Abstract
Comparative genomic hybridization by means of BAC microarrays (array CGH) allows high-resolution profiling of copy-number aberrations in tumor DNA. However, specific genetic lesions associated with small but clinically relevant tumor areas may pass undetected due to intra-tumor heterogeneity and/or the presence of contaminating normal cells. Here, we show that the combination of laser capture microdissection, φ29 DNA polymerase-mediated isothermal genomic DNA amplification, and array CGH allows genomic profiling of very limited numbers of cells. Moreover, by means of simple statistical models, we were able to bypass the exclusion of amplification distortions and variability prone areas, and to detect tumor-specific chromosomal gains and losses. We applied this new combined experimental and analytical approach to the genomic profiling of colorectal adenomatous polyps and demonstrated our ability to accurately detect single copy gains and losses affecting either whole chromosomes or small genomic regions from as little as 2 ng of DNA or 1000 microdissected cells.
INTRODUCTION
Chromosomal instability (CIN) plays a central role in the progression and malignant transformation of solid tumors (1–3). However, evaluation of genome-wide CIN is often hampered by technical and sample limitations. Tumor cytogenetics is often not feasible even from freshly isolated tumors and requires short-term culturing of parenchymal cells. Moreover, different areas within a tumor may show heterogeneous CIN patterns associated with different malignant potentials. Also, tumor specimens are invariably contaminated with varying amounts of surrounding and/or infiltrating normal cells interfering with the analysis. Thus, a method is ideally required that enables the detection of chromosomal aberrations in small numbers of microdissected tumor cells (4,5). Such a method should also be sufficiently sensitive to detect single copy gains and losses, the most frequent genomic alterations in most tumor types (6).
Comparative genomic hybridization (CGH) by means of microarrays containing large-insert genomic clones such as bacterial artificial chromosomes (BACs) provides a sensitive and quantitative approach to assess DNA copy-number aberrations in tissue samples (6–8). Classical array CGH requires hundreds of nanograms of genomic DNA for fluorescent labeling, a prohibitive amount when working with microdissected samples. Several methods to amplify total genomic DNA have been applied to array CGH, most of them based on thermocycling protocols, such as degenerate oligonucleotide-primed (DOP) PCR (9) and ligation-mediated (LM) PCR (10). These methods produce relatively low molecular weight DNA that may not be representative of the entire genome (11). In contrast, multiple strand displacement amplification (MDA) generates thousands of high-molecular weight copies of genomic DNA in a robust simple protocol without the use of thermocycling or ligation of DNA adaptors (12,13). This amplification mechanism favors equal representation of sequences because each priming event is propagated over very long distances in the genome. Array CGH with MDA amplified DNA performs comparable to unamplified array CGH (14), and allows reliable detection of high-level copy-number changes (gene-dosage alterations of 3-fold or more) on cDNA microarrays (15) and synthetic oligonucleotide arrays (16). We aimed at significantly improving this method to allow reliable detection of low-range alterations and apply it to microdissected neoplastic lesions.
We developed an experimental approach combining laser capture microdissection (LCM), isothermal genomic DNA amplification and array CGH on BAC microarrays, which have a better signal-to-noise ratio compared to cDNA microarrays. To validate the method, we have generated a series of amplified CGH profiles from normal female and male DNA, and from cell lines with known gains on chromosome X or 20. Using two simple statistical methods, we were able to avoid the exclusion of genomic regions affected by amplification distortions and high variability, and to detect single copy changes varying in length from a few clones to a whole chromosome. Finally, we applied the combined experimental and statistical approach to the analysis of microdissected dysplastic cells from colorectal adenomatous polyps derived from patients affected by familial adenomatous polyposis (FAP), an autosomal dominant genetic predisposition to the development of hundreds to thousands benign adenomas in the distal gastro-intestinal tract, caused by germline mutations in the APC gene on chromosome 5q (17). We demonstrate the sensitivity of our LCM-CGH array approach by the detection of a small 5q deletion subsequently validated by PCR-based loss-of-heterozygosity (LOH) analysis. These results underline the usefulness of our approach in the study of chromosomal imbalances in small subpopulations of microdissected tumor cells or in other cases where limited amounts of DNA are available.
MATERIALS AND METHODS
Genomic clone library and generation of array probes
The human 3600 BAC/PAC genomic clone set, covering the full genome at 1 Mb-spacing used for the production of our arrays was obtained from the Welcome Trust Sanger Institute (http://www.sanger.ac.uk/). Information on this clone set can be obtained at the BAC/PAC Resources Center Web Site (http://bacpac.chori.org). Degenerated oligonucleotide PCR-products were prepared for spotting on CodeLink® slides (Amersham Biosciences) according to detailed protocols (5) with some modifications (18).
DNA samples for array CGH
The data set comprised the measurements from 17 independent array CGH experiments. Normal male and female DNA samples used as controls and as reference in the hybridization were obtained from a commercial source and consisted of pools of individuals (Promega). The genomic DNA from 5cl20 sample was selected for testing, since it contained a known trisomy over five clones on chromosome 20 (position 12639830 to 16682277 Mb at 20p12.1). The lymphoblastoid cell line used contains a known X trisomy. Normal epithelial (D5_14) and dysplastic adenoma (D5_1) cells were microdissected from human colon tissue specimens from a male individual affected by FAP. Three normal mucosa samples were microdissected from healthy control individuals, one male (NC1) and two females (NC2 and NC3). The frozen colon specimens used for microdissection were collected in Tissue-Tec® (Sakura) embedding medium and snap-frozen in 2-methylbutane (Sigma) and dry ice. Tissue sections (10 μm) were sliced with a Shandon Cryotome® Cryostat (Shandon), and directly transferred to a UV-cross-linked PEN membrane (P.A.L.M. Microlaser Technologies) mounted on a PALM® Membrane Slide (1 mm). Slides with mounted sections were immersed immediately and stored up to one week in 70% ethanol at 4°C.
Microdissection and DNA extraction
Following the staining of the tissue sections with Mayer's Hematoxilin and Eosin Y solutions, 1000 parenchymal cells (600 000 μm2) were microdissected and laser pressure catapulted using the PALM® MicroBeam microscope system (P.A.L.M. Microlaser Technologies). The genomic DNA was extracted using the protocol from Isola et al. (19), adapted to small DNA amounts. Briefly, the adaptations included the resuspension of the microdissected cells in 100 μl of DNA extraction buffer, an overnight digestion with 0.6 mg/ml of proteinase K and a replacement of glycogen by 10 μg of GenElute™ linear polyacrylamide (Sigma) in the precipitation step. All samples were resuspended in 10 μl of TE buffer (10 mM Tris–HCl pH 7.4, 0.1 mM EDTA). Tests performed with DNA amounts that could be assessed on conventional agarose gels indicated that the average size of genomic DNA purified using this procedure was >20 kb.
GenomiPhi amplification and purification
A volume of 1 μl out of 2 ng/μl dilutions from cell lines and control DNA samples and from all microdissected samples (concentrated by speedvac from 8 μl) was used as starting material for the amplification. The φ29-amplification was carried out according to the GenomiPhi kit manufacturer's instructions (Amersham Biosciences), using an incubation time of 16 h. GenomiPhi reactions were checked on a 0.6% agarose gel. An amplification was considered successful when a smear of DNA fragments, ranging from 1 to 20 kb, was visible. Samples were purified using Microcon® YM-100 spin columns (Millipore) according to the manufacturer's protocol and concentrated to 25 μl in water.
TH01 diagnostic PCR
In order to roughly access the amount of DNA isolated from microdissected samples, we have used a PCR, specific for the TH01 human marker (20). Each 2 μl of microdissected sample was compared to 1 μl of serial dilutions of female and male control DNAs (10, 5, 2, 1, 0.5, 0.1, 0.01, 0.001 ng/μl). The 25 μl reactions contained 1X GeneAmp® PCR Buffer (Applied Biosystems), 1.5 mM MgCl2, 0.2 mM dNTPs Mix, 500 nM oligonucleotides TH01-1 (forward: 5′-GTGGGCTGAAAAGCTCCCGATTAT-3′) and TH01-2 (reverse: 5′-ATTCAAAGGGTATCTGGGCTCTGG-3′) and 1 unit of AmpliTaq® DNA Polymerase (Applied Biosystems). The cycling conditions were as follows: 1 min at 94°C; 35 cycles of 20 s at 94°C, 20 s at 55°C and 20 s at 72°C; 10 min at 72°C and then held at 25°C. TH01 amplification products (±200 bp) were checked on a 2% agarose gel or on a Bioanalyzer 2100 (Agilent) using a DNA 1000 LabChip kit, according to manufacturer's instructions. Due to a frequently observed background synthesis in the GenomiPhi amplified water control, the above-described PCR methodology was routinely used as an extra quality control on 1 μl (1/50 dilution) of unpurified φ29-amplified DNA. The TH01 specific band should be present in all GenomiPhi amplified samples and absent from the GenomiPhi water control.
