Abstract
The ability to associate mutations in cancer genes with the disease and its subtypes is critical for understanding oncogenesis and identifying biomarkers for clinical diagnosis. A two-step mutation scanning method that sequentially used endonuclease V (EndoV) to nick at mismatches and DNA ligase to reseal incorrectly or nonspecifically nicked sites was previously developed in our laboratory. Herein we report an optimized single-step assay that enables ligase to proofread EndoV cleavage in real-time under a compromise between buffer conditions. Real-time proofreading results in a dramatic reduction of background cleavage. A universal PCR strategy that employs both unlabeled gene-specific primers and labeled universal primers, allows for multiplexed gene amplification and precludes amplification of primer dimers. Internally labeled PCR primers eliminate EndoV cleavage at the 5′ terminus, enabling high-throughput capillary electrophoresis readout. Furthermore, signal intensity is increased and artifacts are reduced by generating heteroduplexes containing only one of the two possible mismatches (e.g. either A/C or G/T). The single-step assay improves sensitivity to 1:50 and 1:100 (mutant:wild type) for unknown mutations in the p53 and K-ras genes, respectively, opening prospects as an early detection tool.
INTRODUCTION
Multiple somatic alterations lie at the root of cancer development and tumor heterogeneity. Of cancer genes reported in the current databases, ∼90% have somatic mutations, 20% have germline mutations and 10% have both (1). A major challenge in cancer biology is to unravel the molecular anatomy of individual cancers and tumor subtypes, which would allow a better understanding of the role of genetic alterations in tumor progression, and provide diagnostic and prognostic markers.
The molecular basis for the development and progression of colorectal cancers is well developed in the scientific literature. Colorectal tumors have been classified as either demonstrating chromosomal instability (CIN) or microsatellite instability (MIN). The former is associated with sequential sporadic mutations in the APC (70%), K-ras (40%) and p53 (50%) genes, while the latter contains additional sporadic mutations in the TGFBRII (90%) and BAX (50%) genes (2–6). Mutational patterns may reflect alternative routes to activating/inactivating signaling, growth and apoptosis pathways. APC mutations disrupt the association of APC and β-catenin, resulting in excessive amounts of β-catenin and overactivation of the Wingless/Wnt signaling pathway. Oncogenic mutations in β-catenin were also observed in some colorectal cancers that lacked APC mutations (3). Likewise, colorectal tumors lacking K-ras mutations may have their ras pathway activated through B-raf mutations (7).
In addition to these shared mutations, a recent study showed a high frequency of mutations of the PIK3CA gene (a subunit of the phosphatidylinositol 3-kinase; PI3K) in colon cancers, making this gene a potential biomarker for early detection and/or prognosis of the disease (8). Similarly, the discovery of mutations in various tyrosine kinase and tyrosine phosphatase genes in colorectal cancers suggests that these could become targets for personalized therapy (9,10). As the number of genes linked to cancer grows, there is an increasing demand for the development of new methods for high-throughput detection of unknown mutations.
Different mutation detection technologies have been developed to identify known mutations: these include DNA microarrays (11–13), the polymerase chain reaction/ligase detection reaction (PCR/LDR) (14), now used in combination with the universal DNA microarray (15–17) and primer extension assays (18). Different technologies are used for detection of unknown mutations: hybridization analysis using high-density oligonucleotide arrays (12), denaturing high-performance liquid chromatography (DHPLC) (19), capillary electrophoresis-based single strand conformation polymorphism (CE-SSCP) (20), denaturing gradient gel electrophoresis (DGGE) (21) and heteroduplex analysis (HA) (22) [for review, see (13,23,24)]. However, these techniques either lack high sensitivity and are unable to determine the approximate position of the polymorphism, or suffer from low throughput (19,22,25–28). Finally, dideoxy-sequencing has difficulty in detecting heterozygous mutations, and is of limited utility in the analysis of solid tumors where mutant DNA may represent as little as 15% of the total. Other methods include in vitro transcription/translation-based approaches (29–33), chemical and enzymatic mismatch cleavage detection (e.g. Cleavase, RNase, T4 endonuclease VII, MutS enzymes and CEL I). In vitro mismatch cleavage methods encounter variability in signal intensity compared with background bands (23,34–43).
We recently developed a two-step mutation scanning assay based on enzymatic mismatch cleavage by thermostable Tma Endonuclease V, followed by a proofreading step with thermostable DNA ligase to suppress noise arising from the background of matched cleavage (44). Briefly, 5′ fluorescently labeled mutant and wild-type PCR amplicons are heteroduplexed, EndoV recognizes and cleaves DNA one base to the 3′ side of the mismatch, in addition to nicking matched DNA at low levels. Ligase reseals these background nicks (see steps 3 and 4 in Figure 1). Fluorescent products are separated on a DNA sequencing gel, which reveals the approximate position of the mutation. Subsequent DNA sequencing of the specific region identifies the exact sequence variation. This method has been successfully applied to detect mutations and small insertions/deletions in p53, VHL, K-ras, APC, BRCA1 and BRCA2 (17,44), except for certain GC-rich sequences that are refractory to cleavage present at a frequency of only 2% in human DNA. However, background cleavage by EndoV of some exons makes it difficult to distinguish the correct mutation cleavage signal, especially with pooled samples (17,44).
Figure 1.
Universal PCR to prepare heteroduplexed DNA substrates for EndoV/DNA ligase mutation scanning: split label, denaturation, renaturation or lambda exonuclease treatment. Mutant and normal DNA are PCR amplified separately using Taq DNA polymerase (solid diamond) and gene-specific and universal primers, as detailed in the scheme. Substrates for EndoV/ligase are prepared from the PCR products by either of the following methods. (A) ‘Split label, denaturation, renaturation’ procedure. (B) ‘Split label, lambda exonuclease’ procedure: lambda exonuclease (Pacman icon) degrades 5′ phosphorylated DNA, allowing the newly generated single-stranded DNA to anneal and generate labeled heteroduplexed fragments. EndoV preferentially nicks DNA one base 3′ to the mismatch (large triangle), but also generates nonspecific nicks with minor activity (small triangle). DNA ligase (solid circle) is used either subsequently or concurrently with EndoV to reseal these background nicks. Separation of products by capillary electrophoresis under various assay conditions is illustrated: first with a standard 5′ fluorescent label (5′ Fl-Oligo) or 2′-O-methyl cytosine linkage added to the label [5′ Fl-(2′OMeC)2-Oligo]. In both these cases, EndoV cleaves the label off the PCR product, which results in false signal migrating at about 100 nt (cleaved label). Attachment of the fluorescent label internally in the primer sequence via the C6 position of a cytosine (5′ CCGC(C-Fl)-Oligo) renders labeled products resistant to EndoV cleavage; intensity of the mutation signal is stronger when using the lambda exonuclease method than in the denaturation, renaturation method.
Here, we describe a series of improvements resulting in substantial reduction in background and thus improved mutation scanning. These improvements include a universal amplification strategy with nuclease resistant labeling, a single-step EndoV/real-time ligase proofreading reaction, and separation on a capillary electrophoresis array. Optimizing the single-step reaction conditions represents a critical advance given that these two enzymes have opposing buffer requirements. EndoV possesses a variety of endonuclease activity toward deaminated DNA bases deoxyinosine, deoxyxanthine, deoxyoxanosine, deoxyuridine and other aberrant sites such as apurinic and urea sites (45–51), and in general, non-physiological reaction conditions are required to elicit cleavage at mismatched sites (50,52). The challenge in developing real-time proofreading was to find conditions that allowed EndoV to nick at mismatched sites, while simultaneously permitting the ligase to proofread by predominantly resealing nicks at matched sites. The sensitivity of the present method allows detection of 1 mutant in 100 wild-type sequences, making it amenable to sample pooling and early detection strategies.
MATERIALS AND METHODS
Materials
All routine chemical reagents were purchased from Sigma Chemicals (St. Louis, MO, USA) or Fisher Scientific (Fair Lawn, NJ, USA). GeneScan™ –500 (LIZ™) Size Standard, Hi-Di formamide, polymer POP-7, AmpliTaq and AmpliTaq Gold DNA polymerases, and deoxyribonucleoside triphosphates (dNTPs) were purchased from Applied Biosystems (Foster City, CA). BSA and ATP were purchased from Boehringer-Mannheim (Indianapolis, IN, USA). Proteinase K was purchased from QIAGEN (Valencia, CA, USA). Lambda exonuclease and T4 polynucleotide kinase were purchased from New England Biolabs (Beverly, MA). Unlabeled deoxyoligonucleotides were purchased from Integrated DNA Technologies Inc. (Coralville, IA, USA), while HPLC-purified VIC- and NED-labeled deoxyoligonucleotides were obtained from Applied Biosystems. Thermotoga maritima Endonuclease V and Thermus species AK16D DNA ligase were purified as previously described (50,53). Tumor and normal tissue were obtained from surgical resection of colon cancers at Memorial Sloan Kettering Cancer Center, which were snap-frozen in liquid nitrogen within 15 min of tumor removal. DNA from these tissues was extracted and purified using a proteinase-K/lithium chloride/ethanol protocol as described (QIAGEN). Tumor samples known to contain Q192Ter (C→T) or Y205F (A→T) mutations in exon 6 of p53 were used, as well as their matched normal colonic mucosa. Genomic DNA from cell lines was extracted using DNeasy tissue kit from QIAGEN. HT-29 cell line contains the wild-type K-ras gene, while SW480 and SW620 contain pure exon 1 G12V (G→T) mutation. LoVo cell line contains wild-type p53 gene, while HT-29, SW480 and SW620 cell lines contain exon 8 R273H (G→A) mutation.
