Abstract
DNA sequences containing long adjacent inverted repeats (palindromes) are inherently unstable and are associated with many types of chromosomal rearrangements. The instability associated with palindromic sequences also creates difficulties in their molecular analysis: long palindromes (>250 bp/arm) are highly unstable in Escherichia coli, and cannot be directly PCR amplified or sequenced due to their propensity to form intra-strand hairpins. Here, we show that DNA molecules containing long palindromes (>900 bp/arm) can be transformed and stably maintained in Saccharomyces cerevisiae cells lacking a functional SAE2 gene. Treatment of the palindrome-containing DNA with sodium bisulfite at high temperature results in deamination of cytosine, converting it to uracil and thus reducing the propensity to form intra-strand hairpins. The bisulfite-treated DNA can then be PCR amplified, cloned and sequenced, allowing determination of the nucleotide sequence of the junctions. Our data demonstrates that long palindromes with either no spacer (perfect) or a 2 bp spacer can be stably maintained, recovered and sequenced from sae2Δ yeast cells. Since DNA sequences from mammalian cells can be gap repaired by their co-transformation into yeast cells with an appropriate vector, the methods described in this manuscript should provide some of the necessary tools to isolate and characterize palindromic junctions from mammalian cells.
INTRODUCTION
DNA sequences containing palindromes are associated with a high degree of genomic instability in mouse (1–4), yeast (5,6) and bacteria (7,8). The instability associated with palindromes includes increased recombination (5,9–11), gene amplification (12–15), translocations (16) and deletions (17,18). Furthermore, closely spaced inverted repeats are less frequent than expected in the sequenced human (19) and bacterial genomes (20), suggesting that they are either unclonable and therefore missing from the genome databases or that they are inherently deleterious to cells. A unique property of palindromes and closely spaced (<10 bp) inverted repeats is the ability to extrude from normal B-form DNA into a cruciform structure (7). One way in which palindromes could contribute to genomic instability is that cruciform extrusion may create barriers to DNA replication or serve as a target for endonucleases that can disrupt strand integrity (21).
We show here that Saccharomyces cerevisiae cells deleted for the SAE2/COM1 gene allow the stable propagation of plasmids carrying long palindromes. Previously, we found that sae2Δ cells led to high levels of palindrome formation after the induction of a double-strand break (DSB) between chromosomal inverted repeats, presumably due to break-induced replication, followed by some form of non-homologous end-joining (22). Sae2p-deficient yeast cells have also been found to enhance the stability of inverted Alu repeats that are separated by 12 bp (21). Several lines of evidence suggest that Sae2p functions by regulating the activity of the Mre11p/Rad50p (MR) nuclease complex. This is a highly conserved nuclease complex with homologs from mammals (MR) to bacteria [SbcCD; (23)]. In vitro, these complexes have a 3′→5′ exonuclease activity as well as single- and double-strand endonuclease activities, including the ability to cleave hairpin DNA molecules (24–26). In yeast, null mutants of MRE11 or RAD50 have severe defects in a number of DNA metabolic functions including slow growth, an impaired DNA damage response, shortened telomeres and deficiencies in non-homologous end-joining (23,27). Although sae2 mutant cells do not share the severe phenotype associated with null alleles of the MR complex, their phenotype is thus far indistinguishable from certain hypomorphic alleles of mre11 or rad50 (termed ‘s’ for separation of function). For example, as in mre11s and rad50s mutants, Spo11p-induced meiotic DSBs are not processed, and retain Spo11p at their 5′ termini in sae2Δ cells (28–31). Also, mre11s and rad50s mutants have a similar increase in palindrome formation and propagation as do sae2Δ mutants (22).
The ability to clone and stably maintain long palindromes in sae2Δ yeast has opened the opportunity for their molecular analysis. Recently, sodium bisulfite modification of DNA was used to PCR amplify, clone and sequence a nearly palindromic inverted repeat created by DSB-induced replication of 79 bp inverted repeats with a 29 bp non-palindromic center in Chinese hamster ovary (CHO) cells (14) and palindromes created by ligation of a hairpin oligonucleotide (with a 6 bp loop) to the ends of short (150–200 bp) DNA fragments (32). Sodium bisulfite treatment of DNA results in deamination of cytosine converting it to uracil. It has been primarily used to identify the location of methylated cytosine residues, as they are resistant to deamination (33,34). Here, we describe conditions that allow us to use this method to sequence long perfect palindromes (>900 bp/arm) from sae2 mutant S.cerevisiae cells. This is only the second report of a verified perfect palindrome in a eukaryote. Unlike the previous report (4), the method presented here is a general one that does not require prior knowledge of the palindromic sequence. Recently, it was shown that mammalian DNA could be gap-repaired into yeast by transformation-associated recombination (TAR) cloning (35). Taken together, these methods should provide the tools to clone, isolate and sequence long perfectly palindromic regions of the human genome directly from yeast cells.
