Abstract
Using the rat insulinoma cell line INS-1 we generated β-cell clones that are most efficient for gene transfer, as they contain an FRT site for Flp recombinase-mediated, site-directed integration of a single copy transgene. Therefore, the gene-of-interest can be introduced by DNA transfection without the need to select individual cell clones. Additionally, the clones contain the tetracycline repressor allowing tetracycline induction of the transgene. By oligonucleotide microarray we define the β-cell specific phenotype of the Flp-In T-REx cell clones. Using a clone expressing the HNF6, HNF4α and HNF1β transcription factors at a limited level, we introduced the expression vectors encoding these factors. We show efficient tetracycline induction of these transcription factors by western blots and immunocytochemistry. Microarrays reveal that these three factors affect a similar number of genes with only few genes regulated in common. Statistical analysis reveals that the three transcription factors affect genes categorized to different biological processes. Furthermore, we document the usefulness of these Flp-In T-REx cells for the functional analysis of mutated HNF1β transcription factors found in human MODY5 patients. We show that the expression of the mutant P328L329del and A263insGG affects only very few transcripts and these are predominantly distinct from those induced by wild-type HNF1β.
INTRODUCTION
Pancreatic β-cells represent a highly differentiated cell type with specific properties, most notably regulated secretion of insulin. Analysis of specific gene activities in β-cells within the entire organism is difficult, as β-cells are one of the four principal endocrine cell types of the islets of Langerhans. Furthermore, these endocrine cell clusters are scattered throughout the pancreas requiring purification for detailed biochemical analysis (1).
As an alternative, several cell lines have been established that retain β-cell specific properties (2). These homogeneous cell populations are a most useful model to analyze the function of specific genes in differentiated β-cells. In addition, these differentiated β-cell lines are also a most valuable system to analyze β-cell renewal, since it has been shown recently that β-cell lineage renewal may involve mature β-cells and not only undifferentiated pancreatic stem cells as previously suggested (3). As disturbed renewal of β-cells in the pancreas may be a major step leading to diabetes mellitus, differentiated β-cells are a valid model to study β-cell dysfunction. The functional analysis of MODY genes whose dysfunction in β-cells causes maturity onset diabetes of the young is of special interest. Five of the six MODY genes identified are the transcription factors, HNF4α (MODY1), HNF1α (MODY3), PDX1/IPF1 (MODY4), HNF1β (MODY5) and NeuroD1/Beta2 (MODY6) (4–6). These transcription factors are assumed to affect the activity of many genes in the pancreatic β-cells, the majority of which have not yet been identified. Most recently, a systematic analysis has been reported in human pancreatic islets using chromatin immunoprecipitation combined with promoter microarrays (7). This study revealed several hundred gene promoters occupied by the HNF1α, HNF4α and HNF6 transcription factors. However, this approach is limited, as it does not give any information on whether a transcription factor acts positively or negatively on the corresponding promoter.
So far, functional analysis of gene regulation has relied mainly on the description of the expression level of a few selected genes. Recently, microarray techniques have been developed that allow a genome-wide analysis of gene function by the simultaneous measurement of thousands of RNA transcripts (8). Thus, the influence of a given transcription factor on the activity of thousands of genes can be analyzed. Furthermore, it allows the comparison of gene expression patterns induced by different transcription factors. However, this analysis requires a cell population of high purity to get a sensitive and meaningful assay.
Stable integration of transcription factors into cells is a widely used approach to analyze gene function in mammalian cells. As the introduction of genes by DNA transfection or viral transduction targets usually only a minority of the cells, the selection of cell lines with stably integrated transgenes is required to get a pure cell population. There are several major drawbacks to this technique. First, the expression of a gene may inhibit cell multiplication, precluding the establishment of stable cell lines. This can be circumvented by the use of conditional systems, whereby the gene is introduced in a silent state and activated after the establishment of the cell line (9). Second, using standard techniques of DNA transfection, a heterogenous population of cells is obtained that differs in the amount of integrated copies and thus in the level of transgene expression (10). This disadvantage can be overcome by using retroviral vectors that result in single-copy integration, but this approach involves more work. Third, DNA transfection as well as retroviral transduction result in random integration of the transgene and the chromosomal sites differ from cell to cell (10). Since the chromosomal context of the integration site largely defines the activity of the integrated gene, cell clone dependent activity of the introduced gene is observed. Therefore, in many applications the analysis of several cell clones is needed to get significant data resulting in a most labour intensive study. This major drawback can be overcome by using the Flp-In method, an Flp recombinase-mediated, site-directed transgene integration (Invitrogen). In this approach an FRT site sequence is integrated into the host cell that is recombined by Flp recombinase with the FRT site of the vector containing the gene to be inserted.
INS-1 is a frequently used pancreatic β-cell line that has been isolated from a rat insulinoma. This cell line has retained many properties of β-cells including glucose induced insulin secretion (11). The cell line has been used extensively to introduce specific genes under the control of the tetracycline repressor allowing conditional expression of the gene-of-interest (GOI) (12–19). However, as the transgenes were inserted by DNA transfection at random, extensive clonal differences occur. To improve the value of the INS-1 cells for functional gene assays, we have established the Flp-In system in the insulinoma cell line INS-1. We generated four independent cell clones containing a single FRT site and representing highly differentiated β-cells. One of these cell clones was used for the conditional expression of transcription factors known to play a role in β-cells. By microarray analysis we compared the pattern of genes affected by the HNF6, HNF4α and HNF1β transcription factors in pancreatic β-cells. In addition, we explored the effect of two HNF1β mutations on the gene expression pattern.
MATERIALS AND METHODS
Plasmid constructs
The Flp expression vector used (pCSFLPe) has been described previously (20), whereas the pFRT/lacZeo2, pcDNA6/TR and pcDNA5/FRT/TO vectors were obtained from Invitrogen.
To assay the tetracycline repressor activity in the Flp-In cell clones the luciferase gene was taken from the pGL3-Basic vector (Promega) as NheI/XbaI fragment and cloned into the HindIII and BamHI sites of pcDNA5/FRT/TO using as intermediate a multiple cloning site vector containing BglII and HindIII sites. This FRluc plasmid was used in transient transfection assays and the luciferase activity was measured as described previously (21).
For the site specific integration of the genes encoding the HNF6, HNF4α and HNF1β transcription factors the corresponding open reading frame (ORF) of the human cDNA was inserted into the pcDNA5/FRT/TO vector. The human HNF6 ORF was amplified by PCR from cDNA made on HepG2 RNA using the oligonucleotides 5′-CCCAAGCTTTGATAGACATGAACGCGCAGCTGAC-3′ and 5′-ATAGTTTAGCGGCCGCAAGCTTTTCCACCGAGG-3′ as forward and reverse primers, respectively. Subsequently, the ORF was cloned as a HindIII fragment into the pcDNA5/FRT/TO vector containing a histidine tag. The histidine tag was inserted as a DNA linker by two oligonucleotides (5′-AGCTATCGATACCATGCGGGGTTCTCATCATCATCATCATCATGGTGA-3′ and 5′-AGCTTCACCATGATGATGATGATGATGAGAACCCCGCATGGTATCGAT-3′) that recreated a HindIII site at the C-terminus of the tag exclusively. The sequence of the final construct was verified by sequencing.
For HNF4α, the human cDNA representing the splice variant α2 (22) was derived as NotI fragment from the pOP-mycHNF4α expression vector (23) and cloned into the NotI site of the pcDNA5/FRT/TO vector to produce the myc tagged HNF4α.
For HNF1β, the human cDNA sequence was first inserted as EcoRI/XbaI fragment from the myc-Rc/CMV expression vector (24) into the pCS2+MT expression vector (25). Subsequently, the myc tagged HNF1β sequence was cloned as BlnI/XbaI fragment into pBluescript II SK+ (Stratagene) and the NotI/BamHI fragment inserted into the pDNA5/FRT/TO vector. The vectors encoding the P328L329del and the A263insGG mutations of HNF1β were prepared correspondingly using the myc-Rc/CMV expression vectors (24,26).
Establishment of the INS-1 Flp-In T-REx cells
The INS-1 cell line was obtained from Andreas Pfeiffer and grown at 37°C in RPMI-1640 medium supplemented with 10% heat inactivated fetal calf serum, penicillin (100 U/ml), streptomycin (100 U/ml), 1 mM sodium pyruvate, 10 mM HEPES, 2 mM glutamine and 50 μM mercaptoethanol. The method to establish the cell clones was essentially done as described in the user's manual of the Flp-In T-REx Core Kit (Invitrogen Catalog no. K6500-01) using transfection with lipofectamine (Invitrogen). pFRT/lacZeo2 was linearized with ScaI and Zeocin™ selection was done at 200 μg/ml. pcDNA6/TR was linearized with SapI and blastidin selection was done at 10 μg/ml in the presence of Zeocin™ (200 μg/ml). Flp-mediated integration of the pcDNA5/FRT/TO vector was selected with hygromycin at 150 μg/ml in the presence of blastidin (10 μg/ml) and in the absence of Zeocin™.