Labeling and hybridization
By default, Cy3 and Cy5 fluorescent channels were used to label test and reference DNA, respectively, except when otherwise specified. The labeling and hybridization protocols described by Fiegler et al. (5) were used, with some modifications in the labeling. Briefly, 130.5 μl of solution containing 5 μl of φ29-amplified or 450 ng of unamplified DNA, 60 μl of BioPrime® DNA Labeling System random primers solution (Invitrogen) and water was incubated for 10 min at 100°C, and subsequently cooled on ice. After the addition of 15 μl of 10× dNTPs labeling mix (1 mM dCTP, 2 mM dATP, 2 mM dGTP and 2 mM dTTP), 1.5 μl of 1 mM Cy3- or Cy5-dCTP (Amersham Biosciences) and 120 U of BioPrime® DNA Labeling System Klenow fragment (Invitrogen), the mixture was gently mixed and subsequently incubated overnight at 37°C. The addition of 15 μl of BioPrime® DNA Labeling System Stop Buffer (Invitrogen) ended the reaction. In one tube, Cy3-labeled sample and Cy5-labeled reference DNAs were mixed together, and 135 μg of human Cot-1 (Roche), 55 μl of 3 M sodium acetate (pH 5.2) and 1 ml of cold 100% ethanol were added. In a second tube, 80 μg of denatured herring sperm DNA (Sigma) was mixed with 135 μg of human Cot-1 (Roche), 23 μl of 3 M sodium acetate (pH 5.2) and 400 μl of cold 100% ethanol. After gentle mixing of the two labeling mixes, the labeled nucleic acids were precipitated overnight at −20°C. Hybridizations were performed as described in (5). Sixteen-bit fluorescent images were acquired with a DNA Microarray Scanner (Agilent) and the resulting TIFF images were analyzed with the GenePix Pro 4.0 software (Axon Instruments). Per array, a GenePix results file (.gpr) with the extracted Cy3 and Cy5 spot and background raw intensities was generated.
Computational and statistical analysis
Data analysis was performed with a set of functions implemented in R (21) (http://www.r-project.org/). Briefly, gpr files were directly loaded into the R environment using the marrayTools package. Intensity data were normalized with the local robust regression function Lowess contained in the package marrayNorm. The resulting log2 Cy3/Cy5 transformed ratios from each set of triplicate spots were subsequently averaged to produce a unique ratio for each BAC clone. After this step, spots designated as empty, blank and Cy-dye controls, and spots with uncertain chromosome locations (clone id >35650 in the data set) were removed. The final data set, used in subsequent analysis, consisted of 3615 independent observations per sample. For all experiments, data was organized per chromosome and the clones were ordered from chromosome 1 to chromosome Y according to their Golden Path Mb position (http://genome.ucsc.edu/). Although data from all chromosomes was analyzed, for illustration purposes of the methods, the data from chromosomes 5, 20, X and Y only are shown. The data were analyzed separately per chromosome, given that chromosomes are not directly related and some display more variability than others. Histograms of the normalized log2 clone-specific associated residuals were also produced and they indicated that the error measurements approximately follow a normal distribution.
Linear regression model
The explanatory factors of interest are Fx and Fy, which are qualitative variables with levels (k) defined by the columns Fx and Fy level (Table 1). For any given chromosome with n clones, the model used for the copy-number ratios, observed for the level k of the factor under study and clone j, Yjk was
Table 1. Experimental data sets.
| Array | Sample (Cy3) | Known chromosome rearrangements | Refa (Cy5) | X chromosome | Y chromosome | ||
|---|---|---|---|---|---|---|---|
| Expected ratios | Fx level | Expected ratios | Fy level | ||||
| Amplified pilot subset | |||||||
| CGH3_1 | XXa | — | XY | 2:1 | 1 | 0:1 | −1 |
| CGH3_3 | XXa | — | XY | 2:1 | 1 | 0:1 | −1 |
| CGH4_74 | XXX | X | XX | 3:2 | +0.5 | 0:0 | 0 |
| CGH4_75 | XXX+XXb | X | XX | 2.5:2 | +0.25 | 0:0 | 0 |
| CGH4_73 | 5cl20, XY | gain 5 cl 20p12.1 | XY | 1:1 | 0 | 1:1 | 0 |
| CGH3_2 | XYa | — | XY | 1:1 | 0 | 1:1 | 0 |
| CGH3_4 | XYa | — | XY | 1:1 | 0 | 1:1 | 0 |
| CGH5_26 | XY+XXb | — | XX | 1.5:2 | −0.25 | 0.5:0 | +0.5 |
| CGH5_27 | XYa | — | XX | 1:2 | −0.5 | 1:0 | +1 |
| Amplified microdissected samples subset | |||||||
| CGH4_59 | NC1, XY | — | XY | 1:1 | 3 | 1:1 | 0 |
| CGH4_60 | NC2, XX | — | XY | 2:1 | 4 | 0:1 | −1 |
| CGH4_61 | NC3, XX | — | XY | 2:1 | 5 | 0:1 | −1 |
| CGH3_23 | D5_1, XY | ? | XY | 1:1 | 6 | 1:1 | 0 |
| CGH3_40 | D5_14, XY | — | XY | 1:1 | 7 | 1:1 | 0 |
aCommercial DNA pools.
bMixture of equal amounts.
![]()
where αk and βj represent the factor-specific and clone-specific effect respectively, μ is the intercept (level 0, no change) based on the two normal male versus male comparisons (CGH3_2 and 4), and εjk is the error with mean zero and constant variance. The term βj in the model takes care of individual clones varying technical performances. The model in Equation 1 was first fitted to X chromosome clones from the pilot and microdissected subsets (Table 1) and subsequently, the same approach was used for the remaining chromosomes. Finally, we extracted the P-values for the factor under study from the ANOVA table for the fitting of the model in Equation 1. These P-values were not used as probabilities but merely to compare model fits for different chromosomes. Factor effects plots were drawn with the qvcalc R package. Comparable error bars were computed using, instead of the computed variances, quasi-variances (22), which consists of variances adjusted for the covariance structure.
Non-parametric Smith–Waterman dynamic algorithm
To detect gains and deletions over a few neighboring clones and simultaneously test their statistical significance against a reference baseline, we used the Smith–Waterman analytical algorithm (23), implemented in R (24) (http://www.well.ox.ac.uk/~tprice/). The algorithm was first applied to chromosome 20 data from sample 5cl20, to compare its clone ratios to each clone ratio median from the remaining eight samples, using a δ threshold of 3.6. The same algorithm was applied to chromosome 20 and to the other chromosomes data from the remaining samples from the pilot and microdissected data (Table 1).
Loss-of-heterozygosity analysis of 5q dinucleotide repeats
LOH in 5q21.1–5q31.1 was analyzed by amplification of the dinucleotide repeat markers D5S400 (distal), D5S409 (LOH region) and D5S427 (proximal). The localization and the primer sequence of the markers were obtained from the ENSEMBL project (http://www.ensembl.org). Allele sizes were obtained from The Genome Database of the Human Genome Project (http://www.gdb.org). A radioactive PCR was performed using 1 μl of a 1:10 dilution of φ29-amplified tumor and normal mucosa DNA from patient D5. An aliquot of 100 ng of unamplified DNA isolated from ethanol-fixed normal intestinal mucosa from the same patient was used as additional control. Amplifications were carried out in a 10 μl volume, containing 10 mM Tris–HCl, pH 8.9, 50 mM KCl, 2.5 MgCl2, 10% glycerol, 200 μg/ml BSA, 0.01% gelatin, 0.2 mM of each dATP, dGTP and dTTP, 0.05 mM dCTP, 0.25 μCi of dCTP (3000 Ci/mmol), 0.2 U Taq polymerase and 10 pmol of each primer (25). The cycling conditions were as follows: 1 cycle at 94°C for 4 min, followed by 35 cycles at 94°C for 1 min, 55°C for 1 min, 72°C for 1 min and final cycle at 72°C for 6 min. The amplified fragments were resolved in a denaturing 6% polyacrylamide gel and dried on paper, and subsequently scanned on a Typhoon 9200 imager (Amersham Biosciences). The generated TIFF images were subsequently analyzed with ImageQuant v5.2® software (Amersham Biosciences). The allelic imbalance was calculated as described elsewhere (26). LOH is interpreted as significant when the calculated comparative allelic ratio values are >1.5 (loss of the smaller allele) or <0.6 (loss of the larger allele).