PCR amplification and 5′ phosphorylation of deoxyoligonucleotides
DNA sequences of PCR primers used in this study are listed in Table 1. Fifty-microliter PCR reactions with gene-specific primers alone, contained 20 mM Tricine, pH 8.7, 16 mM (NH4)2SO4, 2.5 mM MgCl2, 0.2 mM dNTP, 0.2 μM of each primer and 100 ng genomic DNA. The reaction mixture was incubated at 95°C for 2 min (hot start), followed by the addition of 5 U of AmpliTaq DNA polymerase. PCR amplification conditions were as follows: 35 cycles of 94°C for 20 s, 68°C for 30 s, 72°C for 1 min, followed by a final extension step at 72°C for 7 min.
Table 1. PCR primers used for analysis on the ABI-3730 DNA analyzer.
| Gene | Exon | Primer name | Primer sequence |
|---|---|---|---|
| p53 | Exon 8 | VIC p53Ex8-F72 | 5′ VIC-(2′0MeC)(2′0MeC)-CGCCGCAGGGTGGTTGGGAGTAGATG-3′ |
| NED p53Ex8-R74 | 5′ NED-(2′0MeC)(2′0MeC)-CGCCGCGGTGATAAAAGTGAATCTGAGGCATAAC-3′ | ||
| Exon 8 | VIC p53Ex8-F76 | 5′ VIC-CGCCGCAGGGTGGTTGGGAGTAGATG-3′ | |
| NED p53Ex8-R78 | 5′ NED-CGCCGCGGTGATAAAAGTGAATCTGAGGCATAAC-3′ | ||
| Exon 8 | VIC p53Ex8-F92 | 5′ CCGC(C-c6-VIC)GCAGGGTGGTTGGGAGTAGATG-3′ | |
| NED p53Ex8-R94 | 5′ CCGC(C-c6-NED)GCGGTGATAAAAGTGAATCTGAGGCATAAC-3′ | ||
| Universal | VicUniEV1F | 5′ CGC(C-c6-VIC)GTCACGACACGAAAAC-3′ | |
| Universal | NedUniEV2R | 5′ CGC(C-c6-NED)GTCACGACACGAAACA-3′ | |
| Universal | UniEV1F | 5′ P-CGCCGTCACGACACGAAAAC-3′ | |
| Universal | UniEV2R | 5′ P-CGCCGTCACGACACGAAACA-3′ | |
| Universal | VicUniEV5Fa | 5′ VIC-CCGCCGTCACGACACGAAAAC-3′ | |
| Universal | NedUniEV6Ra | 5′ NED-CCGCCGTCACGACACGAAACA-3′ | |
| Universal | UniEV5F | 5′ P-CCGCCGTCACGACACGAAAAC-3′ | |
| Universal | UniEV6R | 5′ P-CCGCCGTCACGACACGAAACA-3′ | |
| K-ras | Exon 1 | F161 | 5′ CGTCACGACACGAAAACATAGTGTATTAACCTTATGTGTGACATGTTC-3′ |
| R162 | 5′ CGTCACGACACGAAACACAAAATGGTCAGAGAAACCTTTATCTGTATC-3′ | ||
| p53 | Exon 6 | F167 | 5′ CGTCACGACACGAAAACCTCTGATTCCTCACTGATTGCTCTTA-3′ |
| R168 | 5′ CGTCACGACACGAAACAGGCCACTGACAACCACCCTTAAC-3′ | ||
| Exon 8 | F173 | 5′ CGTCACGACACGAAAACCAGGGTGGTTGGGAGTAGATG-3′ | |
| R174 | 5′ CGTCACGACACGAAACAGGTGATAAAAGTGAATCTGAGGCATAAC-3′ | ||
| Exons 8–9 | p53ex8F | 5′ CGTCACGACACGAAAACTGTGGCTTCTCCTCCACCTAC-3′ | |
| p53ex9R | 5′ CGTCACGACACGAAACAGCCCCAATTGCAGGTAAAAC-3′ |
F, forward primer; R, reverse primer. Bases highlighted in bold correspond to the universal sequence.
aUnderlined bases (CCGCC) have a 3′ to 5′ phosphate (inverted) linkage. The fluorescent dye is coupled to the 3′ end of C instead of the 5′ end.
Universal PCR reactions for amplification of specific genes followed by universal primer amplification (50 μl) contained 20 mM Tricine, pH 8.7, 16 mM (NH4)2SO4, 2.5 mM MgCl2, 0.2 mM of dNTP, 0.2 μM of each universal primer, 0.02 μM of each gene-specific primer, 5 U of AmpliTaq Gold DNA polymerase and 150 ng of genomic DNA. Thermo-cycling conditions were as follows: 95°C for 10 min to activate AmpliTaq Gold polymerase, followed by 20 cycles of 94°C for 30 s, 65°C for 1 min, 72°C for 1 min (for gene-specific amplification) and then 30 cycles of 94°C for 30 s, 55°C for 1 min, 72°C for 1 min (for universal amplification), followed by a final extension step at 72°C for 7 min. In the universal PCR reaction, the universal primer pair consisted of a forward VIC-labeled primer and a reverse NED-labeled primer for the standard denaturation/renaturation’ procedure. In the ‘split label, denaturation, renaturation’ procedure, PCR amplification used one VIC- (or NED-) labeled universal primer and one unlabeled universal primer. In the ‘split label, lambda exonuclease’ procedure, PCR amplification used one VIC- (or NED-) labeled universal primer and one 5′ phosphorylated universal primer.
Prior to PCR amplification, primers were phosphorylated at the 5′ end by incubating 200 pmol of each unlabeled primer with 10 U of T4 polynucleotide kinase for 1 h at 37°C in a 25 μl-reaction volume containing 1× T4 polynucleotide kinase buffer (70 mM Tris–HCl, pH 7.6, 10 mM MgCl2, and 5 mM DTT) and 1 mM ATP. The enzyme was then heat inactivated for 20 min at 65°C, and aliquots of the phosphorylation reaction were stored at −20°C.
Preparation of heteroduplexed DNA substrates
In the standard denaturation/renaturation procedure, approximately equal ratios of VIC/NED-labeled wild-type PCR amplicons were mixed with VIC/NED-labeled mutant PCR amplicons in a 12 μl final volume (∼1500 ng total DNA). The wild-type control consisted of VIC/NED-labeled wild-type DNA PCR products alone in a 12 μl final volume (∼1500 ng total DNA). Taq DNA polymerase was inactivated by adding 1 μl of proteinase K (20 mg/ml) to each mixture, and incubating at 65°C for 30 min, followed by a 10 min incubation at 80°C to inactivate proteinase K. For denaturation/renaturation, PCR mixtures were then heated at 95°C for 2 min, and gradually cooled down to room temperature as follows: 95°C for 2 min, 95°C for 15 s, followed by a 0.2°C decrease in temperature every 15 s down to 45°C, and finally a 10 min incubation at 25°C.
For the ‘split label, denaturation, renaturation’ procedure, two distinct PCR mixtures were prepared as follows: approximately equal yields of VIC-labeled wild-type PCR products were mixed with NED-labeled mutant PCR products in a 12 μl final volume (∼1500 ng total DNA; PCR mixture 1); likewise, VIC-labeled mutant PCR products were mixed with NED-labeled wild-type PCR products in another 12 μl reaction (PCR mixture 2). The wild-type control consisted of a mixture of VIC-labeled wild-type PCR products with NED-labeled wild-type PCR products in a 12 μl final volume (∼1500 ng total DNA). As described above, Taq DNA polymerase was inactivated by adding proteinase K and PCR mixtures were subjected to denaturation/renaturation.
In the ‘split label, lambda exonuclease’ procedure, two distinct PCR mixtures were prepared as follows: approximately equal yields of VIC/Phosphate-labeled wild-type PCR products were mixed with Phosphate/NED-labeled mutant PCR products in a 12 μl final volume (∼1500 ng total DNA; PCR mixture 1); likewise, VIC/Phosphate-labeled mutant PCR products were mixed with Phosphate/NED-labeled wild-type PCR products in another 12 μl reaction (PCR mixture 2). The wild-type control consisted of a mixture of VIC/Phosphate-labeled wild-type PCR products with Phosphate/NED-labeled wild-type PCR products in a 12 μl final volume (∼1500 ng total DNA). Taq DNA polymerase was inactivated by adding proteinase K as described above. PCR mixtures were then incubated at 37°C for 1 h with 1 U of lambda exonuclease, which degrades 5′ phosphorylated DNA. Lambda exonuclease was then heat inactivated by incubating the reaction at 75°C for 10 min.