Western blots and immunofluorescence
For Western blots the anti-myc tag antibody (9E10) and the anti-His tag antibody (27) was used for myc and histidine tagged proteins, respectively. Peroxidase coupled sheep anti-mouse immunoglobulin antibody (Amersham, HRP-linked whole Ab, NXA931) was used as secondary antibody and detected using the ECL system (Amersham). For immunofluorescence the Cy3-conjugated rat anti-mouse antibody [Dianova, F(ab′)2-fragment, #415-166-166] or the FITC-conjugated donkey anti-mouse antibody [Dianova, F(ab′)2-fragment, #715-096-151] were used. To detect insulin, the guinea pig anti-insulin antibody (DAKO, A0564) was used together with the donkey anti-guinea pig Cy3 F(ab′)2-fragment (Dianova, #706-166-148).
Oligonucleotide microarray analysis
For microarray analysis, we used the Affymetrix GeneChip platform employing a standard protocol for sample preparation and microarray hybridization that has been described in detail previously (28). Briefly, total RNA was extracted by means of Trizol and converted into double-stranded cDNA using an oligo-deoxythymidine primer containing the T7 RNA polymerase binding site [5′-GCATTAGCGGCCGCGAAATTAATACGACTCACTATAGGGAGA-(dT)24-3′] (MWG Biotech, Munich, Germany) for first strand synthesis. After generation of double-stranded cDNA from the first-strand cDNA, biotinylated cRNA was synthesized by in vitro transcription using the BioArray High Yield RNA Transcript Labeling Kit (Enzo Diagnostics, New York, NY). Labeled cRNA was purified on RNeasy columns (Qiagen, Hilden, Germany), fragmented and hybridized to RAE230A microarrays (Affymetrix, Santa Clara, CA). The arrays were washed and stained according to the manufacturer's recommendation and finally scanned in a GeneArray scanner 2500 (Agilent, Palo Alto, CA).
Arrays images were processed to determine signals and detection calls (present, absent, marginal) for each probeset using the Affymetrix Microarray Suite 5.0 software (MAS 5.0; statistical algorithm). Scaling across all probesets of a given array to an average intensity of 1000 was performed to compensate for variations in the amount and quality of the cRNA samples and other experimental variables of non-biological origin. Pairwise comparisons of induced versus uninduced samples were carried out with MAS 5.0, which calculates the significance (change P-value) of each change in gene expression based on a Wilcoxon ranking test. For each transcription factor introduced into the INS-1 cell line two independent cell lines were analyzed. To limit the number of false positives, we restricted further target identification to those probesets which received at least one present detection call in the uninduced/induced sample pair. Probesets exhibiting a signal log2 ratio >1.32 and a change P-value <0.001 or a signal log2 ratio <−1.32 and a change P-value >0.999 (corresponding to 2.5-fold upregulation or downregulation) were identified by filtering using the Affymetrix Data Mining Tool 3.0 and only those passing these cut-offs in both independent cell lines were considered as targets. The microarray data has been submitted to the GEO repository (http://www.ncbi.nlm.nih.gov/geo/) and the accession numbers are GSE1589, GSE1590 and GSE1591.
Real-time RT–PCR
TaqMan probes for the rat HNF6, HNF4α and HNF1β transcripts were Rn00575362_m1, Rn00573309_m1 and Rn00447453_m1, respectively, from the TaqManR Gene Expression Assays (Applied Biosystems) and used under standard condition according to the manufacturer's recommendation using the ABI PRISMR 7900HT Sequence Detection System (Applied Biosystems). cDNA was made with random primers using the High Capacity cDNA Archive Kit (Applied Biosystems, P/N 4322171) using 1 μg RNA in a 20 μl reaction volume. Real-time PCR were measured in four dilutions in triplicate. At all dilutions a significant increase was observed except for HNF1β probes where only the lowest dilution (1:2.5) gave a response.
RESULTS
Generating inducible INS-1 Flp-In T-REx host cell lines
To generate pancreatic β-cells with stable, inducible expression of defined genes we established the Flp-In™ T-REx™ system from Invitrogen in the rat insulinoma cell line INS-1. As the integration of the GOI is mediated by Flp recombinase at the FRT site of a defined genomic location, cell clones are highly identical and thus there is no need to pick individual cell clones. Furthermore, the presence of the tetracycline repressor in the host cell allows tetracycline induction of the GOI.
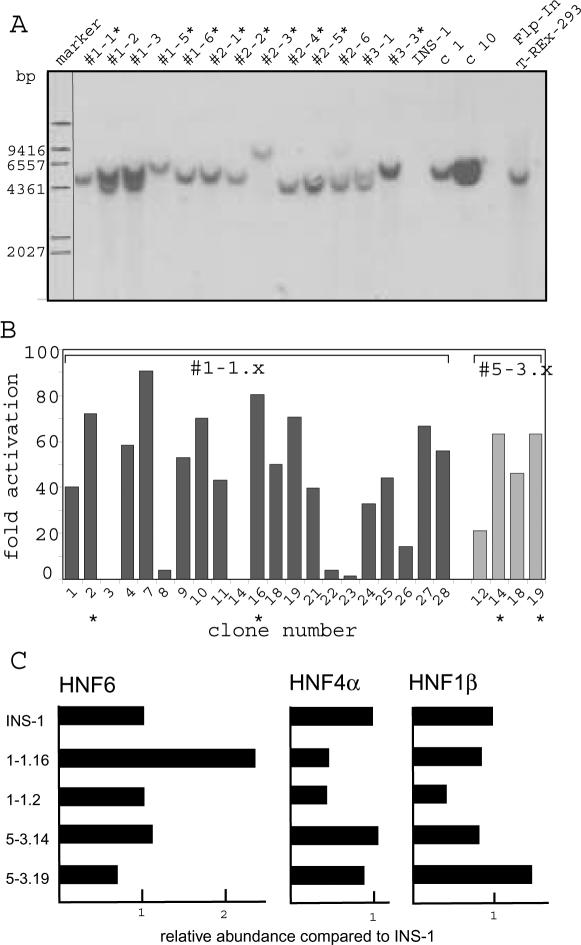
To establish the INS-1 Flp-In T-REx cell lines expressing a defined transcription factor, three rounds of stable transfection were required (Figure 1). In a first step, we introduced the plasmid containing the FRT site (pFRT/lacZeo2) using Zeocin selection. We screened 25 individual Zeocin resistant cell clones for a single integrated FRT site by Southern blot analysis as shown in Figure 2A. Nine cell clones containing a single copy integration (marked by an asterisk) were analyzed for expression of the transfected lacZ–Zeocin fusion gene by the β-galactosidase assay to select clones with a highly and uniformly active integration site (data not shown). Furthermore, we selected clones with high insulin expression to retain the differentiated β-cell phenotype (see below). Using both these criteria we chose clone #1-1 and #5-3 to integrate the tetracycline repressor using the expression vector pcDNA6/TR (step 2 in Figure 1). Cell clones were now selected with blasticidin, and individual clones were screened for tetracycline repressor activity by a luciferase reporter (FRluc) containing a Tet operator in the promoter. As shown in Figure 2B, most clones had an increased luciferase activity upon tetracycline induction. Four clones (#1-1.2, #1-1.16, #5-3.14 and #5-3.19; marked by asterisks in Figure 2B) that showed a high tetracycline-inducible luciferase activity and retained high insulin expression comparable to the original INS-1 cell line were chosen (see below). We refer to these cell clones as INS-1 Flp-In T-REx cells.
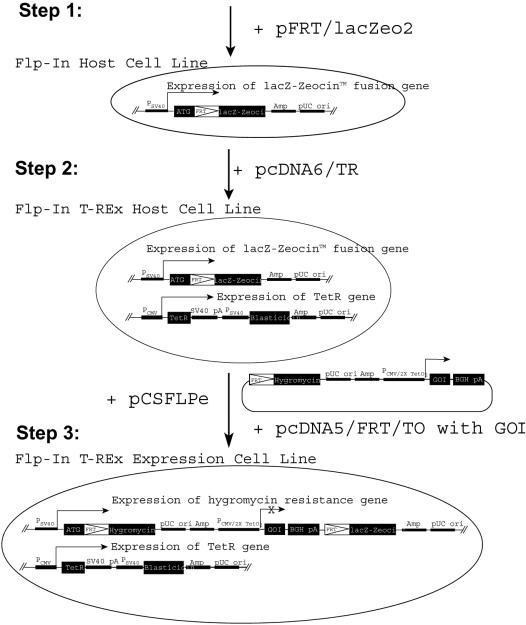
Figure 1.
Generating stable, inducible INS-1 cell lines by Flp recombinase-mediated integration. The generation of the Flp-In T-REx cells involves three integration steps that are schematically illustrated. In the first step, the Flp recognition target (FRT) is integrated by transfection of the FRT/lacZeo2 plasmid. The use of the lacZ–Zeocin fusion gene allows selection for Zeocin resistant cell clones and the lacZ portion is used to characterize the integration site by scoring the β-galactosidase activity. In the second step, an expression vector (pcDNA6/TR) encoding the tetracycline repressor (TetR) is introduced using blasticidin resistance. In the third step, the GOI (gene-of-interest) is introduced by Flp recombinase activity from the pCSFLPe plasmid using the pcDNA5/FRT/TO vector containing the GOI. This construct contains an FRT site in front of the hygromycin ORF that will generate hygromycin resistance only, if a site-directed integration occurs at the FRT site downstream of the ATG of the lacZ–Zeocin cassette. By this recombination, the GOI present in the pcDNA5/FRT/TO vector is integrated downstream of the FRT integration site. As the GOI is driven by the tetracycline operator controlled CMV promoter (PCMV/2TetO2), tetracycline regulation can be obtained. PSV40 (SV40 promoter), Amp (ampiciline resistance), pUCori (bacterial origin of replication), PCMV (CMV promoter), SV40pA (SV40 polyadenylation), BGHpA (bovine growth hormone polyadenylation site). For further details see Flp-In™ system manual (Invitrogen).
