. [DOI] [PubMed] [Google Scholar]
- 20.Ogris M., Walker,G., Blessing,T., Kircheis,R., Wolschek,M. and Wagner,E. (2003) Tumor-targeted gene therapy: strategies for the preparation of ligand-polyethylene glycol–polyethylenimine/DNA complexes. J. Control Release, 91, 173–181. [DOI] [PubMed] [Google Scholar]
- 21.Wagner E., Kircheis,R. and Walker,G.F. (2004) Targeted nucleic acid delivery into tumors: new avenues for cancer therapy. Biomed. Pharmacother., 58, 152–161. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Zhang,Y.-F., Bryant,J., Charles,A., Boado,R.J. and Pardridge,W.M. (2004) Intravenous RNA interference gene therapy targeting the human epidermal growth factor receptor prolongs survival in intracranial brain cancer. Clin. Cancer Res., 10, 3667–3677. [DOI] [PubMed] [Google Scholar]
- 23.Hart S.L., Knight,A.M., Harbottle,R.P., Mistry,A., Hunger,H.D., Cutler,D.F., Williamson,R. and Coutelle,C. (1994) Cell binding and internalization by filamentous phage displaying a cyclic Arg-Gly-Asp-containing peptide. J. Biol. Chem., 269, 12468–12474. [PubMed] [Google Scholar]
- 24.Pasqualini R. and Ruoslahti,E. (1996) Organ targeting in vivo using phage display peptide libraries. Nature, 380, 364–366. [DOI] [PubMed] [Google Scholar]
- 25.Arap W., Pasqualini,R. and Ruoslahti,E. (1998) Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science, 279, 377–380. [DOI] [PubMed] [Google Scholar]
- 26.Janssen M.L., Oyen,W.J., Dijkgraaf,I., Massuger,L.F., Frielink,C., Edwards,D.S., Rajopadhye,M., Boonstra,H., Corstens,F.H. and Boerman,O.C. (2002) Tumor targeting with radiolabeled αvβ3 integrin binding peptides in a nude mouse model. Cancer Res., 62, 6146–6151. [PubMed] [Google Scholar]
- 27.Zitzmann S., Ehemann,V. and Schwab,M. (2002) Arginine-glycine-aspartic acid (RGD)-peptide binds to both tumor and tumor-endothelial cells in vivo. Cancer Res. 62, 5139–5143. [PubMed] [Google Scholar]
- 28.Kim B., Tang,Q., Biswas,P.S., Xu,J., Schiffelers,R.M., Xie,F.Y., Ansari,A.M., Scaria,P.V., Woodle,M.C., Lu,P. and Rouse,B.T. Am. J. Pathol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Neil K.T., Hoess,R.H., Jackson,S.A., Ramachandran,N.S., Mousa,S.A. and DeGrado,W.F. (1992) Identification of novel peptide antagonists for GPIIb/IIIa from a conformationally constrained phage peptide library. Proteins, 14, 509–515. [DOI] [PubMed] [Google Scholar]
- 30.Erbacher P., Bettinger,T., Belguise-Valladier,P., Zou,S., Coll,J.L., Behr,J.P. and Remy,J.S. (1999) Transfection and physical properties of various saccharide, poly(ethylene glycol), and antibody-derivatized polyethylenimines (PEI). J. Gene Med., 1, 210–222. [DOI] [PubMed] [Google Scholar]
- 31.Ogris M., Steinlein,P., Carotta,S., Brunner,S. and Wagner,E. (2001) DNA/polyethylenimine transfection particles: influence of ligands, polymer size, and PEGylation on internalization and gene expression. AAPS PharmSci., 3, E21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu X.M., Chen,Y., Chen,J., Yang,S., Gao,F., Underhill,C.B., Creswell,K. and Zhang,L. (2003) A peptide with three hyaluronan binding motifs inhibits tumor growth and induces apoptosis. Cancer Res., 63, 5685–5690. [PubMed] [Google Scholar]
- 33.Ruoslahti E. (1996) RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol., 12, 697–715. [DOI] [PubMed] [Google Scholar]
- 34.Harbottle R.P., Cooper,R.G., Hart,S.L., Ladhoff,A., McKay,T., Knight,A.M., Wagner,E., Miller,A.D. and Coutelle,C. (1998) An RGD-oligolysine peptide: a prototype construct for integrin-mediated gene delivery. Hum. Gene Ther., 9,1037–1047. [DOI] [PubMed] [Google Scholar]
- 35.Kuwashima N. (1997) Organ-specific adhesion of neuroblastoma cells in vitro: correlation with their hepatic metastasis potential. J. Pediatr. Surg., 32, 546–551. [DOI] [PubMed] [Google Scholar]
- 36.Lemkine G.F. and Demeneix,B.A. (2001) Polyethylenimines for in vivo gene delivery. Curr. Opin. Mol. Ther., 3, 178–182. [PubMed] [Google Scholar]
- 37.Greish K., Fang,J., Inutsuka,T., Nagamitsu,A. and Maeda,H. (2003) Macromolecular therapeutics: advantages and prospects with special emphasis on solid tumour targeting. Clin. Pharmacokinet., 42, 1089–1105. [DOI] [PubMed] [Google Scholar]
- 38.Brekken R.A., Overholser,J.P., Stastny,V.A., Waltenberger,J., Minna,J.D. and Thorpe,P.E. (2000) Selective inhibition of vascular endothelial growth factor (VEGF) receptor 2 (KDR/Flk-1) activity by a monoclonal anti-VEGF antibody blocks tumor growth in mice. Cancer Res., 60, 5117–5124. [PubMed] [Google Scholar]