EndoV/ligase mutation scanning assay
The various buffer conditions tested for optimization of the EndoV/ligase assay are summarized in Table 2. Standard HEPES–Tris buffer conditions as well as Tricine buffer conditions I, II, III, IV, V and VII, all consisted of two-step procedures. In step 1, ∼6.5 μl of each heteroduplex PCR mixture, including the homoduplex wild-type control, were incubated at 65°C (for 40 or 60 min, as indicated) in a 20 μl first step reaction mixture containing the appropriate reagents. Then, in step 2, 15 μl of each step 1 reaction mixture was subjected to the second step reaction in a 20 μl final volume at 65°C (for 30 or 60 min, as indicated), by adding the appropriate reagents. Conditions VI and VIII were single-step reactions, in which ∼6.5 μl of each heteroduplex PCR mixture, including the homoduplex wild-type control, were incubated for 120 min at 65°C in a 20 μl reaction containing the indicated reagents. For determination of the sensitivity of the single-step EndoV/ligase assay, reactions were performed under condition VIII, except that 1 μM EndoV and 12 nM ligase were used instead of 500 nM and 6 nM, respectively.
Table 2. Summary of the various buffer conditions tested for optimization of the EndoV/ligase assay.
| Standard conditions | |
| 40 min | 1. 1× EndoV buffer = 20 mM HEPES, pH 7.5, 1 mM DTT, with 1 μM EndoV |
| 30 min | 2. 1× Ligase buffer = 20 mM Tris, pH 8.5, 50 mM KCl, 10 mM DTT, with 3 nM Ligase + 1 mM NAD |
| Condition I | |
| 60 min | 1. 1× EndoV buffer = 20 mM Tricine, pH 8, 1 mM DTT, with 1 μM EndoV |
| 60 min | 2. 1× Ligase buffer = 40 mM Tricine, pH 8, 10 mM DTT, with 6 nM Ligase + 5 mM NAD |
| Condition II | |
| 60 min | 1. 1× EndoV buffer = 20 mM Tricine, pH 8, 5 mM DTT, with 1 μM EndoV + 6 nM Ligase + 5 mM NAD |
| 60 min | 2. 1× Ligase buffer = 40 mM Tricine, pH 8, 6.25 mM DTT |
| Condition III | |
| 60 min | 1. 1× EndoV buffer = 40 mM Tricine, pH 8, 1 mM DTT, with 1 μM EndoV |
| 60 min | 2. 1× Ligase buffer = 40 mM Tricine, pH 8, 10 mM DTT, with 6 nM Ligase + 5 mM NAD |
| Condition IV | |
| 60 min | 1. 1× EndoV buffer = 40 mM Tricine, pH 8, 5 mM DTT, with 1 μM EndoV + 6 nM Ligase + 5 mM NAD |
| 60 min | 2. 1× Ligase buffer = 40 mM Tricine, pH 8, 6.25 mM DTT |
| Condition V | |
| 60 min | 1. 1× EndoV buffer = 40 mM Tricine, pH 8, 5 mM DTT, with 500 nM EndoV + 6 nM Ligase + 5 mM NAD |
| 60 min | 2. 1× Ligase buffer = 40 mM Tricine, pH 8, 6.25 mM DTT |
| Condition VI | |
| 120 min | 1× EndoV/ligase buffer = 80 mM Tricine, pH 8, 5 mM DTT, with 500 nM EndoV + 6 nM Ligase + 5 mM NAD |
| Condition VII | |
| 60 min | 1. 1× EndoV buffer = 40 mM Tricine, pH 8, 5 mM DTT, with 500 nM EndoV + 6 nM Ligase + 1 mM NAD |
| 60 min | 2. 1× Ligase buffer = 40 mM Tricine, pH 8, 6.25 mM DTT |
| Condition VIII | |
| 120 min | 1× EndoV/ligase buffer = 80 mM Tricine, pH 8, 5 mM DTT, with 500 nM EndoV + 6 nM Ligase + 1 mM NAD |
All 1× EndoV buffers and 1× EndoV/ligase buffers contain 5 mM MgCl2, 5% DMSO, 1.5 M Betain, and 2% glycerol. All 1× Ligase buffers contain 1.25 mM MgCl2 and 20 μg/ml BSA.
Finally, all reactions were terminated by adding EDTA to a final concentration of 10 mM. This inhibited any further EndoV cleavage activity during capillary electrophoresis. One microliter aliquots of the reaction mixtures were denatured at 95°C for 2 min in 9 μl Hi-Di formamide along with 0.4 μl GeneScan™ –500 (LIZ™) Size Standard, and run on the ABI 3730 fluorescence-based capillary electrophoresis instrument (Applied Biosystems, Foster City, CA). Electrophoresis was carried out for 1200 s at 15 kV in POP-7 polymer at 60°C. On completion of the electrophoretic run, a capillary array image was displayed with ABI collection software v1.0. Data analysis was achieved using Gene Mapper fragment analysis software v3.0 (Applied Biosystems, Foster City, CA). Ultimately, capillary array images of the fragment analysis data were displayed with Gel Render, an unpublished program developed in our laboratory to correct for differences in capillary length and fragment mobility in individual capillaries by using internal size standards, and presented as an electrophoretogram in the form of the familiar gel picture.
Signal intensity and signal-to-noise ratio measurement
Fluorescence intensity values of mutation and background cleavage bands obtained from Gene Mapper fragment analysis software v3.0 were normalized to the intensity of the 200 base fragment of the (LIZ™) internal Size Standard for each capillary lane. Signal-to-noise ratios were calculated using the normalized intensity for the brightest VIC- and NED-labeled background bands, which were 144 and 115 bases, respectively.
RESULTS
Reduction of EndoV cleavage of the 5′ fluorescent label off the PCR products and validation of a universal labeling strategy
To achieve high-throughput detection of somatic mutations in tumors, we sought to transfer the EndoV/Ligase scanning assay to an automated capillary electrophoresis format. Preliminary EndoV cleavage data (using standard 5′-end labeled PCR fragments as substrates) revealed the presence of two unanticipated bands migrating at the same mobility as fragments of 102 (VIC) and 94 (NED) bases, respectively. These anomalously migrating bands were removable by filtration with a 10 kDa cutoff. Thus, they most likely resulted from EndoV cleavage near the label, most likely one base 3′ to it, containing the 5′ fluorescent label, a phosphate group (to provide charge), and an additional base. This activity may correspond to the 5′ exonuclease activity reported for Escherichia coli EndoV (54). These bands created two problems: (i) they would obscure authentic signal migrating at the same position, and (ii), this cleavage would reduce the overall intensity of the desired mutation signal.
Three sets of p53 exon 8 PCR primers with differently modified 5′ fluorescent labels (listed in Table 1) were tested for their ability to resist EndoV cleavage one base 3′ to the label. These primers all harbored on their 5′ end a CGCCGC sequence, known to be refractory to EndoV cleavage when this sequence is present in the middle of a fragment (44). The first primer set was 5′-end labeled, the second set had two 2′-O-methylated C residues added to the 5′ end label, and the third carried an internal label, i.e. a C5-dye labeled cytosine inserted within the EndoV resistant sequence. As shown on the capillary array image (Supplementary Material, Figure S1), when using either the 5′-end labeled primers (panel A) or the 2′-O-methylated C primers (panel B), both heteroduplexed (Mut) and homoduplexed (WT) PCR fragments treated with EndoV (both top panels, lanes 2 and 5) showed a dramatic decrease in fluorescence intensity as compared to the untreated ones (lanes 1 and 4). Note that the labeled PCR amplicons are not precisely aligned, due to differences in capillary array lengths. Quenching the EndoV reaction with EDTA post incubation (lanes 3 and 6), resulted in a less dramatic decrease in fluorescence intensity in both cases, suggesting that thermostable EndoV retained residual activity even in high formamide during the capillary run. In contrast, internally labeled PCR fragments treated with EndoV showed only a small decrease in fluorescence intensity (top panel C, lanes 2 and 5) as compared to the untreated ones (lanes 1 and 4). Protection from cleavage was independent of post-incubation EDTA quenching (lanes 3 and 6). These results demonstrate that internally labeled primers eliminate false bands migrating at ∼100 bases, and should increase the intensity of authentic signal.