Figure 2.
Southern blot analysis and tetracycline regulation of Flp-In cell lines. (A) DNA of cell clones obtained after transfection of the FRT site (step 1 in Figure 1) was digested with NcoI and analyzed by Southern blot using a probe specific for FRT/lacZeo2. Clone numbers are given and the INS-1 parental cell line and the Flp-In™ T-REx™ HEK293 cell line (Invitrogen) are included as negative and positive control, respectively. (B) Individual clones containing the tetracycline repressor gene (step 2 in Figure 1) were transfected with a luciferase reporter gene (FRluc) driven by the tetracycline operator controlled CMV promoter (PCMV/2TetO2). The value given represents the fold-activation observed at the addition of tetracycline. Clones, derived from the two independent clones #1-1 and #5-3 from step 1, were analyzed. (C) The abundance of RNA encoding HNF6, HNF4α ad HNF1β in the INS-1 Flp-In cell clones. Using TaqMan probes the amount of RNA was quantified. 18S RNA was used as standard between the various samples.
To compare the gene expression profiles of the four different INS-1 Flp-In T-REx cell clones (#1-1.2, #1-1.16, #5-3.14 and #5-3.19) and the parental INS-1 cell line, we performed an oligonucleotide microarray analysis using the Affymetrix GeneChip RAE230A containing >15 000 probesets. Comparing the expression pattern of the four INS-1 Flp-In T-REx cell lines and the parental INS-1 cells, a similar expression profile was observed. However, clustering of the gene expression data showed that the clones #1-1.2 and #1-1.16, arising from the same INS-1 Flp-In cell clone #1-1, showed a high degree of similarity as well as the clones #5-3.14 and #5-3.19, derived from the INS-1 Flp-In #5-3 clone (data not shown). To validate the four INS-1 Flp-In T-REx cell lines with respect to their properties as differentiated pancreatic β-cells, we have listed in Table 1 the expression of some transcription factors (upper part) and of some other genes (lower part) in these cell lines and the original INS-1 cell line. All cell lines showed significant expression levels of the transcription factors Cdx2, Foxa2 (HNF3β), Foxa3 (HNF3γ), Isl1, NeuroD1 (BETA2), Nkx6a, Pax6, Pdx1 (IPF1) and TCF1 (HNF1α), whereas the genes encoding Foxa1 (HNF3α), Ngn3, Pax4, Ptf1a and TCF2 (HNF1β) were not expressed. In case of HNF4α, probeset- and cell clone-dependent differences were observed with consistent activity in clones #5-3.14 and #5-3.19. The expression of onecut1 (HNF6) transcription factor was low with some significant activity detected in #1-1.16 and #5-3.19. Analyzing the expression of genes encoding proteins distinct from transcription factors, no large qualitative differences between the various cell clones were observed. Although genes known to be expressed in β-cells (Abcc8, Cpe, Gcg, Gck, Glp1r, Ins1, Ins2, Kcnj11 and Slc2a2) were reliably detected, genes known to be expressed in exocrine cells of the pancreas (amy1, Prss1, Prss2) or in δ-cells (somatostatin) were not active. However, we noticed substantial quantitative differences in the extent of expression of some genes between the various cell clones, e.g. glucagon (Gcg) and insulin 2 (Ins2). Considering the absolute transcript level, the expression of the insulin 1 (ins1) gene was the highest of all genes analyzed and similar to the ribosomal protein S23. This illustrates the strong β-cell phenotype of the INS-1 Flp-In T-REx cell lines. In conclusion, all cell clones have retained a β-cell specific expression pattern, but there are clone-specific differences.
Table 1. β-cell-specific phenotype of the INS-1 Flp-In T-REx cells.
| Gene symbol | Probeset ID | INS-1 | # 1-1.16 | # 1-1.2 | # 5-3.14 | # 5-3.19 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cdx2 (Cdx3) | 1387726_at | 253 | P | 278 | P | 278 | P | 265 | P | 370 | P |
| Foxa1 (Hnf3α) | 1369834_at | 5 | A | 21 | A | 14 | A | 5 | A | 11 | A |
| Foxa2 (Hnf3β) | 1368711_at | 4459 | P | 3385 | P | 3095 | P | 3328 | P | 2892 | P |
| Foxa3 (Hnf3γ) | 1387506_at | 204 | P | 436 | P | 313 | P | 410 | P | 498 | P |
| Hnf4a (MODY1) | 1382496_at | 186 | M | 38 | A | 139 | A | 244 | P | 134 | P |
| Hnf4a (MODY1) | 1369289_at | 433 | P | 121 | A | 150 | A | 404 | P | 362 | P |
| Isl1 | 1369681_at | 2544 | P | 1148 | P | 1987 | P | 3029 | P | 2036 | P |
| Neurod1 (BETA2/MODY6) | 1387288_at | 8773 | P | 2790 | P | 6927 | P | 6392 | P | 5773 | P |
| Ngn3 (neurogenin) | 1369853_at | 28 | A | 17 | A | 10 | A | 7 | A | 27 | A |
| Nkx6a | 1368998_at | 2220 | P | 1599 | P | 1053 | P | 1031 | P | 2158 | P |
| Onecut1 (HNF6) | 1387760_a_at | 99 | A | 84 | A | 97 | A | 39 | A | 49 | A |
| Onecut1 (HNF6) | 1371034_at | 94 | M | 157 | P | 51 | A | 76 | A | 82 | P |
| Pax4 | 1370140_a_at | 103 | A | 91 | A | 105 | A | 107 | A | 120 | M |
| Pax6 | 1369242_at | 8395 | P | 4132 | P | 7043 | P | 8947 | P | 9168 | P |
| Pdx1 (IPF1/MODY4) | 1375877_at | 1458 | P | 1518 | P | 1063 | P | 1624 | P | 1641 | P |
| Pdx1 (IPF1/MODY4) | 1369516_at | 2830 | P | 1454 | P | 2343 | P | 2845 | P | 2185 | P |
| Ptf1a (p48) | 1369803_at | 69 | A | 180 | A | 45 | A | 136 | A | 96 | A |
| Tcf1 (HNF1α/MODY3) | 1369148_at | 481 | P | 604 | P | 585 | P | 479 | P | 826 | P |
| Tcf2 (HNF1β/MODY5) | 1369682_at | 23 | A | 30 | A | 43 | A | 20 | A | 44 | A |
| Abcc8 (SUR1) | 1369632_a_at | 2573 | P | 1999 | P | 3012 | P | 2355 | P | 2474 | P |
| Amy1 (amylase) | 1370359_at | 34 | A | 55 | A | 9 | A | 42 | A | 13 | A |
| Amy1 (amylase) | 1369502_a_at | 5 | A | 9 | A | 12 | A | 5 | A | 23 | A |
| Amy1 (amylase) | 1369503_at | 57 | A | 62 | A | 78 | A | 58 | A | 64 | A |
| Cpe (carboxypeptidase H) | 1386921_at | 24769 | P | 19940 | P | 24475 | P | 23725 | P | 23733 | P |
| Gcg (glucagon) | 1369888_at | 6875 | P | 17772 | P | 2064 | P | 20550 | P | 24434 | P |
| Gck (GK/MODY2) | 1387312_a_at | 1165 | P | 767 | P | 787 | P | 635 | P | 1101 | P |
| Glp1r | 1369699_at | 567 | P | 562 | P | 644 | P | 886 | P | 776 | P |
| Ins1 (insulin 1) | 1387815_at | 37360 | P | 34381 | P | 42217 | P | 37973 | P | 37190 | P |
| Ins2 (insulin 2) | 1370077_at | 28816 | P | 5381 | P | 30224 | P | 3953 | P | 4303 | P |
| Kcnj11 (KIR6.2) | 1387698_at | 1963 | P | 1792 | P | 2557 | P | 2891 | P | 2524 | P |
| Kcnj11 (KIR6.2) | 1391007_s_at | 3414 | P | 3042 | P | 3789 | P | 4442 | P | 4118 | P |
| Kcnj11 (KIR6.2) | 1369782_a_at | 1293 | P | 992 | P | 1329 | P | 1635 | P | 928 | P |
| Prss1 (trypsin1) | 1369030_at | 85 | A | 80 | A | 81 | A | 75 | A | 83 | A |
| Prss2 (trypsin2) | 1370126_at | 7 | A | 7 | A | 6 | A | 5 | A | 7 | A |
| Slc2a2 (GLUT2) | 1387228_at | 8926 | P | 6485 | P | 5338 | P | 4153 | P | 10531 | P |
| Sst (somatostatin) | 1367762_at | 83 | A | 30 | A | 13 | A | 15 | A | 72 | A |
P, present; M, marginal; A, absent.
Absolute signal values of a selection of genes expressed in the parental INS-1 cells and the four Flp-In T-Rex cell clones are given. The official gene symbols and in some cases aliases are given in brackets. The probeset IDs are the reference number for the probes of the Affymetrix oligonucleotide microarray (RAE230A).