Genomic microarray data
The genomic microarray data discussed in this publication have been deposited in NCBIs Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE 1841.
RESULTS
Comparative genomic hybridization arrays: quality controls
For this study, we have generated CGH microarrays covering the full human genome by employing the human 3600 BAC/PAC genomic clone set from the Welcome Trust Sanger Institute. Individual BAC clones were amplified by degenerated oligonucleotides and the PCR-products spotted on glass slides (see Materials and Methods). The specificity and sensitivity of our genome-wide 1 Mb-spaced CGH microarray was tested with a series of control hybridizations using unamplified DNA samples (Figure 1A and B). Assuming that a chromosomal region is classified as amplified or deleted when clone ratios fall outside fixed thresholds (+0.26 and −0.26 in log2 scale) for copy-number gain or loss respectively (27–30); for both arrays, >99% of the autosomal BAC clone ratios are contained within these limits (0 ± 0.07). Similar results are obtained for the log2 ratios of clones on the X chromosome (0 ± 0.11) and Y chromosome (0 ± 0.09) in the male versus male comparison (Figure 1A), while the female versus male comparison resulted in log2 ratios of 0.63 ± 0.22 for X-derived and −1.66 ± 1.05 for Y-derived BAC clones (Figure 1B). These results are concordant with the overall results expected for CGH profiling using unamplified control samples (6,8), certifying the quality of our arrays. The log2 ratios observed for X and Y in the female versus male hybridization are smaller than expected for the gene copy differences. This ‘dynamic range compression’ is commonly observed with array CGH, both for unamplified and amplified samples (6,8).
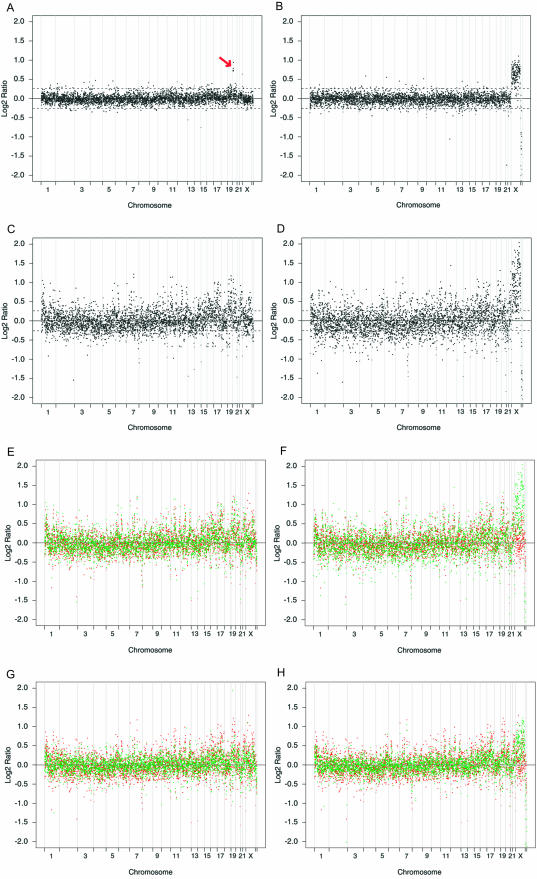
Figure 1.
Reproducibility of φ29-amplified array CGH. (A–D) Comparison of classical array CGH with φ29-amplified array CGH. Normalized copy-number ratios of genomic DNA samples from 5cl20 male, unamplified (A) and φ29-amplified (C), and a control female DNA pool, unamplified (B) and φ29-amplified (D). All samples were compared with an unamplified male reference DNA pool. Data are plotted as the mean log2 ratio of the triplicate spots for each clone after Lowess normalization. The BACs are ordered by position in the genome beginning at 1p and ending at Yq. Borders between chromosomes are indicated by vertical bars, the horizontal solid line indicates a log2 ratio of 0 (no change), and the horizontal dashed lines indicate the +/− 0.26 confidence limits for copy-number gain or loss, respectively. The trisomic region in sample 5cl20 on chromosome 5 is indicated by an arrow in (A). (E–H), co-plotting of sample versus reference array CGH ratios (green) with an ‘average’ amplified normal CGH profile (red; see text), shows the non-random behavior of the φ29-induced effect on the copy-number ratios. Amplified array CGH patterns from the diluted DNA samples 5cl20 male (E) and female control (F) are comparable to those of two normal mucosa DNA samples isolated after microdissection, NC1 male (G) and NC2 female (H).
Linear amplification of genomic DNA
Classical array CGH requires genomic DNA in amounts well above those obtained by LCM tissues. To overcome the problem of limited amounts of starting material, the φ29 polymerase was used to linearly amplify high-molecular weight genomic DNA and LCM-derived templates. It was expected that φ29 polymerase-mediated reactions using small amounts of template would reach high amplification yields (12,15). However, in addition to high amplification yields, φ29-mediated reactions also resulted in the generation of aspecific products both in the presence of non-human DNA (λ genomic DNA) and even when no DNA template was employed (Supplementary Figure A). Apparently, φ29 is capable of primer-directed DNA synthesis in the absence of template via a yet unknown mechanism (15,31). To control for human-specific DNA synthesis, we employed conventional PCR on the φ29-products to amplify the single-copy human marker TH01 (20). As expected, the 200 bp TH01-specific product was observed exclusively when human DNA templates were employed (Supplementary Figure B). Due to the fact that the aspecific amplification contributes to the amount of amplified DNA, conventional DNA quantification methods such as spectrophotometry could not be used. Therefore, we also used the TH01 PCR to semi-quantify the amount of φ29-amplified DNA to be used as input for the labeling reaction. Finally, the TH01 PCR assisted us in the estimation of LCM-derived genomic DNA amounts used as input for the amplification reaction (Supplementary Figure C). The comparison of TH01 amplification products generated from serial dilutions of genomic DNA controls with TH01 amplification products generated using genomic DNA obtained from ∼1000 microdissected colon epithelial cells showed that the microdissection procedure on a tissue section corresponding to an area of approximately 600 000 μm2 yielded 5–10 ng of genomic DNA. The φ29 reaction yielded approximately 1250–6250 ng of amplified DNA from 5 to 10 ng of starting DNA.
Reproducibility of amplification
To test the reproducibility of the φ29 amplification reaction, we co-hybridized φ29-amplified female and male control DNA samples against an unamplified male reference. Apart from the expected overall increase in background noise, a marked and reproducible ratio distortion is observed at specific chromosomal regions (Figure 1C and D). Major affected regions include 1p, and several telomeres (e.g. 7q and 8p) and centromeres, known to be prone to ratio distortions due to the repetitive nature of their sequences (32). The reproducible change in ratio structure becomes evident when replicate normal versus normal hybridizations are compared. The Pearson correlation between the autosomal log2 ratios obtained in two amplified normal female versus male experiments (Table 1, #1, 3) was high (0.90). In addition, we compared the CGH profiles of each individual sample to an ‘average’ of amplified normal CGH profiles (Figure 1E and F; for boxplots of individual BAC clones per chromosome see Supplementary Material). The average normal profile is calculated for autosomal clones as the mean intensity values obtained from pilot subset samples (Table 1) with the exclusion of 5cl20; for X- and Y-specific chromosomal regions, the mean intensity values were calculated from three male samples included in the pilot subset (Table 1, #2, 4, 27). The reproducible wave-like pattern of the ratio distortions is likely to result from variability in the amplification process when repetitive and polymorphic genomic regions are differentially processed by the φ29 polymerase (15). Accordingly, no significant ratio distortion was observed when two normal genomic DNA samples were separately microdissected from the same sections, φ29 amplified and co-hybridized on the same array (Supplementary Figure D). The estimated mean log2 ratios were comparable to those previously observed with unamplified control DNA samples. Furthermore, we compared the ratio variability of φ29-amplified samples co-hybridized with an unamplified or an amplified reference DNA and found no apparent difference (Supplementary Figure E). By including clone-specific characteristics such as median log2 ratio and variability in our downstream statistical analyses, we were able to analyze the amplified array CGH profiles without excluding variability-prone clones or regions.