Since synthesis of new internally labeled primers for each gene is both costly and time consuming, we developed a universal labeling strategy. This strategy would also be amenable to multiplexed amplification of dozens of genes, addressing the practical reality that tumor DNA is often limiting. This strategy employs two sets of primer pairs: unlabeled gene-specific primers that harbor universal tails on their 5′ ends (see Table 1 and Figure 1), and fluorescently labeled or phosphorylated universal primers that can be used for any gene/exon amplification (Figure 1). During the first 20 PCR cycles, an annealing temperature of 65°C allows gene-specific primers at low concentration to anneal selectively to their target DNA template, while universal primers do not hybridize to the gene-specific primer universal tails. By shifting the annealing temperature to 55°C for an additional 30 PCR cycles, the dye-labeled universal primers predominate in amplifying labeled product. The universal primers are identical except for the last two bases on the 3′ end, assuring that each primer amplifies and labels only the intended strand. Since the wrong universal primer may hybridize but not extend, the efficiency of this type of universal amplification is <2-fold per cycle, requiring the additional 30 cycles. Primer dimers and inappropriately short amplicons form hairpins (or panhandles) and are thus unable to amplify. In the course of optimizing reaction conditions, we also improved PCR yield by switching from a standard 10 mM Tris–HCl, pH 8.3 PCR buffer to the 20 mM Tricine, pH 8.7, 16 mM (NH4)2SO4 buffer previously developed in our lab (55).
When testing the internally labeled primers for universal primer PCR amplification of p53 exon 6, we detected two high molecular weight artifacts of ∼600 bases (Supplementary Material, Figure S2A). Thus, we also explored an inverted linkage labeled primer pair (i.e. fluorescent dye-3′-CCGCC-5′-5′-universal sequence-3′; see also Table 1), which did not yield the artifacts, but the fluorescent label was somewhat susceptible to cleavage by EndoV (Supplementary Material, Figure S2B). Despite these distinct features, signal intensity of the EndoV mutation cleavage products is strong and clearly detectable in both cases even in the absence of the ligase proofreading step (Supplementary Material, Figure S2A and S2B, lanes 1 and 3 are wild type, lanes 2 and 4 are heteroduplexes of wild type to mutant). Thus, two different chemistries for attaching fluorescent groups may be used, each one overcoming minor shortcomings in the other.
Reduction of noise resulting from artifact and background bands
Artifactual and background bands could be grouped into the following categories: (i) inappropriate bands after PCR amplification and prior to EndoV treatment (Supplementary Material, Figure S3A, lanes 1 and 3), (ii) multiple EndoV generated bands at near full length, and at slightly larger than primer lengths (Supplementary Material, Figure S3A, lanes 6, 8 and 10), and (iii) specific EndoV generated bands that are not at the position of the mutation (Supplementary Material, Figure S3A, lanes 6, 8 and 10). Detection of the R273H (G→A) mutation in p53 exon 8 was used as a challenging test system for the experiments below. In the original assay, this mutation gives weak signal, and its 350 base substrate generates higher levels of background cleavage (44).
Category (i) artifactual bands are a consequence of the particular dyes used in the PCR amplification, and could be eliminated by labeling each strand in separate amplification reactions as described in Figure 1. This may be achieved by either modifying the standard method (PCR amplification using two labeled primers in one reaction) to eliminate the unlabeled strands using lambda exonuclease (Supplementary Material, Figure S3A, lanes 5, 7 and 9) or by the ‘split label, denaturation, renaturation’ method (Supplementary Material, Figure S3A, lanes 11, 13 and 15). Use of lambda exonuclease provides the added advantage of generating 100% heteroduplexed substrate (i.e. containing only the G/T or A/C mismatch), and consequently doubles the mutation signal intensity (Supplementary Material, Figure S3A, compare lanes 8 and 10 to lanes 14 and 16).
Category (ii) artifactual bands resulted from sequential non-specific EndoV cleavage on labeled and unlabeled strands near the dynamically fraying ends of the DNA. The problem is ameliorated by the concurrent presence of ligase that reseals one strand before the opposite can be cleaved (see next section). In pursuing the source of this background we also determined that shorter PCR amplicons (150–250 bp) give greater background (not shown), while longer PCR amplicons (591 bp) give less background (Supplementary Material, Compare Figure S3A, lane 16 to Figure S3C, lane 10). Longer strands presumably have a reduction in the frequency of unpaired conformations, leading to a slower rate of non-specific EndoV cleavage. To completely eliminate interference of category (ii) artifactual bands with authentic signal, all PCR primers were redesigned to reside at least 60 bases upstream and downstream of the scanning region.
Category (iii) artifactual bands are sequence-specific bands that reflect the ability of that particular sequence to become transiently unpaired under the particular buffer conditions required to achieve cleavage of authentic mismatched bases. Consistent with this hypothesis is the observation that the intensity of such bands varied with changes in buffer pH (see below), as well as with choice of organic co-solvent (5% DMSO + 1.5 M betaine versus 5% tetramethylene sulfone alone, data not shown).
Several other potential causes for these bands were considered and ultimately eliminated following experimentation. One possibility was that PCR errors introduced during amplification were subsequently cleaved after heteroduplex formation. This hypothesis was rejected when no change in these bands was observed following amplification with two different commercially available proofreading polymerases (TaqPlus Precision PCR System, Stratagene, La Jolla, CA or Platinum Taq DNA Polymerase High Fidelity, Gibco BRL Life Technologies, Inc., Gaithersburg, MD; data not shown). A second possible explanation for category (iii) artifactual bands is the formation of multi-stranded structures with unresolved Holliday junctions or Flap structures. These structures are known substrates for EndoV (54). This possibility was also eliminated when there was no reduction in bands following the addition of mesophilic or thermophilic DNA binding proteins (e.g., RecA, T4 gene 32 protein, Aquifex ssb, RuvA and RuvB) to the reaction. Even though these proteins minimize formation of potential tertiary or secondary structures by lambda exonuclease-generated single strands during the heteroduplex formation step, the artifactual bands were unaffected. A third hypothesis to account for the category (iii) bands was that single-stranded fragments form transient secondary Y structures that become substrates for EndoV (54). Although EndoV does cleave single-stranded DNA at non-random positions, the bands differ from those generated by cleavage of heteroduplexed DNA (Supplementary Material, Figure S3B, lanes 2, 4, 6, and 8). Category (iii) artifactual bands are also rectified by combining EndoV cleavage with resealing by ligase in a single step assay (see below).
Real-time Ligase proofreading in a homogeneous single-step EndoV/ligase assay
Optimization of the original EndoV/ligase mutation scanning assay entailed finding artificial buffer conditions for EndoV that minimized cleavage of matched sites while allowing the mismatched site to be cleaved (44,50). This was followed by shifting the reaction to conditions that maximized ligase resealing of nicks at matched sites, while allowing mismatched sites to remain unligated (44). The task is exacerbated by EndoV cleavage one base 3′ to the mismatch. Such substrates with penultimate mismatches are more difficult to discriminate by ligases than substrates with mismatches at the 3′ end (53). If ligase could be made to work during the EndoV reaction, it could continuously proofread and reseal nicks at perfect matches. Signal would be optimized when the desired EndoV cleavage at mismatches minus their unwanted re-ligation exceeds the unwanted EndoV cleavage at matches minus their desired ligation. We have previously demonstrated that EndoV shows optimal mismatch cleavage activity in high pH and no salt, minimal ionic strength buffer conditions (50), whereas the ligase enzyme exhibits higher fidelity at lower pH and 50–100 mM KCl (53). The inherent challenge for a one-step reaction was to find conditions that allow EndoV to cleave at the mismatch (in 5% DMSO, 1.5 M betaine), while allowing ligase to exhibit suitable fidelity and reactivity under the same conditions.