As the aim of our work was to introduce the HNF6 (onecut1), HNF4α and HNF1β (TCF2) transcription factors into the INS-1 cell line, we verified the amount of expression of these three factors using real-time RT–PCR. As summarized in Figure 2C, the level of these three transcription factors differs in the various cell clones as already suggested by the microarray analysis (Table 1). As the cell clone #1-1.2 has the lowest levels for all three transcription factors (Figure 2C), we chose this clone for the introduction of the transcription factors. A second reason to choose the #1-1.2 cell clone is our observation that this cell clone has the lowest level of glucagons (up to 12-fold compared to #5-3.19, see Table 1), and the highest insulin 1 and 2 level (Table 1). Therefore, this cell clone can be considered as the cell clone most close to the normal β-cell phenotype (29).
HNF6, HNF4α and HNF1η affect a distinct pattern of target genes in INS-1 cells
To identify target genes of the transcription factors HNF6, HNF4α and HNF1β in the insulinoma INS-1 cells, expression plasmids encoding these transcription factors were constructed. To allow Flp-mediated integration, we inserted the ORF of these transcription factors into the expression vector pcDNA5/FRT/TO (Figure 1) that contains the tetracycline inducible promoter (CMV 2xTetO2). In the case of the HNF4α factor we chose the splice variant HNF4α2 that has predominantly been investigated and is known to be more active on most promoters (30–32). We ignored the splice variant HNF4α8 that has been reported to be predominantly expressed in β-cells in some reports (33,34), as this exclusivity has been challenged by others (31). The pcDNA5/FRT/TO vectors containing the transcription factors HNF6, HNF4α or HNF1β were introduced into the #1-1.2 cell line by cotransfection with the expression vector pCSFLPe that encodes Flp recombinase. Using hygromycin we selected the cell clones that had properly integrated the pcDNA/FRT/TO vector by Flp recombination downstream of the chromosomal FRT site (step 3 in Figure 1). For each transcription factor two independent pools of cell clones were established. As the integration of the GOI disrupts the SV40 promoter-ATG sequence from the remaining ORF of the lacZ–Zeocin gene (Figure 1), successful site-directed integration was verified by the absence of blue cells after β-galactosidase staining.
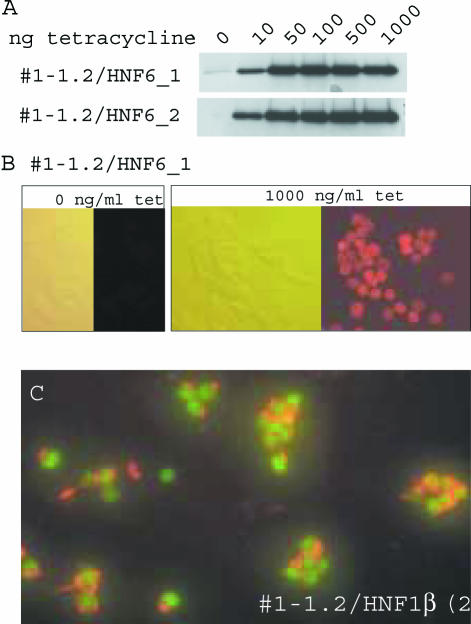
The two independent cell lines for each transcription factor were induced by tetracycline to monitor the induction of the corresponding transcription factor. As exemplified for HNF6, a tetracycline dose dependent induction of HNF6 was observed in western blots 24 h after induction (Figure 3A). By immunofluorescence we observed that >90% of the cells contained nuclear HNF6 24 h after induction with tetracycline (Figure 3B). Notably, in the two independent cell lines the kinetics and extent of HNF6 induction was most similar. Identical results were found for the cell lines expressing HNF4α or HNF1β conditionally (data not shown). To illustrate the homogenous insulin expression in the INS-1 Flp-In cell clones, Figure 3C depicts the double immunostaining of the clone #1-1.2/HNF1β(2b) 24 h after tetracycline induction. The green nuclear staining reflects the induced HNF1β protein in cells that show extensive red staining of the cytoplasmic insulin store.
Figure 3.
Tetracycline induction of HNF6. (A) Two independent cell lines containing the histidine tagged HNF6 transcription factor as GOI were treated for 24 h with tetracycline with the concentrations given. Total protein extracts were probed by western blots using a histidine tag specific antibody. (B) Immunofluorescence of untreated cells or cells treated for 24 h with 1000 ng/ml tetracycline using a histidine specific antibody. Phase contrast (left) and fluorescence pictures (right) are given. Counting the immunopositive cells in the two independent cell lines for each transcription factor gave the following values (untreated/tetracycline treated): HNF6: <1%, <1%/95%, 91%. HNF4α: 3%, 5%/92%, 93%. HNF1β <1%, <1%/95%, 98%. (C) Immuno fluorescence of INS-1 cells expressing HNF1β upon 24 h tetracycline induction (1000 ng/ml). HNF1β was detected using a myc-tag specific primary antibody and a FITC-coupled secondary antibody (green), whereas insulin was detected with a primary insulin antibody and a Cy3-coupled secondary antibody (red).
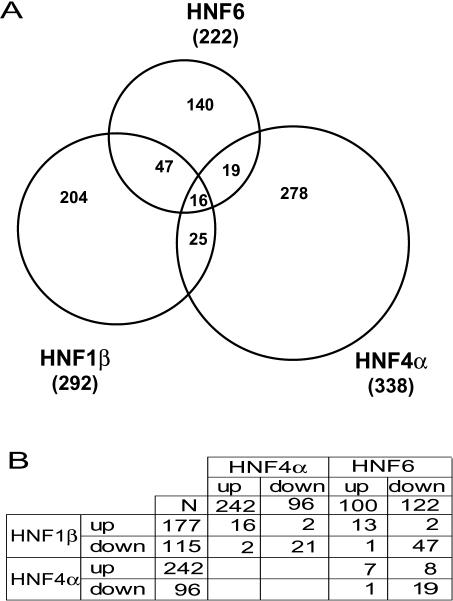
To monitor the genes affected by the expression of these three transcription factors, each cell line was treated with tetracycline (1000 ng/ml) for 24 h and the RNA was assayed by oligonucleotide microarrays (Affymetrix RAE230A) using the RNA of untreated cells as a reference. For each transcription factor the hybridization data of the two independent cell lines were analyzed and the probesets showing a >2.5-fold change in both cell types and at least one present detection call either in the induced or uninduced state were scored (for further details see Materials and Methods). Using these filter criteria we obtained 222, 338 and 292 changed probesets for HNF6, HNF4α and HNF1β, respectively. Based on the information given by Affymetrix (http://www.affymetrix.com/analysis/index.affx) ∼50% of the probesets can be annotated to specific genes. The Venn diagram given in Figure 4A documents that only a limited overlap of the genes affected by these transcription factors exists. Sixteen probesets were affected by all three transcription factors and the corresponding values are given in Table 2. The fold change seen for these 16 probesets was quite similar between the two independent cell lines of each transcription factor. This was a common finding throughout our analysis (data not shown), indicating the reproducibility of the two independent cell lines. Analyzing the targets regulated by all three transcription factors shows that three transcripts (Pzp, Ugt2b12 and probeset ID no. 1375934) were upregulated, whereas 12 probesets were downregulated and only one gene (Cyp4f14) was down or up regulated depending on the transcription factor induced. The numbers of probesets affected by two transcription factors were 19, 25 and 47 for HNF6/HNF4α, HNF4α/HNF1β and HNF1β/HNF6, respectively (Figure 4A). Analyzing the up and down regulated transcripts (Figure 4B), the majority of genes was affected in the same direction by the different transcription factors. To reveal the biological processes affected by these three transcription factors, we have listed in Table 3, the distribution of the probesets to the various categories as defined by the gene ontology annotations (35). The transcription factors influence many different processes including distinct metabolic pathways, cell proliferation, transport mechanism, signaling cascades between cells, cell differentiation and morphogenesis. Analyzing the distribution of the genes to the various categories, we found in several categories a statistically significant overrepresentation that depended on the transcription factor (Table 3). Whereas HNF4α affects predominantly some specific metabolic processes (energy pathway, amino acid and derivative metabolism, organic acid metabolism, amine metabolism and alcohol metabolism), HNF6 and HNF1β rather influence the activity of genes involved in development. This indicates that the three transcription factors have distinct properties concerning their biological role. The entire list of probesets affected by the transcription factors is included as Supplementary Material (Table SI–SVI). The data are arranged according to the area of the Venn diagram in Figure 4A and the fold-induction refers to the average of the two independent cell lines for each transcription factor. The original microarray data have been deposited in the GEO data repository (http://www.ncbi.nlm.nih.gov/geo/).
Figure 4.
Pattern of genes affected by HNF6, HNF4α and HNF1β. (A) Venn diagram showing the probesets affected by the transcription factor HNF6, HNF4α or HNF1β after 24 h of tetracycline induction. Numbers in the overlapping segments refer to the common probesets and numbers in brackets indicate the total number of probesets affected for each transcription factor. (B) Cross table showing the overlaps between up and down regulated targets of the three different transcription factors.