MATERIALS AND METHODS
Strains, plasmids, oligonucleotides, media and general methods
For Escherichia coli transformations, we used chemically competent DH5α cells [F−φ80d lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17(rk−, mk+) phoA supE44 λ− thi-1 gyrA96 relA1; Invitrogen] that were grown in Luria–Bertani (LB) medium supplemented with Ampicillin and X-gal where necessary (36). Subcloning and ligation were done by standard techniques (36). All yeast experiments were done with a sae2 deletion derivative of strain BY4742 (MAT α his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YGL175c::kanMX4; Open Biosystems). S.cerevisiae cells were grown in YEPD or the appropriate synthetic drop-out media (37). Competent yeast cells were prepared by the lithium acetate transformation procedure (38). The Saccharomyces kluyveri HIS3 gene was isolated from plasmid (p)KW301 (39), and cloned into pRS426 (40) as described below. The oligonucleotides used in this study are shown in Table 1. Oligonucleotides HindIII-vec-top and HindIII-vec-bottom were synthesized by Integrated DNA Technologies. All other oligonucleotides were synthesized by Invitrogen.
Table 1. Oligonucleotides.
| Name | Oligonucleotide sequence | Native sequence |
|---|---|---|
| BiSuHis3SK-A | GATGATTTGTGATTGAGTAGTTTGG | GATGACCTGTGACTGAGTAGCTTGG |
| BiSuHis3SK-B | GGGTTGGTTTTGTTATTTTTATGG | GGGCTGGTTCTGTCATTTCTATGG |
| BiSuHis3SK-C | CTTTAATATCAAATTCCTACAACCCA | CTTTGATATCGAATTCCTGCAGCCCG |
| BiSuHis3SK-D | ACAAAAAATAAACACCTTACACTCC | ACGGGAGATAAACGCCTTGCGCTCC |
| BiSuHis3SK-E | CATATAATCCAAAAAACCAACACC | CATATGATCCAAAAAGCCAACACC |
| M13-F | GTAAAACGACGGCCAG | |
| M13-R | CAGGAAACAGCTATGAC | |
| HindIII-vec-top | GAGAGGGAAGAGAGCAGGCAAGGAATGG-AAGCTGTCTGTCGCAGGAGAGGAAG | |
| HindIII-vec-bottom | AGCTCTTCCTCTCCTGTCGCTAAGAGCATG-CTTGCCAATGCTAAGCTCTTCCCTCTC | |
| Bubble-amplify | AGCGATTCTCGTACGAACGGTTACGATTCG | |
| UMHis3SK-F | TTTGAACGCTTGCCCTAATG | |
| UMHis3SK-G | TGTGAATATCGATGACCTGTG | |
| UMHis3SK-H | CGCTCCTGAACAGTTTGTTT |
Southern blot analysis
Yeast DNA for Southern blot analysis was prepared by the glass bead method (41). Approximately 5 μg of DNA was digested with the appropriate endonucleases according to manufacturer's instructions (New England Biolabs). One-half of each sample was then incubated in a boiling water bath for 5 min, and immediately transferred to ice. The DNA samples were electrophoresed in neutral agarose gels, and subsequently soaked in 0.4 N NaOH for 30 min. Southern transfers were done in 0.4 N NaOH onto Hybond N+ membranes (Amersham) by capillary transfer according to manufacturer's instructions. After transfer, the membrane was cross-linked by exposure to UV light in a Stratalinker (Stratagene), and then neutralized by soaking in 10× SSC (36). Hybridization was done according to the protocol suggested for hybridization of Hybond N+ membranes. The probe was an XbaI–Kpn1 fragment from pKW301 that had been labeled with 32P using the Redi-Prime II Kit (Amersham). Blots were analyzed on a Typhoon scanner (Storm) and visualized with ImageQuant software.
Sodium bisulfite modification
Plasmid DNA was enriched from 100 ml of yeast cells using Qiagen columns as described by Singh and Weil (42). The DNA was digested with an appropriate restriction enzyme to reduce the palindromic fragment to ∼1 kb. Digested DNA (0.2–2 μg) was resuspended in 60 μl of H2O, denatured by adding fresh NaOH to 0.3 N and incubating at 37°C for 15 min. Subsequently, 50 μl of the DNA was treated with sodium bisulfite using 350 μl of Reagent I from the CpG Genome Modification Kit (Serologicals Corp.) that had been adjusted to pH 5.0 with 3 N NaOH. The samples were overlaid with mineral oil and incubated at 95°C for 30 min. The bisulfite was removed by cleaning the DNA with the GeneClean Spin Kit (Bio 101). The eluate was adjusted to 0.3 N NaOH and incubated at 37°C for 15 min to desulfonate the DNA. One microliter of either CpG Genome Modification Kit Reagent IV, or of Glycogen (Fermentas), was added, and the sample was ethanol precipitated. The DNA was resuspended in 100 μl 1× TE and used for PCR with the indicated primers. PCR was performed using 1–10 μl of modified DNA in a 50 μl reaction containing 1× Titanium buffer (BD Biosciences), 200 nM dNTPs, 50 pM of the appropriate primers and 1× Titanium Taq polymerase (BD Biosciences). PCR conditions were as follows: 1 cycle of 95°C/3 min; 5 cycles of 94°C/1 min, 50°C/2 min, 72°C/3 min; 25 cycles of 94°C/30 s, 54°C/1 min, 72°C/2 min; and 1 cycle of 72°C/5 min. The PCR products were electrophoresed in agarose, and the bands were eluted using the GeneClean Spin Kit. DNA from each band was then cloned into pCR2.1 using the TopoTA cloning kit (Invitrogen), transformed into DH5α chemically competent cells (Invitrogen), and plated onto LB ampicillin X-gal plates. About 3–6 independent white colonies were sequenced for each PCR product with oligonucleotides M13-F and M13-R (Table 1). Sequencing was performed by the Laboratory of Molecular Technology (SAIC Corp), and sequence analysis was carried out using Sequencher software (Gene Codes Corp).