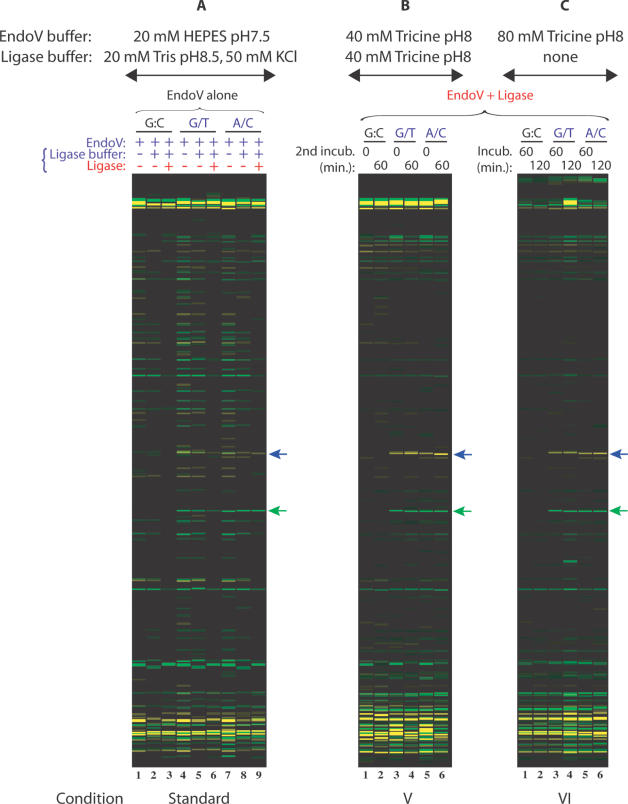
We tested a variety of buffer and incubation conditions (see Table 2) in over 500 capillary electrophoresis runs for the ability to detect the challenging p53 R273H G→A mutation. When EndoV and ligase are used together, a dramatic reduction in bands from background cleavage results (see Figure 2, lanes 16–24, and Figure 3B and 3C). Compared to the sequential use of EndoV and ligase in a two-step reaction (see Figure 2, lanes 3, 5, 8, 10, 13 and 15 and Figure 3A, lanes 3, 6 and 9), fewer background bands are created. An additional incubation with higher concentrations of Tricine buffer to enhance ligation fidelity (see Figure 2, lanes 20, 21, 23 and 24 and Figure 3B, lanes 2, 4 and 6) does not provide an enhancement in mutation signal intensity nor any further reduction of background cleavage bands compared to a single incubation with EndoV and ligase (see Figure 2, lanes 16, 19 and 22 and Figure 3C, lanes 1–6). While increasing the pH from 7.5 to 8.0 initially creates more EndoV cleavage events (compare Figure 3A, lanes 4 and 7 to Figure 2, lanes 6 and 11), the higher ionic strength in the buffer also allows ligase to continuously proofread and reseal these undesired nicks (Figure 2, lanes 17, 18, 20, 21, 23 and 24). Increasing the pH from our original conditions to a pH of 8 provides an effective compromise between optimal conditions for the two enzymes, at the cost of some increase in background EndoV cleavage (readily religated by ligase). There is also some loss of ligation fidelity at the compromise pH, resulting in the disappearance of some correctly-cleaved bands after ligase treatment (Supplementary Material, Figure S4C, compare mutation signal intensity in Conditions I and III to Conditions II and IV). Nevertheless, reduction in category (iii) background bands by simultaneous enzyme incubation is readily apparent by comparing lanes 1–15 with lanes 16–24 in Figure 2.
Figure 2.
Improvement of signal-to-noise ratio in the EndoV/ligase assay when combining both enzymes in a single incubation step. Universal PCR primers (VicUniEV1F/NedUniEV2R), along with gene-specific primers F173 and R174, were used to amplify 350 bp fragments of p53 exon 8 carrying R273H (G→A) mutation. VIC- and NED-labeled mutation cleavage products are 158 nt (green arrow) and 195 nt (blue arrow) in size. The ‘split label, denaturation, renaturation’ method was used to generate heteroduplexes of either top strand wild-type:bottom strand mutant (G/T mismatch) or top strand mutant:bottom strand wild-type (A/C mismatch). Wild-type PCR product mixtures were used as controls (G:C match). Tested EndoV/ligase assay conditions are briefly indicated on top and bottom of the figure and detailed in Table 2. Condition III is a standard two-step reaction, while condition IV combines both enzymes in the first incubation step [EndoV + ligase], followed by a second incubation in ligase buffer. Reaction mixtures were electrophoresed on the ABI 3730 fluorescence-based capillary electrophoresis instrument. Data were analyzed using Gene Mapper fragment analysis software.
Figure 3.
Comparison of a single-step assay, a standard two-step assay, and a combined two-step EndoV/ligase assay in Tricine buffer. Universal PCR primers (VicUniEV1F/NedUniEV2R), along with gene-specific primers F173 and R174, were used to amplify 350 bp fragments of p53 exon 8 carrying R273H (G→A) mutation. VIC- and NED-labeled mutation cleavage products are 158 nt (green arrow) and 195 nt (blue arrow) in size. The ‘split label, denaturation, renaturation’ method was used to generate heteroduplexes of either top strand wild-type:bottom strand mutant (G/T mismatch) or top strand mutant:bottom strand wild-type (A/C mismatch). Wild-type PCR product mixtures were used as controls (G:C match). EndoV/ligase assay conditions tested are briefly indicated on top of the figure and detailed in Table 2. (A) Standard EndoV/ligase procedure; (B) Condition V is a two-step reaction combining both enzymes in the first incubation step [EndoV + ligase], followed by a second incubation in ligase buffer; (C) Condition VI is a single-step reaction with both EndoV and ligase. Reaction mixtures were electrophoresed on the ABI 3730 fluorescence-based capillary electrophoresis instrument. Data were analyzed using Gene Mapper fragment analysis software.
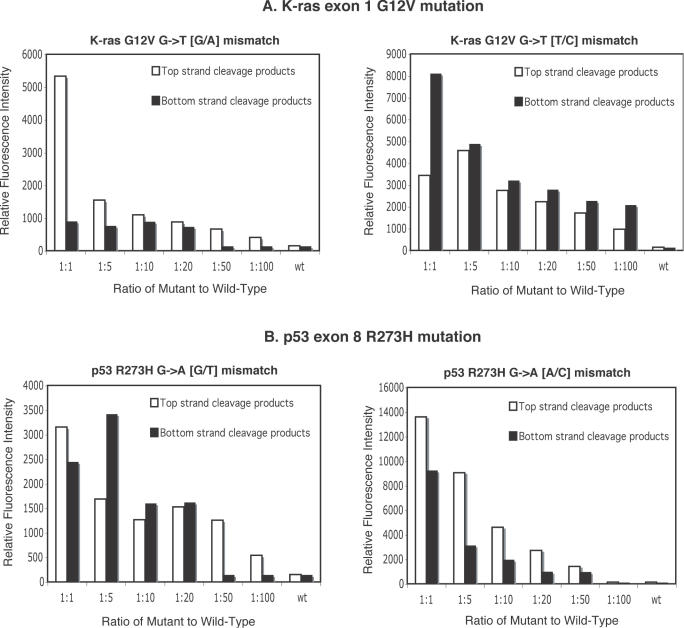
The intensity of each authentic mutation signal was quantified, and the brightest background band for each fluorescent dye was chosen for estimating a conservative, worst case signal-to-noise ratio for various buffer and incubation conditions (Supplementary Material, Figures S4 and S5). In each case, incubation of proofreading ligase during the EndoV cleavage step provided a 5- to 13-fold increase in signal intensity (Supplementary Material, Figure S4, panels C and D, and S5, panels C and D), with significant improvement of signal-to-noise compared to the original conditions. Dilution experiments with known mutations were performed to test for the ability to detect tumor mutations in the presence of an excess of wild-type DNA (see Figure 4). In the case of K-ras exon 1, the G12V G→T mutation could be detected in a 100-fold excess of wild-type DNA on three out of four strands of the two heteroduplexes and in a 20-fold excess for the fourth strand. Even for the more difficult p53 R273H G→A mutation, the combined single-step assay can detect the mutation in a 50-fold excess of wild-type DNA in 3 out of 4 strands. These results represent a 5- to 10-fold improvement over the original two-step assay (44).
Figure 4.
Sensitivity of the EndoV/ligase single-step assay. EndoV/ligase assay conditions are detailed in Materials and Methods. (A) Relative fluorescence intensities of cleavage products for K-ras exon 1 G12V mutation with their respective mutant-to-wild type dilution ratios. Universal PCR primers (VicUniEV1F/NedUniEV2R), along with gene-specific primers F161 and R162, were used to amplify 300 bp PCR fragments of K-ras exon 1 carrying mutation G12V (G→T). The ‘split label, denaturation, renaturation’ method was used to generate heteroduplexes with either a G/A mismatch or a T/C mismatch for various mutant:wild-type ratios. Wild-type PCR product mixtures (wt) were used as controls. Relative fluorescence intensity is defined as the area under a signal's peak as determined by Gene Mapper fragment analysis software. (B) Relative fluorescence intensities of cleavage products of p53 exon 8 R273H mutation with their respective mutant-to-wild type dilution ratios. Universal PCR primers (VicUniEV1F/NedUniEV2R), along with gene-specific primers F173 and R174, were used to amplify p53 exon 8 fragments carrying R273H mutation (G→A). The ‘split label, denaturation, renaturation’ method was used to generate heteroduplexes with either a G/T mismatch or a A/C mismatch for various mutant:wild-type ratios. Wild-type PCR mixtures (wt) were used as controls.