Table 2. Probesets affected by HNF6, HNF4α as well as HNF1β.
| Gene symbol | Probeset ID | HNF6_1 | HNF6_2 | HNF4α_1 | HNF4α_2 | HNF1β_1a | HNF1β_2b |
|---|---|---|---|---|---|---|---|
| Pzp | 1370547_at | 41.9 | 93.7 | 16.2 | 25.8 | 3.3 | 3.7 |
| Ugt2b12 | 1368397_at | 10.7 | 8.9 | 9.1 | 14.4 | 52.0 | 34.1 |
| — | 1375934_at | 4.6 | 4.2 | 3.2 | 2.7 | 4.3 | 3.9 |
| Cyp4f14 | 1368467_at | 0.35 | 0.31 | 4.8 | 3.2 | 14.4 | 14.1 |
| — | 1377404_at | 0.30 | 0.31 | 0.24 | 0.31 | 0.18 | 0.18 |
| Amigo2 | 1373345_at | 0.29 | 0.28 | 0.19 | 0.19 | 0.16 | 0.17 |
| Cyp4a12 | 1368607_at | 0.20 | 0.18 | 0.23 | 0.27 | 0.16 | 0.15 |
| — | 1371541_at | 0.18 | 0.25 | 0.20 | 0.34 | 0.16 | 0.16 |
| — | 1371731_at | 0.17 | 0.18 | 0.12 | 0.12 | 0.20 | 0.18 |
| — | 1379380_at | 0.16 | 0.16 | 0.09 | 0.19 | 0.25 | 0.26 |
| Kcnj11 (KIR6.2) | 1391007_s_at | 0.14 | 0.15 | 0.33 | 0.31 | 0.09 | 0.07 |
| Isl1 | 1369681_at | 0.13 | 0.12 | 0.34 | 0.32 | 0.39 | 0.31 |
| — | 1388598_at | 0.13 | 0.18 | 0.11 | 0.10 | 0.26 | 0.26 |
| Idb4 | 1375120_at | 0.13 | 0.25 | 0.16 | 0.09 | 0.17 | 0.15 |
| — | 1372308_at | 0.10 | 0.15 | 0.13 | 0.14 | 0.27 | 0.23 |
| Ngfr | 1368148_at | 0.08 | 0.11 | 0.05 | 0.05 | 0.11 | 0.06 |
The fold change for the probesets affected by all three transcription factors (see Figure 4A) is listed. For each transcription factor the values obtained for the two independent cell lines are given. The probeset IDs are the reference number for the probes of the Affymetrix oligonucleotide microarray (RAE230A). Where available (Affymetrix, release April 2004) the official gene symbols and in some cases aliases are given in brackets. — indicates that no gene symbol is available.
Table 3. Distribution of the probesets to categories of the GO biological processes.
| HNF6 | HNF4α | HNF1β | RAE230A | |
|---|---|---|---|---|
| Probesets with annotated biological processes | 60 | 95 | 91 | 2652 |
| Metabolism | 28 | 50 | 46 | 1257 |
| Protein metabolism | 7 | 14 | 12 | 456 |
| Transcription | 11* | 5 | 12 | 265 |
| Phosphorus metabolism | 6 | 6 | 5 | 183 |
| Energy pathway | 3 | 8** | 2 | 59 |
| Amino acid and derivative metabolism | 1 | 7* | 4 | 70 |
| Organic acid metabolism | 1 | 11** | 4 | 116 |
| Electron transport | 1 | 6 | 3 | 91 |
| Amine metabolism | 1 | 7* | 4 | 70 |
| Alcohol metabolism | 4 | 7* | 1 | 75 |
| Biosynthesis | 4 | 9 | 6 | 268 |
| Carbohydrate metabolism | 2 | 6 | 1 | 86 |
| Catabolism | 1 | 12 | 7 | 222 |
| Organismal physiological process | 6 | 10 | 9 | 371 |
| Response to stimulus | 5 | 10 | 10 | 328 |
| Cellular physiological process | 27 | 34 | 44* | 1022 |
| Cell motility | 5** | 2 | 4 | 59 |
| Cell proliferation | 6 | 5 | 6 | 185 |
| Transport | 11 | 21 | 26 | 583 |
| Cell organization and biogenesis | 2 | 6 | 2 | 151 |
| Regulation of cellular process | 6* | 3 | 6 | 118 |
| Cell communication | 19 | 26 | 20 | 807 |
| Intracellular signaling cascade | 7 | 7 | 9 | 235 |
| Cell surface receptor linked signal transduction | 12 | 10 | 10 | 331 |
| Cell adhesion | 0 | 6 | 3 | 118 |
| Development | 22*** | 17 | 27** | 452 |
| Cell differentiation | 6* | 2 | 6 | 97 |
| Morphogenesis | 17*** | 12 | 21** | 318 |
| Neurogenesis | 10** | 8 | 10* | 145 |
Based on the ‘Gene Ontology Standard Vocabulary’ the probesets were sorted according to the ‘Biological Process’ into the various categories. Probesets corresponding to the same gene were counted only once. The number of probesets with annotated biological processes is given and corresponds to ∼30% of the probesets affected. All categories that contained at least five probesets for either of the three transcription factors are listed in the table. Categories are given in bold face, if they are above the following categories. Note that the sum of the probeset number does not equal to the number of individual probesets, as probesets can be members of different categories and categories with less than five probesets are not shown. As a reference the distribution of all probesets of the Affymetrix microarray RAE230A to the categories listed is given. Probesets representing the same gene were deleted before categorization. For details see the Affymetrix Gene Ontology Mining Tool Manual (https://www.affymetrix.com/support/technical/manual/go_manual.affx). To analyze whether in some categories genes are overrepresented to what would be expected by chance from a randomly selected group of genes, we made a hypergeometric distribution (http://genome-www5.stanford.edu/help/GO-TermFinder/GO_TermFinder_help.shtml). Values that represent an overrepresentation are indicated with *, ** and *** for P-value cut-offs of P < 0.05, P < 0.01 and P < 0.001, respectively.
In conclusion, our data shows that HNF6, HNF4α and HNF1β affect a similar number of genes with HNF6 affecting the lowest number. The overlap of genes affected is restricted and the extent of overlap between the three transcription factors is similar, with 63, 82 and 70% probesets restricted to HNF6, HNF4α and HNF1β, respectively.
The gene pattern affected by HNF1η transcription factor mutants from MODY5 patients
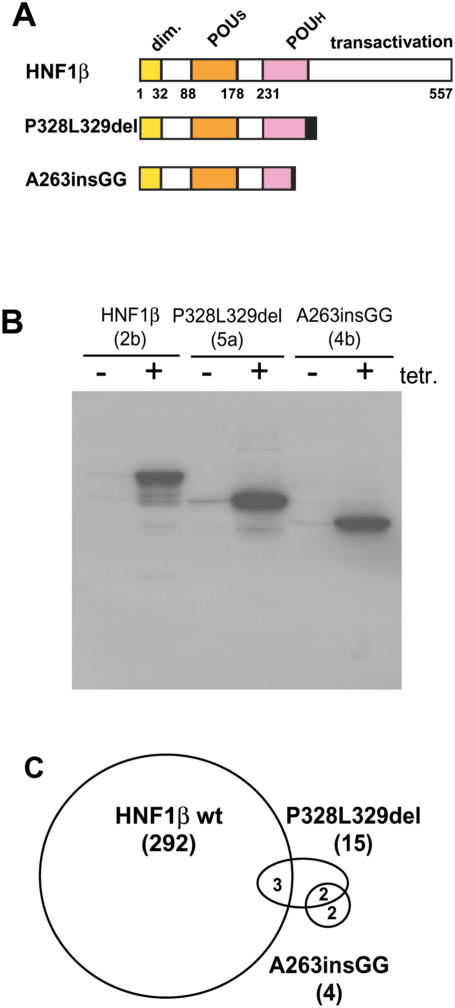
In previous experiments, we have characterized the different properties of mutated HNF1β transcription factors present in human patients with MODY5 and renal defects (26). Analyzing the effect of these mutated transcription factors on kidney formation in Xenopus embryos, we identified two distinct types of mutations. One group led to a reduced pronephric size, whereas the other group resulted in an enlargement of the pronephros. We have introduced a member of each group into the INS-1 Flp-In T-REx cell clone #1-1.2 using Flp-mediated integration. The mutation P328L329del that leads to a reduced pronephric size retains all sequences required for specific DNA binding (Figure 5A), whereas the mutation A263insGG that leads to an enlarged pronephros lacks part of the POUH domain (Figure 5A) and thus cannot bind to DNA. Western blots of cell lines stably containing genes encoding these mutated transcription factors, revealed the tetracycline-dependent expression of truncated proteins of the expected size (Figure 5B). The amount of protein induced within 24 h was quite similar for the two HNF1β derivatives as well as for the wild-type HNF1β protein. The amount of induced HNF1β derivative was also identical between the two independent cell lines produced for each construct (data not shown). To monitor the effect of these two mutants on the gene expression profile, we performed an oligonucleotide microarray analysis using RNA of the two independent cell lines for each mutated HNF1β construct. Using the same filter criteria as for the wild-type HNF1β protein, we identified only 15 and 4 changed signals for P328L329del and A263insGG, respectively. This is a much smaller number compared to the wild-type HNF1β transcription factor that affected 292 probesets. As illustrated in Figure 5C, only three probesets between P328L329del and wild-type HNF1β were in common, and the four probesets identified by A263insGG were not affected by wild-type HNF1β. Comparing the probesets between the P328L329del and the A263insGG mutant, two probesets were in common. Table 4 lists the probesets affected by the mutated HNF1β proteins with the corresponding induction levels obtained for the two independent cell lines. In all three cases where the mutated HNF1β transcription factor affected a probeset that was also targeted by wild-type HNF1β, the direction of response was the same.