Vectorette PCR
Vectorette PCR was done essentially as described (43). Briefly, the oligonucleotides HindIII-vec-top and HindIII-vec-bottom (Table 1) were annealed by mixing ∼5 pM of each oligonucleotide in 50 μl H2O and heating to 65°C for 5 min. MgCl2 was added to 2.5 mM and the oligonucleotides were allowed to cool slowly to room temperature. An aliquot of 1 μl of this mixture was ligated overnight to ∼2 μg of Hind III digested total genomic DNA from cells carrying the plasmid pAL301 candidates. PCR was carried out using 25 pM each of an anchor oligonucleotide, designated UMHis3SK-F (Table 1), and primer ‘bubble-amplify’ (Table 1). Primer ‘bubble-amplify’ can only anneal to the complement of the HindIII-vec-top primer. PCR conditions were as follows: 1 cycle of 95°C/5 min; 35 cycles of 94°C/30 s, 68°C/30 s, 72°C/45 s; and 1 cycle of 72°C/5 min. PCR amplified DNA was gel purified and the correct structure verified by performing hemi-nested PCR with the primers ‘bubble-amplify’ and UMHis3SK-G or UMHis3SK-H (Table 1). Gel purified DNA from the original PCR amplification reaction was cloned into the pCR2.1 Topo TA vector, and sequenced as described above.
RESULTS AND DISCUSSION
Many of the adverse biological consequences associated with palindromes are likely to be related to their propensity to form cruciform structures in vivo. These cruciform structures may pose significant barriers to normal DNA replication, and they may also be targets of specialized endonucleases such as the MR nuclease (see Introduction). Inverted repeats with very short (or no) spacers have a greater propensity to extrude and are therefore more likely to create genomic instability. Indeed, they are not stably maintained in E.coli cells (7). DNA binding proteins may stabilize palindromes to some extent in vivo, but the increased likelihood of intra-strand versus inter-strand annealing of a palindrome is a major barrier to in vitro molecular studies: no amount of exogenous primer can overcome the local concentration provided by the proximity of the adjacent repeat. Here, we describe experiments that have allowed us to overcome these barriers to the molecular analysis of long DNA palindromes.
Construction of palindromic plasmids
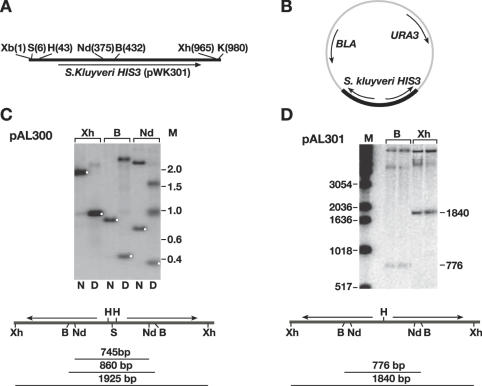
Figure 1A depicts the HIS3 region of pKW301 (39) used in the construction of the inverted repeats. S.kluyveri His3p can complement S.cerevisiae His3p, but the nucleotide sequences are not sufficiently homologous to allow recombination with the remaining endogenous his3-Δ1 sequence, a potential problem with unstable sequences. We constructed two related plasmids by in vitro ligation, depicted in Figure 1B–D. pAL300 (Figure 1C), containing an interrupted palindrome (961 bp/arm with a 2 bp spacer) was constructed by a three-body ligation between XhoI–SpeI and XbaI–KpnI fragments of pKW301 and XhoI–KpnI digested pRS426 (40). A related plasmid, pAL301 (Figure 1D), containing a perfect palindrome (922 bp/arm, no spacer) was similarly constructed by a three-body ligation between XhoI–HindIII and HindIII–KpnI fragments of pKW301 ligated to XhoI–KpnI digested pRS426. pAL301 is identical to pAL300 except that it is joined at the HindIII sites (designated ‘H’ in Figure 1), resulting in a perfect palindrome.
Figure 1.
Construction and analysis of palindromic plasmids. (A) Schematic representation of S.kluyverii HIS3 gene from pWK301 used in construction of palindromic plasmids. B, BstEII; H, HindIII; K, KpnI; S, SpeI; Xb, XbaI; Xh, XhoI. (B) Schematic representation of palindromic plasmids in vector pRS426. The palindromic region is indicated by heavy dark region. (C) Southern blot of a DNA digest of pAL300 (N, native; D, denatured). White dots denote the expected location of the native and self-annealed fragments for each digest. A schematic of the expected fragment sizes is shown below the blots. (D) Southern blots of two independent pAL301 candidates. For both blots the probe was the XhoI–KpnI fragment of pWK301 (M, 1 kb ladder, Invitrogen).
Propagation of palindromic plasmids in S.cerevisiae sae2 mutants
In E.coli, plasmids bearing long palindromes (>250 bp/arm) are highly unstable (7). The longest palindrome thus far isolated from E.coli was that reported by Malagon and Aguilera (8), where the authors were able to isolate a 1.1 kb/arm palindrome from sbcC mutant E.coli strains. However, the plasmid was not stable upon continued propagation (F. Malagon, personal communication). Although we did initially attempt to isolate palindromic plasmids by direct transformation of the ligation mixtures described above into sbcCD mutant E.coli, the recovery of plasmids with the correct structure was rare, and they were unstable (data not shown).