DISCUSSION
Mutation and polymorphism detection is of increasing importance in the field of molecular genetics, resulting in a profusion of enzymatic, chemical and biophysical based methods for mutation detection (13,23). Few of them can detect previously unknown mutations and at the same time position them accurately. Moreover, current methods often lack high specificity or sensitivity, or are too labor-intensive to be amenable to high-throughput mutation screening (23). This work systematically evaluated and developed the following improvements in all aspects of our original EndoV/ligase mutation scanning assay (44): (i) shifting the assay to automated capillary electrophoresis readout, (ii) development of fluorescent labeling chemistries (5′ internal and inverted linkage labeled) that resist removal by EndoV from the PCR amplicon, (iii) development of universal PCR primers for more cost effective and robust labeling, and (iv) development of real-time ligase proofreading conditions. Often, these improvements worked synergistically or revealed unanticipated benefits. For example, suppressing fluorescent dye removal by EndoV was a necessary precondition in achieving the enhanced signal that resulted from continuous EndoV cleavage and ligase proofreading. The universal primer design was conceived to reduce primer synthesis cost and allow for simultaneous amplification of multiple target regions from valuable and limited tumor DNA samples. This design change also enabled significant reduction in PCR generated background bands, both through panhandle formation of primer dimers and shorter fragments, and through the two split-label protocols (Steps 2A and 2B in Figure 1). The latter allowed us to seamlessly double signal intensity by using lambda exonuclease to form 100% mismatched heteroduplexes. Complementary single-strand specific nuclease-based mismatch detection assays (CEL I and S1) demonstrate nibbling of end-labeled duplexes and nuclease-resistant background (56), and may also benefit from the approaches outlined in this report.
Dilution experiments with K-ras and p53 gene mutations show that our EndoV with real-time ligase proofreading scanning assay can detect mutations at a sensitivity of 1:100 and 1:50 (mutant:wild-type DNA ratio), respectively, in three out of four strands of the two heteroduplexes. While another approach based on ligation-mediated PCR and the use of glycosylases TDG and MutY has similar sensitivity (57), it requires multiple steps and cannot detect A→T or G→C transversion mutations, or single base deletion/insertions (as EndoV/ligase can). When the position of the potential mutation is known, even greater sensitivities may be achieved by PCR/LDR/Universal array (15–17), or by suppression of wild-type DNA amplification using PNA (32,58), although the latter is limited to a single hot-spot region per assay. However, our concern here is with detecting unknown, potentially rare mutations in clinical samples. The current single-step real-time EndoV/ligase assay, represents a 5- to 10-fold improvement over our original two-step conditions (44). EndoV detects 98% of mutations including all insertions and deletions even when the position is unknown. This new higher level of sensitivity allows for additional pooling of the same amplicon from multiple tumor samples within the same EndoV/ligase reaction, beyond what we have previously demonstrated (17). This is critical, since tumor samples that have not been microdissected typically contain 10–50% of stromal infiltration (i.e. normal cells), and the ability to detect mutations in pools of 5 or even 10 samples represents a great advantage over automated fluorescent sequencing. Real-time EndoV/ligase proofreading represents a compromise of sub-optimal buffer conditions for both enzymes, and sensitivities may yet be improved an additional order of magnitude by use of optimized variant or engineered EndoV and/or DNA ligase.
Developing cancer screening tools for clinical samples (biopsies, stool or bodily fluids) present special problems. They are often limited in either the total amount of recovered DNA, or the percentage of authentic neoplastic DNA (33,59–64). Future diagnostic tests will require screening numerous candidate genes. For example, the current list of biologically important genes for colon cancer include APC, β-catenin (3,5), K-ras (2), B-raf (7,65), p53 (66), PI3KCA (8), hCDC4 (67), Tyr kinases (9), phosphatases (10), BAX, TGFBRII and TCF4 (1,6,5). Clinically useful diagnostics will likely incorporate detection of the appearance of somatic mutations in these genes (60). The universal primer labeling strategy in this report is compatible with multiplexed pre-amplification of candidate exons in a single reaction, followed by uniplexed amplification/labeling of individual fragments for mutation scanning. Since DNA is usually limiting, clinical samples are generally not amenable to very narrow assays that selectively amplify or detect mutations in a few codons at a time. Likewise, oligonucleotide array-based assays suffer from low inherent sensitivity due to only minor differences in hybridization kinetics between wild-type and mutant probes (11). They have thus failed to provide comprehensive coverage of all mutations in a gene, e.g. scoring 81% (68), 84% (69) and 92% (70) for p53. Furthermore, the number of potential inactivating frameshift mutations is in vast excess of the number of inactivating missense mutations; hybridization chips do not contain addresses for such frameshift mutations, and hence completely fail to detect insertions and deletions. In contrast, in vitro transcription–translation, followed by either multicolor size-based or MALDI-TOF detection, allows identification of truncating mutations (29–31). The sensitivity of in vitro transcription–translation approaches has been extended to 2.5% for identifying APC gene mutations in fecal DNA samples from colorectal cancer patients (31). This sensitivity is obtained by diluting sample to ∼10 molecules of DNA template in each of dozens to hundreds of individual PCR amplifications. We could combine this ‘digital’ approach in our assay, or most likely just use our EndoV/real-time ligase approach to directly detect mutations that are present in a very small portion of the total cells, especially for pre-symptomatic diagnosis.
The importance of a sensitive method capable of detecting multiple unknown mutations in a cancer gene is emphasized by a recent study showing that polyclonal resistance to Gleevec (a small molecule inhibitor of BCR-ABL kinase) in chronic myeloid leukemia patients is conferred by BCR-ABL point mutations, where two or more distinct mutations are sometimes harbored in a single patient (71,72). In this case, the authors sub-cloned the PCR products and sequenced 10 independent clones per patient, a time-consuming approach that was required when sequencing missed critical mutations (71). Once again, a robust and sensitive assay such as the EndoV/real-time ligase reaction could be used in this context to identify mutations in a minority of cells. As newer directed therapies that overcome most Gleevec resistance mutations become available (73), accurate profiling of the mutational status becomes critical for patient care. This emerging need for unknown mutation detection is recapitulated in the treatment of non-small cell lung cancers (NSCLC), where somatic mutations in the EGF receptor were found only in tumor samples from patients who responded to Iressa therapy, but not in Iressa insensitive tumors (74,75). Here too, detection of mutations directly from sputum or non-microdissected lung biopsies, either by our EndoV/real-time ligase mutation scanning assay, or harmonized with PCR/LDR/Universal array identification of known mutations (76), can help predict therapeutic response.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Donald Bergstrom, Daniel Notterman, Weiguo Cao, and Yu-Wei Cheng for helpful discussion, and Reyna Favis and Daniel Turner for critical reading of the manuscript. We thank Michael O'Donnell for gifts of Aquifex ssb, RuvA and RuvB. We thank Seiki Kuramitsu for gift of thermostable RecA, and Hideo Shinagawa for gifts of thermostable RuvA and RuvB. Work in the Barany laboratory is sponsored in part by a sponsored research grant from Applied Biosystems Inc., for which Francis Barany also serves as a consultant. This work was supported by grants from the National Cancer Institute (P01-CA65930 and RO1-CA81467).