In order to evaluate the quality of φ29-amplified genomic DNA from microdissected cells, we compared the CGH profiles of φ29-amplified diluted normal genomic DNA samples (Figure 1E and F) with the profiles from φ29-amplified DNA obtained from microdissected normal epithelial cells (Figure 1G and H). Since these DNA profiles were comparable, the quality of the LCM-derived amplified DNA apparently was not compromised by the microdissection procedure. Therefore, we conclude that genomic DNA samples derived from laser capture microdissection and amplified by φ29 are suitable for array CGH profiling.
Statistical analysis of copy-number changes
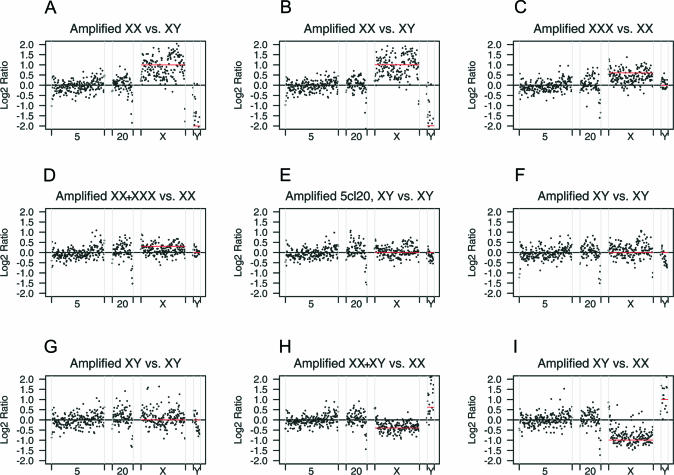
To determine the sensitivity of our amplified array CGH approach in detecting low levels of chromosomal gains and losses, we generated a set of array CGH profiles with DNA samples from cell lines ranging in the number of X and Y chromosomes (Table 1). Since in our experience, ratio distortions introduced by φ29-mediated amplification are systematic, we set to implement an applied and validated statistical model to deal with the variation and signify copy-number changes. Employing chromosome X as a model, we assessed the sensitivity of the CGH array technology in accurately quantifying chromosome-wide changes on φ29-amplified DNA. Using our pilot data set encompassing different chromosome X copy numbers (Table 1), we modeled six different gain or loss scenarios: 2:1, 3:2, 1:1, 2.5:2 (gain of one copy in 50% of the cells), 1.5:2 (loss of one copy in 50% of the cells), and 1:2. The observed log2 ratios for the X and Y chromosomes correlated with the expected chromosome dosage increments for the known karyotypes, while the log2 ratio for autosomal clones was approximately 0 (Figure 2). The graphs in Figure 2 also show that the variability of the measurements increased proportionally to the chromosome X and Y effect size.
Figure 2.
Chromosome X and Y dosage effects in amplified array CGH. (A–I) Normalized array CGH ratios from nine φ29-amplified control hybridizations with varying dosages of X and Y chromosomes are plotted for four illustrative chromosomes (5, 20, X and Y). The genotypes for each sample and reference are given above the plot, mixed DNA samples (1:1) are indicated by XX + XXX, etc. The horizontal red lines represent the expected X and Y chromosome ratios. An example of reproducible under-representation of genomic sequences after amplification is visible for 20q telomeric BACs.
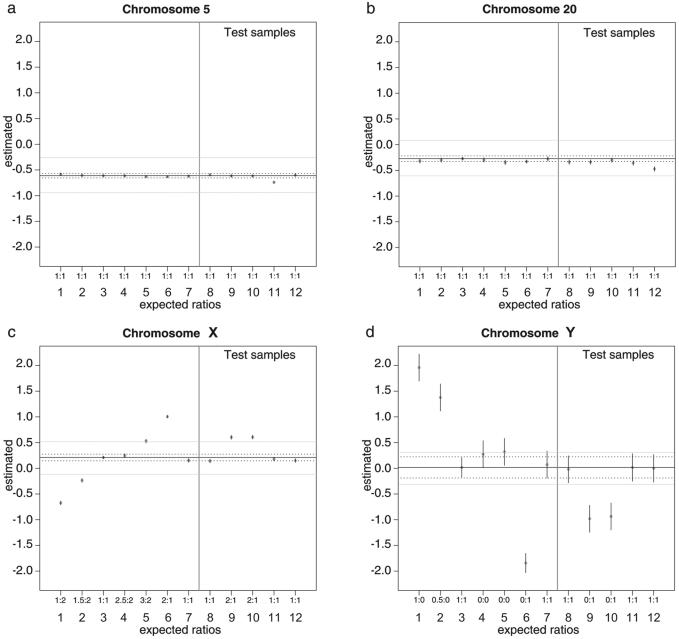
For the detection of gains and losses of whole chromosomes, a linear regression model (see Materials and Methods) was fitted separately per chromosome to all pilot subset samples, using as explanatory factor the class of expected ratios for chromosome X (Fx). As expected, only when data from either chromosomes X or Y was used, the tested variable Fx showed a significant estimated difference compared to the baseline level 0, indicating copy-number changes (Figure 3). Estimated X chromosome ratios of 2:1, 3:2, 1.5:2 and 1:2 were highly significant, as well as the more extreme Y chromosome ratios of 0:1, 0.5:0 and 1:0 (significant values highlighted in bold in Table 2). Similar to classical array CGH, the estimated ratios were compressed compared to the expected ratios: an X chromosomal gene dosage ratio of 2:1, with an expected log2 ratio of 1 was measured as 0.8 difference to the intercept, 3:2 (log2 ratio 0.59) was measured as a difference to the intercept of 0.35, 2.5:2 (log2 ratio 0.32) was measured as 0.06, 1.5:2 (log2 ratio −0.41) as −0.42 and 1:2 (log2 ratio −1) as −0.85. Note, however, that this compression is linear (Figure 3C) and, based on its estimate from the pilot subset samples, it can be corrected. All Fx levels showed a significant estimated effect difference from the intercept (level 0) except for the estimated log2 ratio for 2.5:2 X chromosomes (Fx level 0.25, Table 1). For this chromosome dosage level, the difference to the intercept (0.06) was neither statistically nor biologically meaningful, indicating that the thresholds for the detection of copy-number changes in φ29-amplified genomic DNA are a gain of one copy in all cells, and a loss of one copy in 50% of the cells. Based on this pilot data set, we implemented a threshold for biological relevance in addition to statistical significance (P < 0.00001), requiring a log2 ratio compared to the intercept larger than +0.35 or smaller than −0.35, respectively for gain or loss.
Figure 3.
Whole chromosome copy-number changes estimated by linear regression. (A) Chromosome 5; (B) chromosome 20; (C) chromosome X; (D) chromosome Y. Estimated average copy-number log2 ratios from amplified array CGH hybridizations of control DNA samples 1–7 (left of vertical line in each plot) and microdissected DNA samples 8–12 (right of vertical line in each plot) for illustrative chromosomes 5 (209 BACs), 20 (79 BACs), X (165 BACs) and Y (28 BACs). On the x-axis, the relative amounts of these chromosomes in the test and reference samples are indicated. 1–12 correspond to 1, XY:XX (CGH5_27); 2, XY + XX:XX (CGH5_26); 3, XY:XY (CGH3_2) and XY:XY (CGH3_4) = intercept; 4, XXX + XX:XX (CGH4_75); 5, XXX:XX (CGH4_74); 6, XX:XY (CGH3_1) and XX:XY (CGH3_3); 7, 5cl20 XY:XY (CGH4_73); 8, NC1:XY (CGH4_59); 9, NC2:XY (CGH4_60); 10, NC3:XY (CGH4_61); 11, D5_1:XY (CGH3_23); 12, D5_14:XY (CGH3_40). On the y-axis are the log2 ratios estimated by the linear regression model for each Fx level compared to the intercept (level 0, no change; horizontal solid line; note that the intercept may deviate from log2 ratio 0). The dashed horizontal lines mark the 95% confidence intervals, defined as the level 0 log2 mean estimated copy-number ratio ±2 standard errors. The gray horizontal lines indicate the thresholds for a biologically meaningful copy-number change (intercept ± 0.35). Quasi-error bars (small vertical lines) associated with the estimated effects are only clearly visible for the Y chromosome, where the higher variance is due to the lower number of BACs in the array. Given that the relationship between the estimated and the expected chromosome X values is linear (C), the fitting of the linear regression model (Equation 1) on chromosome X data yielded the function E = −0.243 + 1.23R (R2 = 0.98), which describes the relationship between R, the log2 ratio change estimates and E, their corresponding expected log2 ratio values.