Figure 5.
Pattern of genes affected by HNF1β mutants. (A) Schematic drawing of the structure of the HNF1β protein and of the two truncated proteins encoded by the MODY5 mutants. The dimerization domain (dim), the domains involved in DNA binding (POUS, POU specific domain; POUH, homeodomain) and transactivation are given. The black boxes indicate the additional amino acids present in the truncated protein due to the frame shift mutation. The numbers refer to the amino acid position in HNF1β. P328L329del is P328L329delCCTCT and contains a 5 bp deletion (47) and A263insGG contains a 2 bp insertion (52). (B) Western blots of cell extracts from untreated or treated (18 h, 1000 ng/ml tetracycline) cell lines. (C) Venn diagram of the probesets affected by wild-type HNF1β, P328L329del and A263insGG. Numbers in the overlapping segments refer to the common probesets and numbers in brackets indicate the total number of probesets affected for each construct.
Table 4. Probesets affected by the MODY 5 mutant P328L329del and A263insGG.
| Gene | Probeset ID | del5a | del6a | mut3b | mut4a | wt1a | wt2b |
|---|---|---|---|---|---|---|---|
| Gsta1 | 1367774_at | 6.1 | 5.4 | 9.4 | 17.2 | ||
| — | 1374812_at | 2.6 | 3.0 | 4.8 | 4.6 | ||
| Cyp4a12 | 1368607_at | 0.31 | 0.28 | 0.16 | 0.15 | ||
| — | 1389496_at | 0.33 | 0.26 | 0.39 | 0.36 | ||
| — | 1374657_at | 0.23 | 0.37 | 0.33 | 0.33 | ||
| — | 1376989_at | 3.4 | 3.7 | ||||
| Myc | 1368308_at | 0.22 | 0.18 | ||||
| Bmp3 | 1369773_at | 0.34 | 0.29 | ||||
| — | 1372380_at | 0.32 | 0.30 | ||||
| — | 1373625_at | 0.40 | 0.38 | ||||
| — | 1375898_at | 0.39 | 0.25 | ||||
| Ddc | 1368064_a_at | 0.13 | 0.14 | ||||
| — | 1374932_at | 0.21 | 0.04 | ||||
| Pklr | 1387263_at | 0.37 | 0.37 | ||||
| — | 1393221_at | 0.18 | 0.37 | ||||
| — | 1389066_at | 0.35 | 0.38 | ||||
| — | 1368289_at | 0.39 | 0.31 |
The fold change for the probesets affected by the mutants (Figure 5C) are listed. For each transcription factor the values obtained for the two independent cell lines are given. del5a and del6a express P328L329del, mut3b and mut4a express A263insGG, whereas wt1a and wt2b correspond to the HNF1β clones used in Figure 4. The probeset IDs are the reference number for the probes of the Affymetrix oligonucleotide microarray (RAE230A). Where available (Affymetrix, release April 2004) the official gene symbols (gene) are given. — indicates that no gene symbol is available.
DISCUSSION
We have established β-cell derived cell clones (INS-1 Flp-In T-REx) that allow the site-directed integration of any GOI into differentiated β-cells. This novel approach reduces the variability of transgene expression that is observed using the traditional transfection or transduction protocol leading to random gene integration. Furthermore, the INS-1 Flp-In T-REx cells contain the constitutive active tetracycline repressor allowing the conditional expression of the transgene. The INS-1 Flp-In T-REx cell clones have been selected for high insulin expression to maintain a differentiated β-cell phenotype (Figure 3C). From hybridization analysis using microarrays we confirmed the β-cell phenotype by the presence of transcripts specific for β-cells (Table 1). Most notably, many β-cell-specific transcription factors were expressed, including HNF4α, HNF1α, Pdx1 and NeuroD1 encoded by the MODY1, MODY3, MODY4 and MODY6 genes, respectively. In all four clones analyzed as well as in the parental INS-1 cell line, HNF1β (MODY5) was expressed only at a low level consistent with reports in adult islets (36,37). Nevertheless, the INS-1 cells are highly differentiated and this is consistent with the report that HNF1β predominates in early pancreatic precursor cells (37). Similarly, high HNF6 expression is also restricted to early stages of β-cell differentiation (38,39). The microarray analysis revealed a substantial difference between the four cell clones and the original INS-1 cell line (Table 1). It seems unlikely that this reflects the integration of the FRT site (step 1 in Figure 1) at different chromosomal sites. We rather assume that it represents heterogeneity within the INS-1 cell line.
Analyzing the probesets affected by the three transcription factors, a pattern of induced and decreased transcripts was observed that differs extensively between the three transcription factors. This possibly reflects the completely distinct DNA elements recognized by these three transcription factors (40–42). Although HNF4α and HNF1β affect a similar number of probes (338 and 292, respectively), HNF6 influences a slightly lower number (222). This differs from a recent report analyzing the target genes of HNF6, HNF1α and HNF4α by chromatin immunoprecipitation (ChIP) combined with promoter microarrays (7). Based on this study, HNF4α was bound to ∼5- to 10-times more promoters (1323) than HNF6 (189) and HNF1α (106). This analysis was done on human cells purified from pancreatic islets and thus species-specific differences, the purity of the cell types or changes inherent to the establishment of cell lines could contribute to the different result. However, we assume that the difference mainly reflects the differential assay to score target genes. In the ChIP assay any bound transcription factor will be identified, whereas in our functional expression system the change in promoter activity is recorded. It is most likely that within promoters, that are bound by several transcription factors, the introduction of an additional transcription factor has no effect on promoter activity, as the promoter activity may be saturated or the newly added transcription factor replaces a preexisting factor without altering promoter activity. Therefore, ChIP assays and microarray analysis reveal different patterns of regulated genes and are two complementary approaches. A major advantage of the measurement of transcripts induced by a transcription factor by microarrays is that this assay also reveals the direction of response. Since all three transcription factors are known as positive regulators, it is quite surprising that ∼30% of the transcripts are downregulated (Figure 4B). This might either reveal a property of these transcription factors that has not been observed previously or the fact that each gene is part of a complex regulatory network. This includes mutually exclusive binding of transcription factors to promoters as well as the activation of regulatory cascades. The presence of such regulatory cascades is supported by our finding that the expression of other transcription factors is affected (see Table 2 and Tables in Supplementary Material). The three transcription factors downregulated the transcription factor Isl1 and Idb4 (Table 2), but nevertheless the common targets affected are limited to 16 probesets. Similarly, HNF1β and HNF6 downregulated Pax6 and Pdx1 (Table SVI), but the common genes affected by these two transcription factors is limited to 47 probesets (Figure 4A). These findings imply the existence of a most complex combinatorial network controlling gene expression, whose changes upon the activation of a specific transcription factor cannot be predicted yet.
Several reports have analyzed specific promoters of genes expressed in pancreatic β-cells by transient transfection assays. For instance the insulin promoter was shown to be regulated by HNF4α (43), but based on our analysis no increase of the endogenous transcription of the insulin 1 and 2 transcript was found. Furthermore, we have shown that the P2 promoter of HNF4α is activated by HNF6 and HNF1β (33), but neither transcription factor led to an induction of HNF4α expression. These examples illustrate that reporter assays in transient transfections are not sufficient to predict the outcome of the introduction of a transcription factor on the activity of chromosomal genes. In the case of HNF4α the activation of the endogenous genes encoding Glut2 (Slc2a2), L-PK (Pklr), aldolase B (Aldob), 2-oxoglutarate dehydrogenase E1 subunit (OGDH E1 subunit) and HNF1α (TCF1) has been reported in INS-1 cells (15). We can confirm the induction of Glut2 (Slc2a2, 2.6-fold in Table SII) and aldolase B (aldob, 8.9-fold in Table SIII), whereas L-PK (Pklr) and 2-oxoglutarate dehydrogenase E1 subunit (OGDH E1 subunit) are not induced although both are expressed. The transcription factor HNF1α (TCF1), expressed in both cell lines, is activated by a factor of 1.4 in one clone and therefore does not meet our filter criteria. The differences between our data and the results described previously (15) possibly reflect the selection process in establishing the INS-1 Flp-In T-REx cells.
The list of probesets affected by all three transcription factors includes Kcnj11 (KIR6.2) that is downregulated by HNF6, HNF4α and HNF1β. The membrane protein Kcnj11 is part of the ATP-sensitive potassium channel that plays a crucial role in insulin secretion in β-cells. Interestingly, mutations and polymorphisms in this gene have been proposed to contribute to the susceptibility to type II diabetes in human populations (44,45). Since activating mutations in the Kcnj11 gene have been associated with permanent neonatal diabetes (46), the negative effect of HNF6, HNF4α and HNF1β on Kcnj11 expression could play a crucial role for regulated insulin secretion.