REFERENCES
- 1.Futreal P.A., Coin,L., Marshall,M., Down,T., Hubbard,T., Wooster,R., Rahman,N. and Stratton,M.R. (2004) A census of human cancer genes. Nat. Rev. Cancer, 4, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adjei A.A. (2001) Blocking oncogenic Ras signaling for cancer therapy. J. Natl Cancer Inst., 93, 1062–1074. [DOI] [PubMed] [Google Scholar]
- 3.Grady W.M. and Markowitz,S.D. (2002) Genetic and epigenetic alterations in colon cancer. Annu. Rev. Genomics Hum. Genet., 3, 101–128. [DOI] [PubMed] [Google Scholar]
- 4.Lindblom A. (2001) Different mechanisms in the tumorigenesis of proximal and distal colon cancers. Curr. Opin. Oncol., 13, 63–69. [DOI] [PubMed] [Google Scholar]
- 5.Narayan S. and Roy,D. (2003) Role of APC and DNA mismatch repair genes in the development of colorectal cancers. Mol. Cancer, 2, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duval A. and Hamelin,R. (2002) Mutations at coding repeat sequences in mismatch repair-deficient human cancers: toward a new concept of target genes for instability. Cancer Res., 62, 2447–2454. [PubMed] [Google Scholar]
- 7.Rajagopalan H., Bardelli,A., Lengauer,C., Kinzler,K.W., Vogelstein,B. and Velculescu,V.E. (2002) Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature, 418, 934. [DOI] [PubMed] [Google Scholar]
- 8.Samuels Y., Wang,Z., Bardelli,A., Silliman,N., Ptak,J., Szabo,S., Yan,H., Gazdar,A., Powell,S.M., Riggins,G.J. et al. (2004) High frequency of mutations of the PIK3CA gene in human cancers. Science, 304, 554. [DOI] [PubMed] [Google Scholar]
- 9.Bardelli A., Parsons,D.W., Silliman,N., Ptak,J., Szabo,S., Saha,S., Markowitz,S., Willson,J.K., Parmigiani,G., Kinzler,K.W. et al. (2003) Mutational analysis of the tyrosine kinome in colorectal cancers. Science, 300, 949. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z., Shen,D., Parsons,D.W., Bardelli,A., Sager,J., Szabo,S., Ptak,J., Silliman,N., Peters,B.A., van der Heijden,M.S. et al. (2004) Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science, 304, 1164–1166. [DOI] [PubMed] [Google Scholar]
- 11.Hacia J.G., Brody,L.C., Chee,M.S., Fodor,S.P. and Collins,F.S. (1996) Detection of heterozygous mutations in BRCA1 using high density oligonucleotide arrays and two-colour fluorescence analysis. Nature Genet., 14, 441–447. [DOI] [PubMed] [Google Scholar]
- 12.Hacia J.G. (1999) Resequencing and mutational analysis using oligonucleotide microarrays. Nature Genet., 21, 42–47. [DOI] [PubMed] [Google Scholar]
- 13.Kirk B.W., Feinsod,M., Favis,R., Kliman,R.M. and Barany,F. (2002) Single nucleotide polymorphism seeking long term association with complex disease. Nucleic Acids Res., 30, 3295–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khanna M., Park,P., Zirvi,M., Cao,W., Picon,A., Day,J., Paty,P. and Barany,F. (1999) Multiplex PCR/LDR for detection of K-ras mutations in primary colon tumors. Oncogene, 18, 27–38. [DOI] [PubMed] [Google Scholar]
- 15.Gerry N.P., Witowski,N.E., Day,J., Hammer,R.P., Barany,G. and Barany,F. (1999) Universal DNA microarray method for multiplex detection of low abundance point mutations. J. Mol. Biol., 292, 251–262. [DOI] [PubMed] [Google Scholar]
- 16.Favis R., Day,J.P., Gerry,N.P., Phelan,C., Narod,S. and Barany,F. (2000) Universal DNA array detection of small insertions and deletions in BRCA1 and BRCA2. Nature Biotechnol., 18, 561–564. [DOI] [PubMed] [Google Scholar]
- 17.Favis R., Huang,J., Gerry,N.P., Culliford,A., Paty,P., Soussi,T. and Barany,F. (2004) Harmonized microarray/mutation scanning analysis of TP53 mutations in undissected colorectal tumors. Hum. Mutat., 24, 63–75. [DOI] [PubMed] [Google Scholar]
- 18.Pastinen T., Raitio,M., Lindroos,K., Tainola,P., Peltonen,L. and Syvanen,A.C. (2000) A system for specific, high-throughput genotyping by allele-specific primer extension on microarrays. Genome Res., 10, 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao W. and Oefner,P.J. (2001) Denaturing high-performance liquid chromatography: a review. Hum. Mutat., 17, 439–474. [DOI] [PubMed] [Google Scholar]
- 20.Larsen L.A., Christiansen,M., Vuust,J. and Andersen,P.S. (2000) High throughput mutation screening by automated capillary electrophoresis. Comb. Chem. High Throughput Screen., 3, 393–409. [DOI] [PubMed] [Google Scholar]
- 21.Fodde R. and Losekoot,M. (1994) Mutation detection by denaturing gradient gel electrophoresis (DGGE). Hum. Mutat., 3, 83–94. [DOI] [PubMed] [Google Scholar]
- 22.Nataraj A.J., Olivos-Glander,I., Kusukawa,N. and Highsmith,W.E.,Jr (1999) Single-strand conformation polymorphism and heteroduplex analysis for gel-based mutation detection. Electrophoresis, 20, 1177–1185. [DOI] [PubMed] [Google Scholar]
- 23.Taylor G.R. (1999) Enzymatic and chemical cleavage methods. Electrophoresis, 20, 1125–1130. [DOI] [PubMed] [Google Scholar]
- 24.Syvanen A.C. (2001) Accessing genetic variation: genotyping single nucleotide polymorphisms. Nature Rev. Genet., 2, 930–942. [DOI] [PubMed] [Google Scholar]
- 25.Cooper C.S. (2001) Applications of microarray technology in breast cancer research. Breast Cancer Res., 3, 158–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gross E., Arnold,N., Goette,J., Schwarz-Boeger,U. and Kiechle,M. (1999) A comparison of BRCA1 mutation analysis by direct sequencing, SSCP and DHPLC. Hum. Genet., 105, 72–78. [DOI] [PubMed] [Google Scholar]
- 27.Footz T., Somerville,M.J., Tomaszewski,R., Sprysak,K.A. and Backhouse,C.J. (2003) Heteroduplex-based genotyping with microchip electrophoresis and dHPLC. Genet. Test, 7, 283–293. [DOI] [PubMed] [Google Scholar]
- 28.Andersen P.S., Jespersgaard,C., Vuust,J., Christiansen,M. and Larsen,L.A. (2003) Capillary electrophoresis-based single strand DNA conformation analysis in high-throughput mutation screening. Hum. Mutat., 21, 455–465. [DOI] [PubMed] [Google Scholar]
- 29.Garvin A.M., Parker,K.C. and Haff,L. (2000) MALDI-TOF based mutation detection using tagged in vitro synthesized peptides. Nat. Biotechnol., 18, 95–97. [DOI] [PubMed] [Google Scholar]
- 30.Gite S., Lim,M., Carlson,R., Olejnik,J., Zehnbauer,B. and Rothschild,K. (2003) A high-throughput nonisotopic protein truncation test. Nat. Biotechnol., 21, 194–197. [DOI] [PubMed] [Google Scholar]
- 31.Traverso G., Diehl,F., Hurst,R., Shuber,A., Whitney,D., Johnson,C., Levin,B., Kinzler,K.W. and Vogelstein,B. (2003) Multicolor in vitro translation. Nat. Biotechnol., 21, 1093–1097. [DOI] [PubMed] [Google Scholar]
- 32.Sun X., Hung,K., Wu,L., Sidransky,D. and Guo,B. (2002) Detection of tumor mutations in the presence of excess amounts of normal DNA. Nat. Biotechnol., 20, 186–189. [DOI] [PubMed] [Google Scholar]
- 33.Traverso G., Shuber,A., Levin,B., Johnson,C., Olsson,L., Schoetz,D.J.,Jr, Hamilton,S.R., Boynton,K., Kinzler,K.W. and Vogelstein,B. (2002) Detection of APC mutations in fecal DNA from patients with colorectal tumors. N. Engl. J. Med., 346, 311–320. [DOI] [PubMed] [Google Scholar]
- 34.Cotton R.G., Rodrigues,N.R. and Campbell,R.D. (1988) Reactivity of cytosine and thymine in single-base-pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations. Proc. Natl Acad. Sci. USA, 85, 4397–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mashal R.D., Koontz,J. and Sklar,J. (1995) Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nature Genet., 9, 177–183. [DOI] [PubMed] [Google Scholar]
- 36.Youil R., Kemper,B.W. and Cotton,R.G. (1995) Screening for mutations by enzyme mismatch cleavage with T4 endonuclease VII. Proc. Natl Acad. Sci. USA, 92, 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marshall D.J., Heisler,L.M., Lyamichev,V., Murvine,C., Olive,D.M., Ehrlich,G.D., Neri,B.P. and de Arruda,M. (1997) Determination of hepatitis C virus genotypes in the United States by cleavase fragment length polymorphism analysis. J. Clin. Microbiol., 35, 3156–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldrick M.M., Kimball,G.R., Liu,Q., Martin,L.A., Sommer,S.S. and Tseng,J.Y. (1996) NIRCA: a rapid robust method for screening for unknown point mutations. Biotechniques, 21, 106–112. [PubMed] [Google Scholar]
- 39.Goldrick M.M. (2001) RNase cleavage-based methods for mutation/SNP detection, past and present. Hum. Mutat., 18, 190–204. [DOI] [PubMed] [Google Scholar]
- 40.Babon J.J., McKenzie,M. and Cotton,R.G. (1999) Mutation detection using fluorescent enzyme mismatch cleavage with T4 endonuclease VII. Electrophoresis, 20, 1162–1170. [DOI] [PubMed] [Google Scholar]