Table 2. Regression-fitted estimated log2 ratios for chr 5, 20, X and Y.
| Expected ratios | Arrays per level | Chromosome 5 | Chromosome 20 | Chromosome X | Chromosome Y | ||||
|---|---|---|---|---|---|---|---|---|---|
| Est. | P | Est. | P | Est. | P | Est. | P | ||
| Amplified pilot subset | |||||||||
| XY:XY | CGH3_2 CGH3_4 | 0 | — | 0 | — | 0 | — | 0 | — |
| XX:XY | CGH3_1 CGH3_3 | −0.02 | 0.09 | −0.06 | 0 | 0.80 | <2e−16 | −1.88 | <2e−16 |
| XXX:XX | CGH4_74 | −0.01 | 0.31 | −0.06 | 0.03 | 0.35 | <2e−16 | 0.29 | 0.1 |
| XXX+XX:XX | CGH4_75 | 0 | 0.85 | −0.02 | 0.53 | 0.06 | 1.1e−2 | 0.25 | 0.2 |
| XY+XX:XX | CGH5_26 | 0.01 | 0.43 | −0.01 | 0.69 | −0.42 | <2e−16 | 1.35 | 1.5e−12 |
| XY:XX | CGH5_27 | 0.03 | 0.01 | −0.03 | 0.34 | −0.85 | <2e−16 | 1.93 | <2e−16 |
| XY(5cl20):XY | CGH4_73 | −0.01 | 0.55 | −0.01 | 0.85 | −0.06 | e−2 | 0.05 | 0.74 |
| ANOVA P-value |
|
4e−4 |
|
3e−6 |
|
0 |
|
4.8e−55 |
|
| Amplified microdissected samples subset | |||||||||
| XY:XY | CGH4_59 | 0.01 | 0.29 | −0.07 | 0.01 | −0.07 | 0 | −0.04 | 0.82 |
| XX:XY | CGH4_60 | 0 | 0.81 | −0.07 | 0.01 | 0.39 | <2e−16 | −1 | 2e−9 |
| XX:XY | CGH4_61 | 0 | 0.75 | −0.03 | 0.29 | 0.4 | <2e−16 | −0.96 | 9.3e−9 |
| XY:XY | CGH3_23 | −0.13 | <2e−16 | −0.09 | 0.001 | −0.03 | 0.17 | 0 | 0.99 |
| XY:XY | CGH3_40 | 0.01 | 0.46 | −0.20 | 2e−13 | 0.06 | 0.01 | −0.02 | 0.91 |
The Fx levels were compared to the intercept level 0, based on two control male versus male hybridizations (CGH3_2 and 4). Statistically (P < 0.00001) and biologically significant (estimated log2 ratio < −0.35 or > +0.35) values are highlighted in bold in the columns P and Est., respectively. All microdissected samples were co-hybridized with male control reference DNA. Est., estimated log2 ratio; P, P-value.
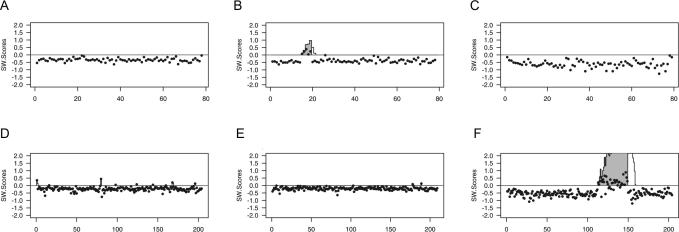
Subchromosomal regions containing copy-number alterations can be difficult to detect, especially when they overlap with highly variable regions. We used the analytical Smith–Waterman algorithm as modified by Karlin et al. (23,33), implemented in R by Price et al. (24) (http://www.well.ox.ac.uk/~tprice/1), to detect subchromosomal gains and losses in our pilot CGH data (Figure 4A–C). After subtracting a user-defined baseline threshold delta (δ) to generate a negative mean for each chromosome, the genome is scanned for contiguous sequences of high positive or low negative scores, which may indicate polysomic or monosomic regions, respectively. High- or low-scoring segments of clones are denominated as ‘islands’, when their scores cannot be increased by shrinkage or expansion of the segment boundaries. The statistical significance of an island is estimated as the proportion of times that a higher-scoring island is found in 1000 random permutations of the coordinates of the scores, based on the premise that successive scores from the permuted data approximate the null distribution. The trisomic region present over five clones on chromosome 20 from sample 5cl20 was correctly predicted with a δ threshold of 3.6 (Figure 4B), with no false-positive islands detected. Applying the same threshold to chromosome 20 data from other samples (Figure 4A and C) did not result in the detection of any significant islands (p < 0.001). Also, chromosome 5 ratios (Figure 4D and E) and ratios from the remaining chromosomes (data not shown) were analyzed using the same approach and, as expected, no significant copy-number alterations were detected.
Figure 4.
Detection of subchromosomal copy-number changes. Comparison of φ29-amplified array CGH data for two illustrative chromosomes, (A–C) 20 and (D–F) 5, analyzed with the Smith–Waterman algorithm to detect subchromosomal gains and deletions. Control male DNA (A and D), 5cl20 DNA (B and E) and microdissected adenoma D5_1 DNA (C and F) were co-hybridized with an unamplified control male reference DNA (Table 1). Chromosomal position of the BAC clones is indicated on the x-axis. The y-axis indicates the SW-scores associated with each clone, produced by subtracting a user-defined threshold delta (δ) from the Lowess normalized log2 ratios of the triplicate spots from each BAC clone. The vertical lines represent the Smith–Waterman algorithm partial sums S and the score of the highest-scoring island (filled in gray) is given by its maximum height: chromosome 20 (δ = 3.6) island in 5cl20 (B), score 0.98 (P = 0) and chromosome 5 (δ = 2.5) island in D5_1 (F), score 6.53 (P = 0.009).
Analysis of laser-capture microdissected epithelial cells from human colon mucosa and adenomatous polyps
To obtain proof of principle for our combined LCM–array CGH approach in the genomic profiling of human tissues, the method was applied to φ29-amplified DNA samples derived from approximately 1000 microdissected human colon cells. Three samples originated from control individuals, one male (NC1) and two females (NC2 and NC3), and two samples were derived from a male individual affected by FAP, normal epithelium (D5_14) and dysplastic adenoma cells (D5_1). Two illustrative normal CGH array profiles (NC1 and NC2) co-plotted with the average normal CGH profile showed no autosomal genomic imbalances (Figure 1G and H).
We fitted the linear model using the Fx levels (Table 1) as factor to these samples to detect whole chromosome copy-number changes (Figure 3, right panel of each plot; Table 2). Except for the sex chromosomes in sex-mismatched hybridizations, none of the chromosomes from the microdissected normal and patient samples showed statistically significant and biologically meaningful changes from the expected number of two copies. Remarkably, the estimated X and Y chromosome average log2 ratios in the female samples versus male reference are lower than the corresponding estimated ratios from diluted control DNA samples. We believe this might be due to variations in the non-measurable low quantities of LCM-derived DNA used as templates in φ29 amplifications.