We previously found that sae2Δ yeast strains had high levels of chromosomal palindrome formation after the introduction of a DSB between inverted repeats (22). Our molecular analysis of these events indicated that the putative palindromes were stably maintained, and suggested that it might be possible to propagate plasmids containing artificially constructed palindromes in sae2 strains. We transformed the ligation mixtures described in the previous section directly into both wild-type and sae2Δ yeast strains, and selected for both the plasmid marker (URA3) and the marker within the palindrome (HIS3). Ura+ His+ transformants were further assessed for their inability to grow on 5-fluoroorotic acid (5-FOA) media lacking histidine. 5-FOA selects for Ura−cells, and as expected if the plasmid carries both the URA3 and HIS3 genes, all the cells that became Ura− were unable to grow on media lacking histidine. Twelve independent transformants for each ligation mixture were chosen for further analysis. Total DNA was isolated by the glass bead disruption method (see Materials and Methods), and examined by Southern blot to verify the palindromic structure. When the ligation mixtures were transformed into SAE2 wild-type cells, none of the few His+ Ura+ transformants had restriction fragments consistent with a palindromic structure, but instead appeared to be rearranged (data not shown). However, when the ligation mixtures were transformed into a sae2Δ strain, all 12 transformants of the ligation mixture for pAL300 (2 bp interrupted palindrome) and 11/12 transformants of the ligation mixture for pAL301 (perfect palindrome) had the expected restriction pattern for recovery of an intact palindromic structure (see Figure 1 and below). Notably, the DNA was isolated from cells that had been grown under conditions that only select for the plasmid backbone, not the insert. Thus, the palindrome is stably maintained in the absence of selection.
An example of a Southern blot analysis of DNA isolated by the glass bead disruption method from one of the pAL300 plasmids is shown in Figure 1C. When the cut DNA is denatured by boiling followed by quick cooling prior to loading on a native agarose gel, the palindromic fragment migrates at about half of the expected size, as would be expected for intra-strand annealing (see white dots in Figure 1C). A Southern blot digest of the native DNA from two pAL301 candidates is shown in Figure 1D.
PCR amplification of sodium-bisulfite-modified DNA
For sequence analysis of the palindromes, we used DNA preparations enriched for plasmid DNA from three independent S.cerevisiae pAL300 transformants and five independent pAL301 transformants (see Materials and Methods). DNA was digested with BstEII (see Figure 1C and D) and treated with sodium bisulfite as described in Materials and Methods.
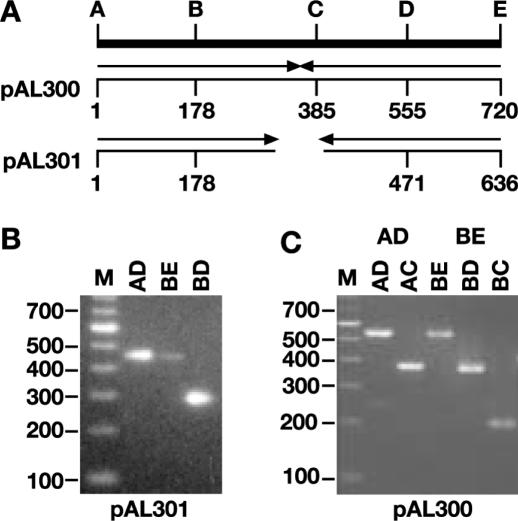
After sodium bisulfite treatment, the DNA was PCR amplified using oligonucleotides designed to prime only from the cytosine deaminated DNA. Table 1 shows the sequence of the oligonucleotides used for PCR amplification. The unmodified sequence at the same location is included in Table 1 for the purpose of comparison. The oligonucleotides are designated BiSuHis3SK-X, where ‘X’ refers to the designations (A–E) shown in Figure 2. Note that oligonucleotides A and B are designed to anneal directly to the modified DNA, whereas oligonucleotides C, D and E can only anneal to DNA that has been extended by primer A or B (Table 1 and see below). An example of the primary PCR amplification of pAL301 bisulfite-treated DNA with three different oligonucleotide pairs is shown in Figure 2B. Each of the PCR fragments was then gel purified and the correct structure further confirmed by PCR amplification with hemi-nested primers. An example of the hemi-nested PCR of pAL300 is shown in Figure 2C. As is evident, the hemi-nested primers do amplify appropriate-sized fragments from the primary PCR reaction.
Figure 2.
PCR amplification of bisulfite-modified DNA. (A) Schematic representation showing the locations of oligonucleotides used for amplification. The positions of the oligonucleotides (designated A–E) are shown for both pAL300 and pAL301 relative to oligonucleotide ‘A’, with the arrowheads indicating the approximate extent of the palindromic sequences. Oligonucleotide sequences are shown in Table 1 as BiSuSK-‘X’ where ‘X’ refers to the corresponding oligonucleotide shown here (A–E). (B) Primary PCR amplification of bisulfite-modified pAL301 with the indicated oligonucleotides (i.e. a PCR amplification with oligonucleotides B and C is designated ‘BC’). (C) Hemi-nested PCR amplification of pAL300 PCR products AD and BE with the indicated primers. (M—100 bp ladder, Invitrogen.)