We used the INS-1 Flp-In T-REx cell line to define the properties of two mutated HNF1β transcription factors we have extensively described in previous reports (24,26). Both mutations, P328L329del (47) and A263insGG (48), have been described in MODY5 patients and are believed to deregulate insulin secretion in β-cells. In fact, mice with β-cell restricted inactivation of the HNF1β gene show a reduced glucose-stimulated insulin release (49). However, both mutations also lead to kidney malformations (47,48). Although the mutation P328L329del retains all domains required for DNA binding (Figure 5A) and acts in transfection assays as a gain-of-function mutation (24), the mutation A263insGG has lost part of the POUH domain essential for DNA binding (Figure 5A) and acts either as a loss-of-function mutation (26) or even as a dominant-negative factor (48,50), depending on the cell type or reporter used. The analysis in the INS-1 Flp-In T-REx cell lines shows that both mutated transcription factors affect only very few genes compared to the wild-type factor (Figure 5C), implying that they both are loss-of-function mutations. As our novel assay measures endogenous genes, we assume that it comes closer to the in vivo function in β-cells. In the case of A263insGG, a competition with the low level of the endogenous HNF1β (Figure 2C) is possible, but it could also compete with the endogenous HNF1α, as HNF1α and HNF1β are known to readily heterodimerize (4). Therefore, the poor effect by A263insGG cannot be easily explained. The mutant A263insGG down-regulates four probesets, whose identities are not yet known and thus are not informative. In contrast, the mutant P328L329del increases three and decreases twelve transcripts. However, most of these probes are not affected by the wild-type factor, implying that P328L329del affects a distinct set of genes. This conclusion is supported by the fact that the three probes, affected by P328L329del as well as the wild-type factor, are changed to a similar level, but all the other probes affected by the wild-type protein (up to a factor of 160-fold, see Table SV, Supplementary Material) are not affected by P328L329del. Compared to wild-type, both mutated HNF1β proteins could be classified as loss-of-function mutations. However, this conclusion contrasts with our recent data that revealed the performance of both mutations in the developing Xenopus kidney. Whereas P328L329del led to a reduced size of the embryonic kidney (pronephros), A263insGG resulted in an enlarged kidney (26), excluding a loss-of-function phenotype. Furthermore, kidney-specific overexpression of A263insGG in transgenic mice generated polycystic renal dysplasia (51). All these data reveal some distinct functional properties of the mutated HNF1β proteins, but these differences could be restricted in its function to renal cell types.
From a series of experiments we know that tetracycline on its own has no effect on gene expression (data not shown). Consistent with this finding, the present study shows that none of the 16 probesets affected by all the three transcription factors (Table 2) is influenced by the expression of the MODY5 mutant A263insGG (Table 4). Therefore, we exclude that tetracycline treatment induces any gene activity.
The INS-1 Flp-In T-REx cell line we have established will be of great value to dissect the function of defined genes in the context of β-cell-specific differentiation. Although the identity of ∼50% of the Affymetrix rat microarray probesets is not known at the moment, the available rat genome sequence will improve this within a short period of time.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We greatly acknowledge the technical assistance of Nadine Esser and Adriane Parchatka for microarray analysis. We thank Tanja Boes for the hypergeometric distribution analysis and Noel Morgan for critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (TH 799/1-1 and RY5/4-5).
REFERENCES
- 1.Edlund H. (2002) Pancreatic organogenesis—developmental mechanisms and implications for therapy. Nature Rev.Genet., 3, 524–532. [DOI] [PubMed] [Google Scholar]
- 2.McClenaghan N.H. and Flatt,P.R. (1999) Engineering cultured insulin-secreting pancreatic B-cell lines. J. Mol. Med., 77, 235–243. [DOI] [PubMed] [Google Scholar]
- 3.Dor Y., Brown,J., Martinez,O.I. and Melton,D.A. (2004) Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature, 429, 41–46. [DOI] [PubMed] [Google Scholar]
- 4.Ryffel G.U. (2001) Mutations in the human genes encoding the transcription factors of the hepatocyte nuclear factor (HNF)1 and HNF4 families: functional and pathological consequences. J. Mol. Endocrinol., 27, 11–29. [DOI] [PubMed] [Google Scholar]
- 5.Fajans S.S., Bell,G.I. and Polonsky,K.S. (2001) Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N. Engl. J. Med., 345, 971–980. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell S. and Frayling,T. (2002) The role of transcription factors in maturity-onset diabetes of the young. Mol. Genet. Metab., 77, 35. [DOI] [PubMed] [Google Scholar]
- 7.Odom D.T., Zizlsperger,N., Gordon,D.B., Bell,G.W., Rinaldi,N.J., Murray,H.L., Volkert,T.L., Schreiber,J., Rolfe,P.A., Gifford,D.K. et al. (2004) Control of pancreas and liver gene expression by HNF transcription factors. Science, 303, 1378–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butte A. (2002) The use and analysis of microarray data. Nature Rev. Drug Discov., 1, 951–960. [DOI] [PubMed] [Google Scholar]
- 9.Gossen M. and Bujard,H. (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA, 89, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colosimo A., Goncz,K.K., Holmes,A.R., Kunzelmann,K., Novelli,G., Malone,R.W., Bennett,M.J. and Gruenert,D.C. (2000) Transfer and expression of foreign genes in mammalian cells. Biotechniques, 29, 314–318, 320–322, 324. [DOI] [PubMed] [Google Scholar]
- 11.Asfari M., Janjic,D., Meda,P., Li,G., Halban,P.A. and Wollheim,C.B. (1992) Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology, 130, 167–178. [DOI] [PubMed] [Google Scholar]
- 12.Hagenfeldt-Johansson K.A., Herrera,P.L., Wang,H., Gjinovci,A., Ishihara,H. and Wollheim,C.B. (2001) Beta-cell-targeted expression of a dominant-negative hepatocyte nuclear factor-1alpha induces a maturity-onset diabetes of the young (MODY)3-like phenotype in transgenic mice. Endocrinology, 142, 5311–5320. [DOI] [PubMed] [Google Scholar]
- 13.Wang H., Maechler,P., Hagenfeldt,K.A. and Wollheim,C.B. (1998) Dominant-negative suppression of HNF-1alpha function results in defective insulin gene transcription and impaired metabolism-secretion coupling in a pancreatic beta-cell line. EMBO J., 17, 6701–6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H., Antinozzi,P.A., Hagenfeldt,K.A., Maechler,P. and Wollheim,C.B. (2000) Molecular targets of a human HNF1alpha mutation responsible for pancreatic beta-cell dysfunction. EMBO J., 19, 4257–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H., Maechler,P., Antinozzi,P.A., Hagenfeldt,K.A. and Wollheim,C.B. (2000) Hepatocyte nuclear factor 4alpha regulates the expression of pancreatic beta-cell genes implicated in glucose metabolism and nutrient-induced insulin secretion. J. Biol. Chem., 275, 35953–35959. [DOI] [PubMed] [Google Scholar]
- 16.Wang H., Maechler,P., Ritz-Laser,B., Hagenfeldt,K.A., Ishihara,H., Philippe,J. and Wollheim,C.B. (2001) Pdx1 level defines pancreatic gene expression pattern and cell lineage differentiation. J. Biol. Chem., 276, 25279–25286. [DOI] [PubMed] [Google Scholar]
- 17.Wang H., Gauthier,B.R., Hagenfeldt,K.A., Iezzi,M. and Wollheim,C.B. (2002) Foxa2 (HNF3beta) controls multiple genes implicated in metabolism-secretion coupling of glucose-induced insulin release. J. Biol. Chem., 277, 17564–17570. [DOI] [PubMed] [Google Scholar]
- 18.Wang H., Maechler,P., Antinozzi,P.A., Herrero,L., Hagenfeldt-Johansson,K.A., Bjorklund,A. and Wollheim,C.B. (2003) The transcription factor SREBP-1c is instrumental in the development of beta-cell dysfunction. J. Biol. Chem., 278, 16622–16629. [DOI] [PubMed] [Google Scholar]
- 19.Yang Q., Yamagata,K., Fukui,K., Cao,Y., Nammo,T., Iwahashi,H., Wang,H., Matsumura,I., Hanafusa,T., Bucala,R. et al. (2002) Hepatocyte nuclear factor-1alpha modulates pancreatic beta-cell growth by regulating the expression of insulin-like growth factor-1 in INS-1 cells. Diabetes, 51, 1785–1792. [DOI] [PubMed] [Google Scholar]