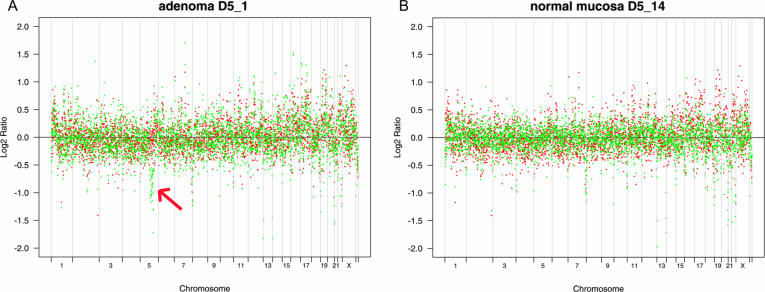
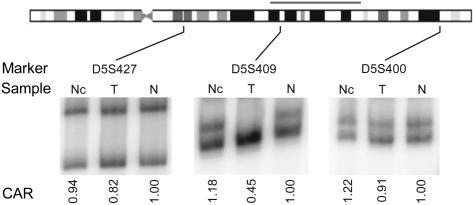
Co-plotting of the adenoma sample CGH ratios (D5_1) with the average normal CGH profile (Figure 5A), suggested a loss of genetic material in a subchromosomal region of chromosome 5 that was not observed in the corresponding normal tissue (Figure 5B). Consequently, using the Smith–Waterman algorithm with a δ threshold of 2.5, we detected a sequence of 37 clones (∼36 Mb), deleted in the region spanning from 5q21.1 to 5q31.1 (base position 100714436 to 135902806) encompassing the APC gene (5q22) causal to FAP. Allelic imbalance at chromosome 5q was confirmed by LOH analysis of dinucleotide repeat markers in the region of interest. The comparative allelic ratio of marker D5S409 (≤0.6), encompassed by the deletion, is indicative of loss of heterozygosity at this locus (Figure 6). Markers D5S400 and D5S427, mapping respectively distal and proximal to the deletion predicted by array CGH, did not reveal allelic imbalances, thus confirming the subchromosomal nature of the 5q deletion.
Figure 5.
Amplified array CGH profiles from microdissected human colon cells. φ29-amplified DNA profiles from 1000 microdissected cells (green), obtained (A) from a low grade dysplastic area of a colorectal adenomatous polyp from a FAP patient (D5_1) and (B) from the corresponding histologically normal colorectal mucosa (D5_14) plotted against an ‘average’ control DNA profile (red; see text). This co-plotting reveals the presence of a deletion on chromosome 5q in the adenoma sample (arrow).
Figure 6.
LOH analysis of microdissected adenoma and corresponding normal mucosa at 5q. Three dinucleotide repeat markers mapped on chromosome 5D5S400 (distal), D5S409 (deleted region) and D5S427 (proximal) were used on genomic DNA from the FAP patient D5. The DNA samples were derived from non-microdissected, unamplified normal mucosa (Nc), from microdissected, φ29-amplified adenomatous tumor (T) and from the corresponding microdissected, φ29-amplified normal mucosa (N). The gray horizontal line above chromosome 5 represents the 37 Mb deletion. In the normal mucosa, the three loci show the expected two alleles, but in the tumor sample one allele of D5S409 marker, internal to the deletion is lost. The comparative allelic ratio (CAR) for this marker indicates the loss of the larger allele (CAR ≤ 0.6).
DISCUSSION
We present a robust, standardized protocol for the analysis of genome copy-number alterations in DNA derived from as few as 1000 microdissected cells from histology specimens. The φ29 polymerase-based MDA generates high-molecular DNA replication products in high yields, and is suitable when processing large numbers of samples. Some representational distortion occurs, which is likely to result from variability in priming density and processing by the φ29 polymerase of repetitive and polymorphic sequences. Similar systematic changes in copy-number structure were reported in other array CGH studies using φ29-amplified DNA (15,16). Some amplification biases can be partially compensated for by using test and reference samples amplified under the same conditions (10,15,34–36). We compared the results of co-hybridization of our test sample with amplified and unamplified reference DNA samples, and surprisingly found similar ratio distributions. Since previous studies used either a different strand displacement polymerase (Bst) and cDNA microarrays (15), or a PCR-based amplification method (10,34–36), these results may not be directly comparable to ours. Moreover, Lage et al. also successfully used an unamplified reference sample in an evaluation experiment on human BAC arrays (15). Taken the fact that the use of an unamplified reference is simpler and less expensive for high-throughput analyses of many samples, we opted for an unamplified reference. As we show, the observed over- and under-representations of specific genomic regions were reproducible and as such can be handled with adequate statistical tools. By taking individual clone-specific effects into account in our statistical analyses, they were dealt with in an effective way without excluding clones based on an arbitrary threshold. This approach allowed us to analyze the whole genome, including the Y chromosome, which is routinely excluded entirely due to variable hybridization results, even in classical array CGH (6,15,36).
Using the X chromosome as a model, we show that our amplified array CGH method detects monosomy and trisomy of the whole chromosomes using the linear regression model. It was even possible to detect a loss of one copy in 50% of the cells, while a similar sensitivity was not reached for a single copy gain. The observed underestimation of the magnitude of copy-number changes has been previously reported in array CGH studies, and most likely results from incomplete suppression of repetitive sequences or errors in background subtraction (6,8). However, for all the analyzed X chromosome dosage levels, the relationship between the estimated and the expected ratios were linear, indicating that the amplitude of compression is constant over these levels. As the copy number departed farther from the genome average, the variance of the ratios measured for the X and Y chromosome clones increased. These ratio variations were reproducible, suggesting that the sequence characteristics of individual clones, possibly differing amounts of sequence shared between the X and Y chromosomes, play a role (6). To establish a limit of accuracy for small regions of gains or deletions, we applied amplified array CGH to a sample known to harbor a trisomic region spanning five BAC clones (5 Mb) on chromosome 20. This aberration was readily detected using a simple, nonparametric Smith–Waterman dynamic algorithm. The lack of assumptions in this method makes its use very convenient. For our purpose, it demonstrated good sensitivity and specificity indicating that the resolution of our amplified array CGH method is at least 5 Mb. Both statistical models have a very low false discovery rate, when log2 ratio thresholds compared to the intercept of −0.35 (0.78 in linear scale) and 0.35 (1.27 in linear scale), representing a biologically meaningful gain or loss, are applied in addition to a significant p-value. Our data also show the importance of using a pilot study, with normal and control samples carrying known copy-number changes. The generated data can be subsequently used for calibration and for sensitivity determination in the analytical approach.
Applying the novel LCM-array CGH protocol to the analysis of normal colon mucosa and colonic adenomatous polyps from FAP patients provided proof of principle. A subchromosomal 5q deletion was detected by LCM-array CGH analysis and confirmed by an independent PCR-based method. Although we have not tested the minimum amount of input DNA for the MDA reaction, the recommended minimal amount is 1 ng, representing approximately 300–500 human genomic equivalents.
As shown here, cryo-preserved tissues provide excellent starting material for φ29 amplification. However, the amplification efficiency is reduced proportionally to a decrease in molecular weight of the starting material, which is problematic for amplification of formalin-fixed archival DNA (15). Recently, several other methods for whole-genome amplification in combination with array CGH were described. Balanced-PCR amplification (36) employs digestion and ligation of a target and control genome with distinct linkers, which are mixed and amplified in a single PCR, thereby avoiding biases associated with PCR saturation and impurities. This procedure showed equivalent performance compared to MDA on cDNA microarrays using intact genomic DNA, but overcomes problems associated with modest DNA degradation in formalin-fixed paraffin-embedded tissues (36). Guillaud-Bataille et al. present an optimized LM-PCR protocol using 1 ng of starting DNA and BAC arrays. This approach preserves the initial ratios observed with BAC array CGH, allowing the reliable detection of one-copy-level variations among the amplified material (10). Although this method has not yet been applied to microdissected samples, the results on cell line DNA are very promising.
cDNA arrays typically consisting of several tens of thousands of features are used for array CGH experiments because of their more common availability and higher resolution (15,36–38). Compared to BAC arrays, however, the signal-to-noise ratio is lower and signals of two to five neighboring clones are averaged to improve signal reproducibility. To reach a similar resolution using BAC arrays, around 10 000–30 000 BACs would be necessary, resulting in a tiling array for the human genome (39). Since the size of the BACs, 150–200 kb, ultimately obscures higher resolution, microarrays containing 25mer (38,40,41) or 60–70mer (42–44) oligonucleotide probes are currently being explored for measuring DNA copy-number changes. The commercially available synthetic 25mer high-density oligonucleotide arrays, which were originally designed to detect single-nucleotide polymorphisms (SNPs) (45), have the advantage of giving genotyping data in conjunction with copy-number analysis (38,40,41). Overall, this platform exhibited more variability than BAC array-based CGH, and while high-level amplifications and homozygous deletions were reliably reported, changes resulting in loss or gain of a single copy were often missed in unamplified tumor cell line DNA (38,41). It is anticipated that as SNP density increases, resolution and the ability to assess subtle copy-number changes will increase. Importantly, in a small-scale study, Wong et al. used SNP arrays in combination with φ29-amplified DNA from two tumor biopsies, and showed good concordance between amplified and unamplified DNA for a high-level amplification and deletion (40). This study shows the feasibility of combining MDA whole-genome amplification with sensitive copy-number analysis at high resolution.