- 41.Smith J. and Modrich,P. (1996) Mutation detection with MutH, MutL, and MutS mismatch repair proteins. Proc. Natl Acad. Sci. USA, 93, 4374–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beaulieu M., Larson,G.P., Geller,L., Flanagan,S.D. and Krontiris,T.G. (2001) PCR candidate region mismatch scanning: adaptation to quantitative, high-throughput genotyping. Nucleic Acids Res., 29, 1114–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oleykowski C.A., Bronson Mullins,C.R., Godwin,A.K. and Yeung,A.T. (1998) Mutation detection using a novel plant endonuclease. Nucleic Acids Res., 26, 4597–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang J., Kirk,B., Favis,R., Soussi,T., Paty,P., Cao,W. and Barany,F. (2002) An endonuclease/ligase based mutation scanning method especially suited for analysis of neoplastic tissue. Oncogene, 21, 1909–1921. [DOI] [PubMed] [Google Scholar]
- 45.Yao M., Hatahet,Z., Melamede,R.J. and Kow,Y.W. (1994) Purification and characterization of a novel deoxyinosine-specific enzyme, deoxyinosine 3′ endonuclease, from Escherichia coli. J. Biol. Chem., 269, 16260–16268. [PubMed] [Google Scholar]
- 46.Yao M., Hatahet,Z., Melamede,R.J. and Kow,Y.W. (1994) Deoxyinosine 3′ endonuclease, a novel deoxyinosine-specific endonuclease from Escherichia coli. Ann. N. Y. Acad. Sci., 726, 315–316. [DOI] [PubMed] [Google Scholar]
- 47.Yao M. and Kow,Y.W. (1997) Further characterization of Escherichia coli endonuclease V. Mechanism of recognition for deoxyinosine, deoxyuridine, and base mismatches in DNA. J. Biol. Chem., 272, 30774–30779. [DOI] [PubMed] [Google Scholar]
- 48.He B., Qing,H. and Kow,Y.W. (2000) Deoxyxanthosine in DNA is repaired by Escherichia coli endonuclease V. Mutat. Res., 459, 109–114. [DOI] [PubMed] [Google Scholar]
- 49.Moe A., Ringvoll,J., Nordstrand,L.M., Eide,L., Bjoras,M., Seeberg,E., Rognes,T. and Klungland,A. (2003) Incision at hypoxanthine residues in DNA by a mammalian homologue of the Escherichia coli antimutator enzyme endonuclease V. Nucleic Acids Res., 31, 3893–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang J., Lu,J., Barany,F. and Cao,W. (2001) Multiple cleavage activities of endonuclease V from Thermotoga maritima: recognition and strand nicking mechanism. Biochemistry, 40, 8738–8748. [DOI] [PubMed] [Google Scholar]
- 51.Hitchcock T.M., Gao,H. and Cao,W. (2004) Cleavage of deoxyoxanosine-containing oligodeoxyribonucleotides by bacterial endonuclease V. Nucleic Acids Res., 32, 4071–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao M. and Kow,Y.W. (1994) Strand-specific cleavage of mismatch-containing DNA by deoxyinosine 3′-endonuclease from Escherichia coli. J. Biol. Chem., 269, 31390–31396. [PubMed] [Google Scholar]
- 53.Tong J., Cao,W. and Barany,F. (1999) Biochemical properties of a high fidelity DNA ligase from Thermus species AK16D. Nucleic Acids Res., 27, 788–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao M. and Kow,Y.W. (1996) Cleavage of insertion/deletion mismatches, flap and pseudo-Y DNA structures by deoxyinosine 3′-endonuclease from Escherichia coli. J. Biol. Chem., 271, 30672–30676. [DOI] [PubMed] [Google Scholar]
- 55.Day J.P., Hammer,R.P., Bergstrom,D. and Barany,F. (1999) Nucleotide analogs and new buffers improve a generalized method to enrich for low abundance mutations. Nucleic Acids Res., 27, 1819–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Till B.J., Burtner,C., Comai,L. and Henikoff,S. (2004) Mismatch cleavage by single-strand specific nucleases. Nucleic Acids Res., 32, 2632–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y., Kaur,M., Price,B.D., Tetradis,S. and Makrigiorgos,G.M. (2002) An amplification and ligation-based method to scan for unknown mutations in DNA. Hum. Mutat., 20, 139–147. [DOI] [PubMed] [Google Scholar]
- 58.Prix L., Uciechowski,P., Bockmann,B., Giesing,M. and Schuetz,A.J. (2002) Diagnostic biochip array for fast and sensitive detection of K-ras mutations in stool. Clin. Chem., 48, 428–435. [PubMed] [Google Scholar]
- 59.Traverso G., Shuber,A., Olsson,L., Levin,B., Johnson,C., Hamilton,S.R., Boynton,K., Kinzler,K.W. and Vogelstein,B. (2002) Detection of proximal colorectal cancers through analysis of faecal DNA. Lancet, 359, 403–404. [DOI] [PubMed] [Google Scholar]
- 60.Dong S.M., Traverso,G., Johnson,C., Geng,L., Favis,R., Boynton,K., Hibi,K., Goodman,S.N., D′Allessio,M., Paty,P. et al. (2001) Detecting colorectal cancer in stool with the use of multiple genetic targets. J. Natl Cancer Inst., 93, 858–865. [DOI] [PubMed] [Google Scholar]
- 61.Su Y.H., Wang,M., Brenner,D.E., Ng,A., Melkonyan,H., Umansky,S., Syngal,S. and Block,T.M. (2004) Human urine contains small, 150 to 250 nucleotide-sized, soluble DNA derived from the circulation and may be useful in the detection of colorectal cancer. J. Mol. Diagn., 6, 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoque M.O., Lee,J., Begum,S., Yamashita,K., Engles,J.M., Schoenberg,M., Westra,W.H. and Sidransky,D. (2003) High-throughput molecular analysis of urine sediment for the detection of bladder cancer by high-density single-nucleotide polymorphism array. Cancer Res., 63, 5723–5726. [PubMed] [Google Scholar]
- 63.Taback B., Chan,A.D., Kuo,C.T., Bostick,P.J., Wang,H.J., Giuliano,A.E. and Hoon,D.S. (2001) Detection of occult metastatic breast cancer cells in blood by a multimolecular marker assay: correlation with clinical stage of disease. Cancer Res., 61, 8845–8850. [PubMed] [Google Scholar]
- 64.Mike Makrigiorgos G. (2004) PCR-based detection of minority point mutations. Hum. Mutat., 23, 406–412. [DOI] [PubMed] [Google Scholar]
- 65.Davies H., Bignell,G.R., Cox,C., Stephens,P., Edkins,S., Clegg,S., Teague,J., Woffendin,H., Garnett,M.J., Bottomley,W. et al. (2002) Mutations of the BRAF gene in human cancer. Nature, 417, 949–954. [DOI] [PubMed] [Google Scholar]
- 66.Beroud C. and Soussi,T. (2003) The UMD-p53 database: new mutations and analysis tools. Hum. Mutat., 21, 176–181. [DOI] [PubMed] [Google Scholar]
- 67.Rajagopalan H., Jallepalli,P.V., Rago,C., Velculescu,V.E., Kinzler,K.W., Vogelstein,B. and Lengauer,C. (2004) Inactivation of hCDC4 can cause chromosomal instability. Nature, 428, 77–81. [DOI] [PubMed] [Google Scholar]
- 68.Ahrendt S.A., Halachmi,S., Chow,J.T., Wu,L., Halachmi,N., Yang,S.C., Wehage,S., Jen,J. and Sidransky,D. (1999) Rapid p53 sequence analysis in primary lung cancer using an oligonucleotide probe array. Proc. Natl Acad. Sci. USA, 96, 7382–7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wikman F.P., Lu,M.L., Thykjaer,T., Olesen,S.H., Andersen,L.D., Cordon-Cardo,C. and Orntoft,T.F. (2000) Evaluation of the performance of a p53 sequencing microarray chip using 140 previously sequenced bladder tumor samples. Clin. Chem., 46, 1555–1561. [PubMed] [Google Scholar]
- 70.Wen W.H., Bernstein,L., Lescallett,J., Beazer-Barclay,Y., Sullivan-Halley,J., White,M. and Press,M.F. (2000) Comparison of TP53 mutations identified by oligonucleotide microarray and conventional DNA sequence analysis. Cancer Res., 60, 2716–2722. [PubMed] [Google Scholar]
- 71.Shah N.P., Nicoll,J.M., Nagar,B., Gorre,M.E., Paquette,R.L., Kuriyan,J. and Sawyers,C.L. (2002) Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell, 2, 117–125. [DOI] [PubMed] [Google Scholar]
- 72.Shah N.P. and Sawyers,C.L. (2003) Mechanisms of resistance to STI571 in Philadelphia chromosome-associated leukemias. Oncogene, 22, 7389–7395. [DOI] [PubMed] [Google Scholar]
- 73.Shah N.P., Tran,C., Lee,F.Y., Chen,P., Norris,D. and Sawyers,C.L. (2004) Overriding imatinib resistance with a novel ABL kinase inhibitor. Science, 305, 399–401. [DOI] [PubMed] [Google Scholar]
- 74.Lynch T.J., Bell,D.W., Sordella,R., Gurubhagavatula,S., Okimoto,R.A., Brannigan,B.W., Harris,P.L., Haserlat,S.M., Supko,J.G., Haluska,F.G. et al. (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med., 350, 2129–2139. [DOI] [PubMed] [Google Scholar]
- 75.Paez J.G., Janne,P.A., Lee,J.C., Tracy,S., Greulich,H., Gabriel,S., Herman,P., Kaye,F.J., Lindeman,N., Boggon,T.J. et al. (2004) EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science, 304, 1497–1500. [DOI] [PubMed] [Google Scholar]
- 76.Fouquet C., Antoine,M., Tisserand,P., Favis,R., Wislez,M., Commo,F., Rabbe,N., Carette,M.F., Milleron,B., Barany,F. et al. (2004) Rapid and sensitive p53 alteration analysis in biopsies from lung cancer patients using a functional assay and a universal oligonucleotide array: a prospective study. Clin. Cancer. Res., 10, 3479–3489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.