Because S.cerevisiae does not have methylated cytosine, in theory all cytosines are substrates for deamination. However, in practice we find that the treatment with sodium bisulfite under the conditions used here results in deamination of 82–95% of all cytosines (see below). Since the modification is not complete, each PCR fragment contains a mixture of DNA molecules with different levels of modification. Therefore, it was necessary to clone the PCR products prior to sequence analysis.
Sequence analysis of sodium-bisulfite-modified DNA
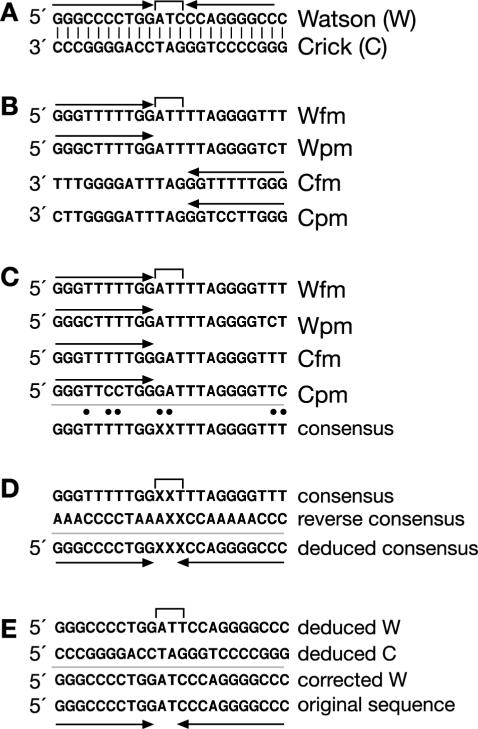
The sequence analysis of the sodium-bisulfite-modified palindromic DNA has some unique properties that both complicate and simplify the determination of the original sequence. An example meant to illustrate some of the issues that arise during the sequence analysis is shown in Figure 3. Figure 3A shows the expected central region of a hypothetical palindromic sequence with a 3 bp spacer. Fully (fm) and partially (pm) modified examples of the sequence are shown in Figure 3B.
Figure 3.
Analysis of bisulfite-modified palindromic DNA. (A) Expected unmodified sequence showing the central region of symmetry of a hypothetical palindromic sequence. The arrows above the sequence indicate the extent of the palindrome; W, Watson strand, C, Crick strand. (B) Examples of the products of PCR amplification of fully (fm) and partially (pm) modified sequence of the W and C strands. (C) Best alignment of sequences. Note that the 5′ ends of both W and C strands align. The ‘best’ consensus sequence of the alignment is shown below. ‘X’ indicates sequences that represent mispairs other than T/C. Dots indicate positions where there are ambiguities in the sequence alignment. (D) Alignment of the consensus sequence with the reverse complement of the consensus sequence allows determination of the original palindromic sequence (deduced sequence). (E) The sequence of the central spacer can be determined by examining the central base pairs in the original alignment (C), and aligning the sequences as either Watson or Crick (W and C chosen arbitrarily).
Normally Watson (W) strands of DNA only align with the complement of the Crick (C) strands. However, when a sequence is palindromic, a primer that can anneal to the right side of the W strand can also anneal to the left side of the C strand since the 5′ ends of both W and C strands are identical. This makes it difficult to determine which of the sequenced clones were originally derived from amplification of the W strand versus the C strand (Figure 3C).
However, one can still produce a ‘best alignment’ of the sequences, and generate a consensus sequence (Figure 3C). Two factors will contribute to heterogeneity in the sequence alignment (identified as dots above the consensus sequence). First, incomplete modification will lead to T/C ambiguities (assuming the clones are aligned by the G-rich strand as shown in Figure 3). Although this misalignment may help determine the residues that were originally C residues, the sequence can still be determined from fully modified DNA (see below). Second, spacer nucleotides between the palindromic sequences may show up as ambiguous bases by aligning residues other than T/C. This is useful in distinguishing clones that were originally of opposite polarities (W versus C; see below). For the purpose of generating the initial consensus sequence, one can designate the non T/C ambiguous residues as ‘X’ (Figure 3C). The consensus sequence thus determined is used to generate a ‘reverse consensus’ sequence (reverse complement of consensus), and the two sequences are aligned via A/G and C/T residues (Figure 3D). The original unmodified sequence is deduced by selecting the G for A/G aligned residues, and the C for C/T aligned residues. Sequences that were originally A (or T) residues will have an A (or T) at the same position in both the consensus and reverse consensus sequence (Figure 3D). The extent of the central spacer will be determined by the base pairs that have ‘X’ residues in the alignment.
The sequence of the central spacer can then be determined by returning to the original alignment (Figure 3C) to identify the residues corresponding to this region. The sequence in this region should be present in both polarities, representing the original W and C clones, allowing one to determine the deduced W and deduced C sequences. Any G/T misalignments then represent G/C residues where the C was modified. Thus, one is able to determine the correct unmodified sequence.