In summary, we show that the strand displacement polymerase φ29 reproducibly amplifies starting amounts of genomic DNA as low as 2 ng or 1000 laser-capture microdissected cells, resulting in the reliable detection of single copy variations on BAC array CGH. This method allows the detection of specific genetic alterations in small neoplastic lesions, including tumor biopsies obtained by non-surgical sampling methods like endoscopies, and in clinically relevant subpopulations of tumor cells, e.g. invading fronts of tumors. The significance of the capability to detect single-copy alterations in tissue samples consisting of only a few thousand cells, lies in the greatly expanded potential for discovery of novel genetic alterations limited to small clonal patches in tumors, or present in small preneoplastic lesions, as demonstrated here by the detection of the subchromosomal 5q deletion in a FAP-derived adenomatous polyp by combined LCM-array CGH analysis. These features will facilitate the identification of novel oncogenes or tumor suppressors mapping to regions of gene gain or loss (6,46). An additional application in medical genetics is the detection of copy-number changes in DNA derived from buccal swaps for diagnosis of congenital chromosomal abnormalities, such as microdeletions and duplications, and unbalanced chromosomal translocations.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Stephan White for kindly providing us DNA from the XXX cell line. This work was supported by grants from the Dutch Cancer Society (2001-2482; J.C. and L.M.), the Center of Biomedical Genetics, the Netherlands (J.M.B.), and the Center of Medical System Biology (CMSB) established by the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research (NGI/NWO).
REFERENCES
- 1.Bardi G., Sukhikh,T., Pandis,N., Fenger,C., Kronborg,O. and Heim,S. (1995) Karyotypic characterization of colorectal adenocarcinomas. Genes Chromosomes Cancer, 12, 97–109. [DOI] [PubMed] [Google Scholar]
- 2.Fearon E.R. and Vogelstein,B. (1990) A genetic model for colorectal tumorigenesis. Cell, 61, 759–767. [DOI] [PubMed] [Google Scholar]
- 3.Ried T., Knutzen,R., Steinbeck,R., Blegen,H., Schrock,E., Heselmeyer,K., Du Manoir,S. and Auer,G. (1996) Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes Cancer, 15, 234–245. [DOI] [PubMed] [Google Scholar]
- 4.Thompson C.T. and Gray,J.W. (1993) Cytogenetic profiling using fluorescence in situ hybridization (FISH) and comparative genomic hybridization (CGH). J. Cell. Biochem. Suppl., 17G, 139–143. [DOI] [PubMed] [Google Scholar]
- 5.Fiegler H., Carr,P., Douglas,E.J., Burford,D.C., Hunt,S., Scott,C.E., Smith,J., Vetrie,D., Gorman,P., Tomlinson,I.P. and Carter,N.P. (2003) DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer, 36, 361–374. [DOI] [PubMed] [Google Scholar]
- 6.Snijders A.M., Nowak,N., Segraves,R., Blackwood,S., Brown,N., Conroy,J., Hamilton,G., Hindle,A.K., Huey,B., Kimura,K., Law,S., Myambo,K., Palmer,J., Ylstra,B., Yue,J.P., Gray,J.W., Jain,A.N., Pinkel,D. and Albertson,D.G. (2001) Assembly of microarrays for genome-wide measurement of DNA copy number. Nature Genet., 29, 263–264. [DOI] [PubMed] [Google Scholar]
- 7.Solinas-Toldo S., Lampel,S., Stilgenbauer,S., Nickolenko,J., Benner,A., Dohner,H., Cremer,T. and Lichter,P. (1997) Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer, 20, 399–407. [PubMed] [Google Scholar]
- 8.Pinkel D., Segraves,R., Sudar,D., Clark,S., Poole,I., Kowbel,D., Collins,C., Kuo,W.L., Chen,C., Zhai,Y., Dairkee,S.H., Ljung,B.M., Gray,J.W. and Albertson,D.G. (1998) High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nature Genet., 20, 207–211. [DOI] [PubMed] [Google Scholar]
- 9.Daigo Y., Chin,S.F., Gorringe,K.L., Bobrow,L.G., Ponder,B.A., Pharoah,P.D. and Caldas,C. (2001) Degenerate oligonucleotide primed-polymerase chain reaction-based array comparative genomic hybridization for extensive amplicon profiling of breast cancers: a new approach for the molecular analysis of paraffin-embedded cancer tissue. Am. J. Pathol., 158, 1623–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guillaud-Bataille M., Valent,A., Soularue,P., Perot,C., Inda,M.M., Receveur,A., Smaili,S., Crollius,H.R., Benard,J., Bernheim,A., Gidrol,X. and Danglot,G. (2004) Detecting single DNA copy number variations in complex genomes using one nanogram of starting DNA and BAC-array CGH. Nucleic Acids Res., 32, e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung V.G. and Nelson,S.F. (1996) Whole genome amplification using a degenerate oligonucleotide primer allows hundreds of genotypes to be performed on less than one nanogram of genomic DNA. Proc. Natl Acad. Sci. USA, 93, 14676–14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean F.B., Hosono,S., Fang,L., Wu,X., Faruqi,A.F., Bray-Ward,P., Sun,Z., Zong,Q., Du,Y., Du,J., Driscoll,M., Song,W., Kingsmore,S.F., Egholm,M. and Lasken,R.S. (2002) Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl Acad. Sci. USA, 99, 5261–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosono S., Faruqi,A.F., Dean,F.B., Du,Y., Sun,Z., Wu,X., Du,J., Kingsmore,S.F., Egholm,M. and Lasken,R.S. (2003) Unbiased whole-genome amplification directly from clinical samples. Genome Res., 13, 954–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes S., Lim,G., Beheshti,B., Bayani,J., Marrano,P., Huang,A. and Squire,J.A. (2004) Use of whole genome amplification and comparative genomic hybridisation to detect chromosomal copy number alterations in cell line material and tumour tissue. Cytogenet. Genome Res., 105, 18–24. [DOI] [PubMed] [Google Scholar]
- 15.Lage J.M., Leamon,J.H., Pejovic,T., Hamann,S., Lacey,M., Dillon,D., Segraves,R., Vossbrinck,B., Gonzalez,A., Pinkel,D., Albertson,D.G., Costa,J. and Lizardi,P.M. (2003) Whole genome analysis of genetic alterations in small DNA samples using hyperbranched strand displacement amplification and array-CGH. Genome Res., 13, 294–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paez J.G., Lin,M., Beroukhim,R., Lee,J.C., Zhao,X., Richter,D.J., Gabriel,S., Herman,P., Sasaki,H., Altshuler,D., Li,C., Meyerson,M. and Sellers,W.R. (2004) Genome coverage and sequence fidelity of phi29 polymerase-based multiple strand displacement whole genome amplification. Nucleic Acids Res., 32, e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powell S.M., Zilz,N., Beazer-Barclay,Y., Bryan,T.M., Hamilton,S.R., Thibodeau,S.N., Vogelstein,B. and Kinzler,K.W. (1992) APC mutations occur early during colorectal tumorigenesis. Nature, 359, 235–237. [DOI] [PubMed] [Google Scholar]
- 18.Knijnenburg J., Szuhai,K., Giltay,J., Molenaar,L., Sloos,W., Poot,M., Tanke,H.J. and Rosenberg,C. Insight from genomic microarrays into structural chromosome rearragments. Am. J. Hum. Genet., in press. [DOI] [PubMed] [Google Scholar]
- 19.Isola J., Devries,S., Chu,L., Ghazvini,S. and Waldman,F. (1994) Analysis of changes in DNA sequence copy number by comparative genomic hybridization in archival paraffin-embedded tumor samples. Am. J. Pathol., 145, 1301–1308. [PMC free article] [PubMed] [Google Scholar]
- 20.Fox J.C., Cave,C.A. and Schumm,J.W. (2003) Development, characterization, and validation of a sensitive primate-specific quantification assay for forensic analysis. Biotechniques, 34, 314–318, 320, 322. [DOI] [PubMed] [Google Scholar]
- 21.Ihaka R. and Gentleman,R. (1996) R: language for data analysis and graphics. J. Comput. Graph. Statist., 5, 299–314. [Google Scholar]
- 22.Firth D. and Menezes,R.X. (2004) Quasi-variances. Biometrika, 91, 65–80. [Google Scholar]
- 23.Smith T.F. and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 24.Price T., Regan,R., Mott,R., Ventress,N., Smith,L., Greenfield,A., Ayyub,H., Daniels,R.J., Tiganescu,A., Broxholme,J., Slahan,A., Pedraza-Diaz,S., Ragoussis,I., Higgs,D.R., Flint,J. and Knight,S.J.L. (2003) A new method to access the statistical significance of copy number changes identified in genomic DNA by array comparative genome hybridization. MSc. Dissertation in Applied Statistics, University of Oxford.