- 20.Werdien D., Peiler,G. and Ryffel,G.U. (2001) FLP and Cre recombinase function in Xenopus embryos. Nucleic Acids Res., 29, e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drewes T., Clairmont,A., Klein-Hitpass,L. and Ryffel,G.U. (1994) Estrogen-inducible derivatives of hepatocyte nuclear factor-4, hepatocyte nuclear factor-3 and liver factor B1 are differently affected by pure and partial antiestrogens. Eur. J. Biochem., 225, 441–448. [DOI] [PubMed] [Google Scholar]
- 22.Drewes T., Senkel,S., Holewa,B. and Ryffel,G.U. (1996) Human hepatocyte nuclear factor 4 isoforms are encoded by distinct and differentially expressed genes. Mol. Cell. Biol., 16, 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lausen J., Thomas,H., Lemm,I., Bulman,M., Borgschulze,M., Lingott,A., Hattersley,A.T. and Ryffel,G.U. (2000) Naturally occurring mutations in the human HNF4alpha gene impair the function of the transcription factor to a varying degree. Nucleic Acids Res., 28, 430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wild W., Pogge v.Strandmann,E., Nastos,A., Senkel,S., Lingott-Frieg,A., Bulman,M., Bingham,C., Ellard,S., Hattersley,A.T. and Ryffel,G.U. (2000) The mutated human gene encoding hepatocyte nuclear factor 1beta inhibits kidney formation in developing Xenopus embryos. Proc. Natl Acad. Sci. USA, 97, 4695–4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rupp R.A., Snider,L. and Weintraub,H. (1994) Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev., 8, 1311–1323. [DOI] [PubMed] [Google Scholar]
- 26.Bohn S., Thomas,H., Turan,G., Ellard,S., Bingham,C., Hattersley,A.T. and Ryffel,G.U. (2003) Distinct molecular and morphogenetic properties of mutations in the human HNF1beta gene that lead to defective kidney development. J. Am. Soc. Nephrol., 14, 2033–2041. [DOI] [PubMed] [Google Scholar]
- 27.Pogge v.Strandmann E., Zoidl,C., Nakhei,H., Holewa,B., Pogge von Strandmann,R., Lorenz,P., Klein-Hitpass,L. and Ryffel,G.U. (1995) A highly specific and sensitive monoclonal antibody detecting histidine- tagged recombinant proteins. Protein Eng., 8, 733–735. [DOI] [PubMed] [Google Scholar]
- 28.Dürig J., Nückel,H., Hüttmann,A., Kruse,E., Hölter,T., Halfmeyer,K., Führer,A., Rudolph,R., Kalhori,N., Nusch,A. et al. (2003) Expression of ribosomal and translation-associated genes is correlated with a favorable clinical course in chronic lymphocytic leukemia. Blood, 101, 2748–2755. [DOI] [PubMed] [Google Scholar]
- 29.Huang H.P. and Tsai,M.J. (2000) Transcription factors involved in pancreatic islet development. J. Biomed. Sci., 7, 27–34. [DOI] [PubMed] [Google Scholar]
- 30.Nakhei H., Lingott,A., Lemm,I. and Ryffel,G.U. (1998) An alternative splice variant of the tissue specific transcription factor HNF4alpha predominates in undifferentiated murine cell types. Nucleic Acids Res., 26, 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eeckhoute J., Moerman,E., Bouckenooghe,T., Lukoviak,B., Pattou,F., Formstecher,P., Kerr-Conte,J., Vandewalle,B. and Laine,B. (2003) Hepatocyte nuclear factor 4alpha isoforms originated from the P1 promoter are expressed in human pancreatic beta-cells and exhibit stronger transcriptional potentials than P2 promoter-driven isoforms. Endocrinology, 144, 1686–1694. [DOI] [PubMed] [Google Scholar]
- 32.Torres-Padilla M.E. and Weiss,M.C. (2003) Effects of interactions of hepatocyte nuclear factor 4alpha isoforms with coactivators and corepressors are promoter-specific. FEBS Lett., 539, 19–23. [DOI] [PubMed] [Google Scholar]
- 33.Thomas H., Jaschkowitz,K., Bulman,M., Frayling,T.M., Mitchell,S.M., Roosen,S., Lingott-Frieg,A., Tack,C.J., Ellard,S., Ryffel,G.U. et al. (2001) A distant upstream promoter of the HNF-4alpha gene connects the transcription factors involved in maturity-onset diabetes of the young. Hum. Mol. Genet., 10, 2089–2097. [DOI] [PubMed] [Google Scholar]
- 34.Hansen S.K., Parrizas,M., Jensen,M.L., Pruhova,S., Ek,J., Boj,S.F., Johansen,A., Maestro,M.A., Rivera,F., Eiberg,H. et al. (2002) Genetic evidence that HNF-1alpha-dependent transcriptional control of HNF-4alpha is essential for human pancreatic beta cell function. J. Clin. Invest., 110, 827–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coffinier C., Barra,J., Babinet,C. and Yaniv,M. (1999) Expression of the vHNF1/HNF1beta homeoprotein gene during mouse organogenesis. Mech. Dev., 89, 211–213. [DOI] [PubMed] [Google Scholar]
- 37.Maestro M.A., Boj,S.F., Luco,R.F., Pierreux,C.E., Cabedo,J., Servitja,J.M., German,M.S., Rousseau,G.G., Lemaigre,F.P. and Ferrer,J. (2003) Hnf6 and Tcf2 (MODY5) are linked in a gene network operating in a precursor cell domain of the embryonic pancreas. Hum. Mol. Genet., 12, 3307–3314. [DOI] [PubMed] [Google Scholar]
- 38.Rausa F., Samadani,U., Ye,H., Lim,L., Fletcher,C.F., Jenkins,N.A., Copeland,N.G. and Costa,R.H. (1997) The cut-homeodomain transcriptional activator HNF-6 is coexpressed with its target gene HNF-3 beta in the developing murine liver and pancreas. Dev. Biol., 192, 228–246. [DOI] [PubMed] [Google Scholar]
- 39.Gannon M., Ray,M.K., Van Zee,K., Rausa,F., Costa,R.H. and Wright,C.V. (2000) Persistent expression of HNF6 in islet endocrine cells causes disrupted islet architecture and loss of beta cell function. Development, 127, 2883–2895. [DOI] [PubMed] [Google Scholar]
- 40.Sheng W., Yan,H., Rausa,F.M.,III, Costa,R.H. and Liao,X. (2004) Structure of the hepatocyte nuclear factor 6alpha (HNF-6alpha) and its interaction with DNA. J. Biol. Chem., 279, 33928–33936. [DOI] [PubMed] [Google Scholar]
- 41.Tronche F., Ringeisen,F., Blumenfeld,M., Yaniv,M. and Pontoglio,M. (1997) Analysis of the distribution of binding sites for a tissue- specific transcription factor in the vertebrate genome. J. Mol. Biol., 266, 231–245. [DOI] [PubMed] [Google Scholar]
- 42.Sladek F.M. and Seidel,S.D. (2001) Hepatocyte nuclear factor 4alpha. In Burris,T.B. and McCabe,E.R.B. (eds), Nuclear Receptors and Genetic Disease. Academic Press, San Diego, pp. 309–361. [Google Scholar]
- 43.Bartoov-Shifman R., Hertz,R., Wang,H., Wollheim,C.B., Bar-Tana,J. and Walker,M.D. (2002) Activation of the insulin gene promoter through a direct effect of hepatocyte nuclear factor 4alpha. J. Biol. Chem., 277, 25914–25919. [DOI] [PubMed] [Google Scholar]
- 44.Nichols C.G. and Koster,J.C. (2002) Diabetes and insulin secretion: whither KATP? Am. J. Physiol. Endocrinol. Metab., 283, e403–e412. [DOI] [PubMed] [Google Scholar]
- 45.Huopio H., Shyng,S.L., Otonkoski,T. and Nichols,C.G. (2002) K(ATP) channels and insulin secretion disorders. Am. J. Physiol. Endocrinol. Metab., 283, e207–e216. [DOI] [PubMed] [Google Scholar]
- 46.Gloyn A.L., Pearson,E.R., Antcliff,J.F., Proks,P., Bruining,G.J., Slingerland,A.S., Howard,N., Srinivasan,S., Silva,J.M., Molnes,J. et al. (2004) Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N. Engl. J. Med., 350, 1838–1849. [DOI] [PubMed] [Google Scholar]
- 47.Bingham C., Ellard,S., Allen,L., Bulman,M., Shepherd,M., Frayling,T., Berry,P.J., Clark,P.M., Lindner,T., Bell,G.I. et al. (2000) Abnormal nephron development associated with a frameshift mutation in the transcription factor hepatocyte nuclear factor-1beta. Kidney Int., 57, 898–907. [DOI] [PubMed] [Google Scholar]
- 48.Tomura H., Nishigori,H., Sho,K., Yamagata,K., Inoue,I. and Takeda,J. (1999) Loss-of-function and dominant-negative mechanisms associated with hepatocyte nuclear factor-1beta mutations in familial type 2 diabetes mellitus. J. Biol. Chem., 274, 12975–12978. [DOI] [PubMed] [Google Scholar]
- 49.Wang L., Coffinier,C., Thomas,M.K., Gresh,L., Eddu,G., Manor,T., Levitsky,L.L., Yaniv,M. and Rhoads,D.B. (2004) Selective deletion of the HNF1{beta} (MODY5) gene in {beta} cells leads to altered gene expression and defective insulin release. Endocrinology, 145, 3941–3949. [DOI] [PubMed] [Google Scholar]
- 50.Bai Y., Pontoglio,M., Hiesberger,T., Sinclair,A.M. and Igarashi,P. (2002) Regulation of kidney-specific Ksp-cadherin gene promoter by hepatocyte nuclear factor-1beta. Am. J. Physiol. Renal Physiol., 283, F839–F851. [DOI] [PubMed] [Google Scholar]
- 51.Hiesberger T., Bai,Y., Shao,X., McNally,B.T., Sinclair,A.M., Tian,X., Somlo,S. and Igarashi,P. (2004) Mutation of hepatocyte nuclear factor-1beta inhibits Pkhd1 gene expression and produces renal cysts in mice. J. Clin. Invest., 113, 814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishigori H., Yamada,S., Kohama,T., Tomura,H., Sho,K., Horikawa,Y., Bell,G.I., Takeuchi,T. and Takeda,J. (1998) Frameshift mutation, A263fsinsGG, in the hepatocyte nuclear factor- 1beta gene associated with diabetes and renal dysfunction. Diabetes, 47, 1354–1355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.