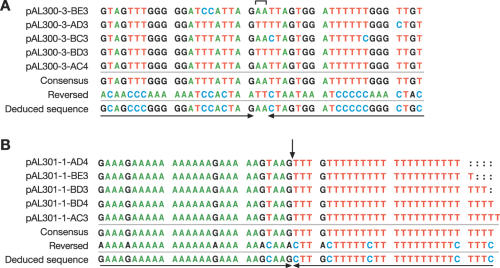
The DNA from three independently generated pAL300 candidates was treated with sodium bisulfite, and 3–5 independent Topo cloned bisulfite PCR products were sequenced for each clone. An example of five independent Topo clones of one of the pAL300 candidates is shown in Figure 4A. The bracket above the sequence alignment indicates the region of interrupted symmetry at the center of the palindrome. The consensus sequence is shown below the alignment, as is the reverse complement of the consensus sequence. The deduced sequence from this alignment was identical to the expected sequence for all three independent pAL300 isolates (not shown) indicating that the palindromes were indeed stably maintained in sae2Δ S.cerevisiae cells.
Figure 4.
Sequence alignment of sodium-bisulfite-modified palindrome clones. (A) Central portion of sequence from five clones of the bisulfite-modified PCR fragment of one of the independent pAL300 candidates. Only one strand is represented for each clone. The bracket at the top of the sequence alignment indicates the central non-palindromic bases. The clone designations indicate the pAL300 candidate number sequenced (i.e. clone 3) with the primers (A–E) used for amplification as designated in Figure 2, followed by the designated number of the particular Topo clone sequenced. A consensus sequence derived from the alignment of the various clones is shown below (consensus). By aligning the consensus with the reverse complement of the consensus sequence, we were able to correctly deduce the starting sequence of the palindrome. (B) Central portion of sequence from independent Topo clones for pAL301 candidate 1 aligned as described in (A). The arrow defines the center of symmetry.
A similar alignment for one of the pAL301 candidates is shown in Figure 4B. In this case, the center of symmetry (indicated by the arrow) is adjacent to a sequence that, prior to bisulfite modification, is a T-rich tract interrupted by occasional C residues (or on the opposite arm, an A-rich tract interrupted by G residues). When fully modified, this results in a stretch of T residues that is 23 bp long. Upon sequencing, we found that the clones had a variable number of T residues in this region (Figure 4B). We believe that the variability in the length of the T-tract is due to slippage during PCR amplification rather than actual changes in the sequence of the palindrome itself. As described above and in Figure 3, about one-half of the sequenced PCR products should be from the left arm of the W strand, and one-half should be from the right arm of the C strand. Notably, no alterations were detected in the length or sequence of the A-tract that is complementary to the slippery T-tract, presumably because it is stabilized by the presence of the G residues.
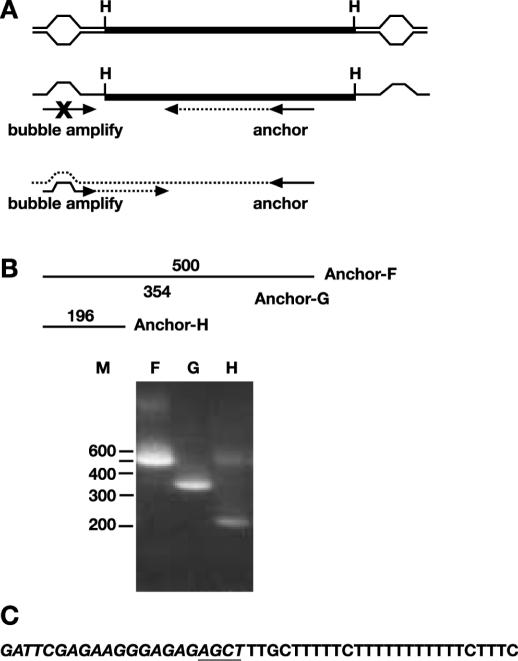
To further confirm that the pAL301 transformants did not contain alterations in the T-tract, we digested the DNA with HindIII (located at the center of symmetry) and cloned each of the palindrome arms by vectorette PCR as described in Materials and Methods. The vectorette oligonucleotides include mismatched bases in the central region (Figure 5A). PCR amplification by this method uses a specific oligonucleotide primer within the sequence of choice (anchor), and another primer that extends from the vectorette (bubble-amplify) (43). The vectorette PCR primer (bubble-amplify) is designed such that synthesis along the complementary strand of the vectorette (from the specific ‘anchor’ primer) is required before extension can proceed from the vectorette primer (Figure 5A). We further confirmed that the PCR fragments obtained by vectorette PCR had the expected structure by performing hemi-nested PCR with internal ‘anchor’ primers. An example of the hemi-nested PCR is shown in Figure 5B, indicating that the sequence does have the expected structure. Six independent colonies from the primary vectorette PCR reactions were sequenced for each of the three independent pAL301 transformants. In all cases, the sequence was identical to the expected sequence, the central portion of which is shown in Figure 5C. These data indicate that there is no variability in the length of the T-tract in the palindromic plasmids isolated from yeast. This method will be useful for confirming the sequences at the center of unknown palindromes; however, it does require that there be a restriction site present in the central portion of the palindrome. Importantly, these results indicate that a plasmid bearing a perfect palindrome is stably maintained in sae2Δ S.cerevisiae strains. To our knowledge, this is the first description of stable maintenance of a perfect palindrome.
Figure 5.