- 25.Nycz C.M., Dean,C.H., Haaland,P.D., Spargo,C.A. and Walker,G.T. (1998) Quantitative reverse transcription strand displacement amplification: quantitation of nucleic acids using an isothermal amplification technique. Anal. Biochem., 259, 226–234. [DOI] [PubMed] [Google Scholar]
- 26.Smits R., Kartheuser,A., Jagmohan-Changur,S., Leblanc,V., Breukel,C., De Vries,A., Van Kranen,H., Van Krieken,J.H., Williamson,S., Edelmann,W., Kucherlapati,R., Khanpm and Fodde,R. (1997) Loss of Apc and the entire chromosome 18 but absence of mutations at the Ras and Tp53 genes in intestinal tumors from Apc1638N, a mouse model for Apc-driven carcinogenesis. Carcinogenesis, 18, 321–327. [DOI] [PubMed] [Google Scholar]
- 27.Du Manoir S., Schrock,E., Bentz,M., Speicher,M.R., Joos,S., Ried,T., Lichter,P. and Cremer,T. (1995) Quantitative analysis of comparative genomic hybridization. Cytometry, 19, 27–41. [DOI] [PubMed] [Google Scholar]
- 28.Du Manoir S., Speicher,M.R., Joos,S., Schrock,E., Popp,S., Dohner,H., Kovacs,G., Robert-Nicoud,M., Lichter,P. and Cremer,T. (1993) Detection of complete and partial chromosome gains and losses by comparative genomic in situ hybridization. Hum. Genet., 90, 590–610. [DOI] [PubMed] [Google Scholar]
- 29.Lundsteen C., Maahr,J., Christensen,B., Bryndorf,T., Bentz,M., Lichter,P. and Gerdes,T. (1995) Image analysis in comparative genomic hybridization. Cytometry, 19, 42–50. [DOI] [PubMed] [Google Scholar]
- 30.Mitelman F. (ed.) (1995) ISCN (1995): An International System for Human Cytogenetics Nomenclature. S.Karger, Basel. [Google Scholar]
- 31.Dean F.B., Nelson,J.R., Giesler,T.L. and Lasken,R.S. (2001) Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res., 11, 1095–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirchhoff M., Gerdes,T., Rose,H., Maahr,J., Ottesen,A.M. and Lundsteen,C. (1998) Detection of chromosomal gains and losses in comparative genomic hybridization analysis based on standard reference intervals. Cytometry, 31, 163–173. [PubMed] [Google Scholar]
- 33.Karlin S., Blaisdell,B.E. and Brendel,V. (1990) Identification of significant sequence patterns in proteins. Meth. Enzymol., 183, 388–402. [DOI] [PubMed] [Google Scholar]
- 34.Huang Q., Schantz,S.P., Rao,P.H., Mo,J., Mccormick,S.A. and Chaganti,R.S. (2000) Improving degenerate oligonucleotide primed PCR-comparative genomic hybridization for analysis of DNA copy number changes in tumors. Genes Chromosomes Cancer, 28, 395–403. [PubMed] [Google Scholar]
- 35.Voullaire L., Wilton,L., Slater,H. and Williamson,R. (1999) Detection of aneuploidy in single cells using comparative genomic hybridization. Prenat. Diagn., 19, 846–851. [PubMed] [Google Scholar]
- 36.Wang G., Brennan,C., Rook,M., Wolfe,J.L., Leo,C., Chin,L., Pan,H., Liu,W.H., Price,B. and Makrigiorgos,G.M. (2004) Balanced-PCR amplification allows unbiased identification of genomic copy number changes in minute cell and tissue samples. Nucleic Acids Res., 32, e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollack J.R., Perou,C.M., Alizadeh,A.A., Eisen,M.B., Pergamenschikov,A., Williams,C.F., Jeffrey,S.S., Botstein,D. and Brown,P.O. (1999) Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nature Genet., 23, 41–46. [DOI] [PubMed] [Google Scholar]
- 38.Zhao X., Li,C., Paez,J.G., Chin,K., Janne,P.A., Chen,T.H., Girard,L., Minna,J., Christiani,D., Leo,C., Gray,J.W., Sellers,W.R. and Meyerson,M. (2004) An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res., 64, 3060–3071. [DOI] [PubMed] [Google Scholar]
- 39.Ishkanian A.S., Malloff,C.A., Watson,S.K., Deleeuw,R.J., Chi,B., Coe,B.P., Snijders,A., Albertson,D.G., Pinkel,D., Marra,M.A., Ling,V., Macaulay,C. and Lam,W.L. (2004) A tiling resolution DNA microarray with complete coverage of the human genome. Nature Genet., 36, 299–303. [DOI] [PubMed] [Google Scholar]
- 40.Wong K.K., Tsang,Y.T., Shen,J., Cheng,R.S., Chang,Y.M., Man,T.K. and Lau,C.C. (2004) Allelic imbalance analysis by high-density single-nucleotide polymorphic allele (SNP) array with whole genome amplified DNA. Nucleic Acids Res., 32, e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bignell G.R., Huang,J., Greshock,J., Watt,S., Butler,A., West,S., Grigorova,M., Jones,K.W., Wei,W., Stratton,M.R., Futreal,P.A., Weber,B., Shapero,M.H. and Wooster,R. (2004) High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res., 14, 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brennan C., Zhang,Y., Leo,C., Feng,B., Cauwels,C., Aguirre,A.J., Kim,M., Protopopov,A. and Chin,L. (2004) High-resolution global profiling of genomic alterations with long oligonucleotide microarray. Cancer Res., 64, 4744–4748. [DOI] [PubMed] [Google Scholar]
- 43.Carvalho B., Ouwerkerk,E., Meijer,G.A. and Ylstra,B. (2004) High resolution microarray comparative genomic hybridisation analysis using spotted oligonucleotides. J. Clin. Pathol., 57, 644–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lucito R., Healy,J., Alexander,J., Reiner,A., Esposito,D., Chi,M., Rodgers,L., Brady,A., Sebat,J., Troge,J., West,J.A., Rostan,S., Nguyen,K.C., Powers,S., Ye,K.Q., Olshen,A., Venkatraman,E., Norton,L. and Wigler,M. (2003) Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res., 13, 2291–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuzaki H., Loi,H., Dong,S., Tsai,Y.Y., Fang,J., Law,J., Di,X., Liu,W.M., Yang,G., Liu,G., Huang,J., Kennedy,G.C., Ryder,T.B., Marcus,G.A., Walsh,P.S., Shriver,M.D., Puck,J.M., Jones,K.W. and Mei,R. (2004) Parallel genotyping of over 10,000 SNPs using a one-primer assay on a high-density oligonucleotide array. Genome Res., 14, 414–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruder C.E., Hirvela,C., Tapia-Paez,I., Fransson,I., Segraves,R., Hamilton,G., Zhang,X.X., Evans,D.G., Wallace,A.J., Baser,M.E., Zucman-Rossi,J., Hergersberg,M., Boltshauser,E., Papi,L., Rouleau,G.A., Poptodorov,G., Jordanova,A., Rask-Andersen,H., Kluwe,L., Mautner,V., Sainio,M., Hung,G., Mathiesen,T., Moller,C., Pulst,S.M., Harder,H., Heiberg,A., Honda,M., Niimura,M., Sahlen,S., Blennow,E., Albertson,D.G., Pinkel,D. and Dumanski,J.P. (2001) High resolution deletion analysis of constitutional DNA from neurofibromatosis type 2 (NF2) patients using microarray-CGH. Hum. Mol. Genet., 10, 271–282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.