Vectorette PCR of pAL301. (A) Schematic representation of vectorette PCR showing location of anchor-F primer (UMHis3SK-F, Table 1). Note that the primer ‘bubble-amplify’ only hybridizes to the complement of the top strand of the vectorette bubble, and therefore can only be amplified after DNA synthesis (dotted lines) has been extended from the anchor primer. (B) Schematic representation showing approximate locations of internal primers. Primer sequences are shown in Table 1 designated UMHis3SK-X, where X refers to the primer shown here (F, G or H). Gel showing hemi-nested PCR of the product derived from PCR with the primers ‘anchor F’ and ‘bubble-amplify’. (C) Relevant sequence of vectorette clones. The sequence of the vectorette bubble primers are shown in italics, with the AGCT of the HindIII (H) site used for ligation of the bubble primers underlined.
CONCLUSIONS
We show here that long palindromes, either with or without a short central spacer, can be stably maintained in S.cerevisiae strains lacking the sae2/com1 gene. Although the sequences used here are not mammalian in origin, the observation that closely spaced Alu repeats are stabilized in sae2Δ cells (21) indicates that there is unlikely to be any species specificity to the nature of the sequences stabilized in sae2Δ cells.
In this study, we have analyzed artificially constructed palindromes to develop a method for sequencing long palindromic DNAs. The presence of the selectable marker was useful in identifying the palindrome-containing plasmids. However, once a palindromic plasmid was identified, the palindromic plasmids were stably maintained in absence of selection for the marker (see above). Therefore, by using TAR cloning methodology (35), which does not require selection for the insert, it should be possible to clone palindromic sequences in the absence of selection.
Treatment of plasmid DNA isolated from yeast cells with sodium bisulfite has allowed us to PCR amplify and sequence palindromic regions. Taken together, these techniques should provide the necessary tools to characterize palindromic regions from mammalian cells. This may allow the elucidation of events that lead to genome amplification, or perhaps even the ability to isolate and sequence some of the gaps from the human genome.
Acknowledgments
ACKNOWLEDGEMENTS
I wish to thank Susanna Lewis for suggesting this approach and to Jeff Strathern for providing his enthusiastic support for this project. I also thank Francisco Malagon, Sharon Moore and Anne Welcker for critical review of the manuscript, and Marge Mills for administrative assistance. This work was sponsored by the National Cancer Institute, Department of Health and Human Services.
REFERENCES
- 1.Collick A., Drew,J., Penberth,J., Bois,P., Luckett,J., Scaerou,F., Jeffreys,A. and Reik,W. (1996) Instability of long inverted repeats within mouse transgenes. EMBO J., 15, 1163–1171. [PMC free article] [PubMed] [Google Scholar]
- 2.Akgun E., Zahn,J., Baumes,S., Brown,G., Liang,F., Romanienko,P.J., Lewis,S. and Jasin,M. (1997) Palindrome resolution and recombination in the mammalian germ line. Mol. Cell. Biol., 17, 5559–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis S., Akgun,E. and Jasin,M. (1999) Palindromic DNA and genome stability. Further studies. Ann. N.Y. Acad. Sci., 870, 45–57. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham L.A., Cote,A.G., Cam-Ozdemir,C. and Lewis,S.M. (2003) Rapid, stabilizing palindrome rearrangements in somatic cells by the center-break mechanism. Mol. Cell. Biol., 23, 8740–8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lobachev K.S., Shor,B.M., Tran,H.T., Taylor,W., Keen,J.D., Resnick,M.A. and Gordenin,D.A. (1998) Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics, 148, 1507–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordenin D.A., Lobachev,K.S., Degtyareva,N.P., Malkova,A.L., Perkins,E. and Resnick,M.A. (1993) Inverted DNA repeats: a source of eukaryotic genomic instability. Mol. Cell. Biol., 13, 5315–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leach D.R. (1994) Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays, 16, 893–900. [DOI] [PubMed] [Google Scholar]
- 8.Malagon F. and Aguilera,A. (1998) Genetic stability and DNA rearrangements associated with a 2 × 1.1-Kb perfect palindrome in Escherichia coli. Mol. Gen. Genet., 259, 639–644. [DOI] [PubMed] [Google Scholar]
- 9.Nag D.K. and Kurst,A. (1997) A 140-bp-long palindromic sequence induces double-strand breaks during meiosis in the yeast Saccharomyces cerevisiae. Genetics, 146, 835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farah J.A., Hartsuiker,E., Mizuno,K., Ohta,K. and Smith,G.R. (2002) A 160-bp palindrome is a Rad50.Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics, 161, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nasar F., Jankowski,C. and Nag,D.K. (2000) Long palindromic sequences induce double-strand breaks during meiosis in yeast. Mol. Cell. Biol., 20, 3449–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ford M. and Fried,M. (1986) Large inverted duplications are associated with gene amplification. Cell, 45, 425–430. [DOI] [PubMed] [Google Scholar]
- 13.Hyrien O., Debatisse,M., Buttin,G. and de Saint Vincent,B.R. (1987) A hotspot for novel amplification joints in a mosaic of Alu-like repeats and palindromic A + T-rich DNA. EMBO J., 6, 2401–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka H., Tapscott,S.J., Trask,B.J. and Yao,M.C. (2002) Short inverted repeats initiate gene amplification through the formation of a large DNA palindrome in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 8772–8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Z.H., Akgun,E. and Jasin,M. (2001) Repeat expansion by homologous recombination in the mouse germ line at palindromic sequences. Proc. Natl Acad. Sci. USA, 98, 8326–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edelmann L., Spiteri,E., Koren,K., Pulijaal,V., Bialer,M.G., Shanske,A., Goldberg,R. and Morrow,B.E. (2001) AT-rich palindromes mediate the constitutional t(11;22) translocation. Am. J. Hum. Genet., 68, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henthorn P.S., Mager,D.L., Huisman,T.H. and Smithies,O. (1986) A gene deletion ending within a complex array of repeated sequences 3′ to the human beta-globin gene cluster. Proc. Natl Acad. Sci. USA, 83, 5194–5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran H.T., Degtyareva,N.P., Koloteva,N.N., Sugino,A., Masumoto,H., Gordenin,D.A. and Resnick,M.A. (1995) Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and the RAD50 and RAD52 genes. Mol. Cell. Biol., 15, 5607–5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobachev K.S., Stenger,J.E., Kozyreva,O.G., Jurka,J., Gordenin,D.A. and Resnick,M.A. (2000) Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J., 19, 3822–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Achaz G., Coissac,E., Netter,P. and Rocha,E.P. (2003) Associations between inverted repeats and the structural evolution of bacterial genomes. Genetics, 164, 1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lobachev K.S., Gordenin,D.A. and Resnick,M.A. (2002) The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell, 108, 183–193. [DOI] [PubMed] [Google Scholar]
- 22.Rattray A.J., McGill,C.B., Shafer,B.K. and Strathern,J.N. (2001) Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics, 158, 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Connelly J.C. and Leach,D.R. (2002) Tethering on the brink: the evolutionarily conserved Mre11–Rad50 complex. Trends Biochem. Sci., 27, 410–418. [DOI] [PubMed] [Google Scholar]
- 24.Trujillo K.M. and Sung,P. (2001) DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J. Biol. Chem., 276, 35458–35464. [DOI] [PubMed] [Google Scholar]
- 25.Paull T.T. and Gellert,M. (1998) The 3″ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell, 1, 969–979. [DOI] [PubMed] [Google Scholar]
- 26.Moreau S., Ferguson,J.R. and Symington,L.S. (1999) The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol., 19, 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Amours D. and Jackson,S.P. (2002) The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nature Rev. Mol. Cell Biol., 3, 317–327. [DOI] [PubMed] [Google Scholar]
- 28.Keeney S. and Kleckner,N. (1995) Covalent protein–DNA complexes at the 5′ strand termini of meiosis-specific double-strand breaks in yeast. Proc. Natl Acad. Sci. USA, 92, 11274–11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKee A.H. and Kleckner,N. (1997) A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics, 146, 797–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prinz S., Amon,A. and Klein,F. (1997) Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics, 146, 781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neale M.J., Ramachandran,M., Trelles-Sticken,E., Scherthan,H. and Goldman,A.S. (2002) Wild-type levels of Spo11-induced DSBs are required for normal single-strand resection during meiosis. Mol. Cell, 9, 835–846. [DOI] [PubMed] [Google Scholar]
- 32.Laird C.D., Pleasant,N.D., Clark,A.D., Sneeden,J.L., Hassan,K.M., Manley,N.C., Vary,J.C., Jr., Morgan,T., Hansen,R.S. and Stoger,R. (2004) Hairpin-bisulfite PCR: assessing epigenetic methylation patterns on complementary strands of individual DNA molecules. Proc. Natl Acad. Sci. USA, 101, 204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark S.J., Harrison,J., Paul,C.L. and Frommer,M. (1994) High sensitivity mapping of methylated cytosines. Nucleic Acids Res., 22, 2990–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warnecke P.M., Stirzaker,C., Song,J., Grunau,C., Melki,J.R. and Clark,S.J. (2002) Identification and resolution of artifacts in bisulfite sequencing. Methods, 27, 101–107. [DOI] [PubMed] [Google Scholar]
- 35.Noskov V.N., Kouprina,N., Leem,S.H., Ouspenski,I., Barrett,J.C. and Larionov,V. (2003) A general cloning system to selectively isolate any eukaryotic or prokaryotic genomic region in yeast. BMC Genomics, 4, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K.(eds) (1994) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY. [Google Scholar]
- 37.Sherman F., Fink,G. and Hicks,J. (1986) Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 38.Ito H., Fukada,Y., Murata,K. and Kimura,A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol., 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinstock K.G. and Strathern,J.N. (1993) Molecular genetics in Saccharomyces kluyveri: the HIS3 homolog and its use as a selectable marker gene in S. kluyveri and Saccharomyces cerevisiae. Yeast, 9, 351– 361. [DOI] [PubMed] [Google Scholar]
- 40.Sikorski R. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffman C. and Winston,F. (1987) A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene, 57, 262–272. [DOI] [PubMed] [Google Scholar]
- 42.Singh M.V. and Anthony Weil,P. (2002) A method for plasmid purification directly from yeast. Anal. Biochem., 307, 13–17. [DOI] [PubMed] [Google Scholar]
- 43.Riley J., Butler,R., Ogilvie,D., Finniear,R., Jenner,D., Powell,S., Anand,R., Smith,J.C. and Markham,A.F. (1990) A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res., 18, 2887–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]