SUMMARY
The SCF/c-Kit pathway has crucial roles in controlling hematopoietic stem cell (HSC) renewal. However, little is known about the intracellular regulation of the SCF/c-Kit pathway in HSCs. We report here that Slug, a zinc-finger transcription repressor, functions as a direct transcriptional repressor of c-Kit in HSCs. Conversely, SCF/c-Kit signaling positively regulates Slug through downstream c-Myc and FoxM1 transcription factors. Intriguingly, c-Kit expression is induced by SCF/c-Kit signaling in Slug-deficient HSCs. The balance between Slug and c-Kit are critical for maintaining HSC repopulating potential in vivo. Together, our studies demonstrate that Slug functions in a novel negative-feedback regulatory loop in the SCF/c-Kit signaling pathway in HSCs.
Keywords: HSC, Self-renewal, SCF, c-Kit, c-Myc, Slug, FoxM1
INTRODUCTION
Hematopoietic stem cells (HSCs) are quiescent and multipotent tissue-specific stem cells with extensive self-renewal capacity and potential to differentiate into all hematopoietic lineages 1, 2. HSCs represent only approximately 0.001–0.01% of total murine bone marrow (BM) cells and consist of two functionally defined populations: long-term repopulating HSCs (LT-HSCs) and short-term repopulating HSCs (ST-HSCs) with more restricted self-renewal capacity 3–5. BM provides a unique microenvironment or “niche” consisting of various positive and negative regulators. These regulators establish a complex and dynamic molecular crosstalk with HSCs in the niche and direct HSC fate. As an extrinsic regulator, stem cell factor (SCF) and its transmembrane tyrosine kinase receptor c-Kit play critical roles in HSC self-renewal and differentiation. Mutations in either SCF or c-Kit locus result in hematopoietic deficiency and anemia 6. Loss-of-function mutations in c-Kit impair the self-renewal of HSCs 7. Conditional deletion of c-Kit causes hematopoietic failure and splenic atrophy both at steady state and after BM ablation, leading to the demise of injured adult mice 8. Recently, it was shown that HSCs with low levels of surface c-Kit expression exhibit enhanced self-renewal and long-term reconstitution potential, whereas HSCs with high expression levels of c-Kit show restricted self-renewal capacity with an intrinsic megakaryocytic lineage bias 9.
Signaling downstream of receptor tyrosine kinase c-Kit has been studied in several different types of cells, including mast cells and immortalized hematopoietic cell lines 10. Activation of c-Kit by SCF triggers MEK/ERK and PI3K/Akt kinase cascades 10, which could lead to upregulation of various downstream targets, including c-Myc 11 and FoxM1 12. c-Myc is a well-known positive regulator for cell-cycle progression, and plays a critical role in regulation of HSC self-renewal 13. FoxM1 is a member of the FOX family of transcription factors and plays a critical role in cell cycle progression. FoxM1 expression and activity are regulated by PI3K/AKT and Raf/MEK/MAPK signaling pathways 14. Despite its well known profound effects on HSC self-renewal and differentiation, the molecular mechanism through which SCF/c-Kit signaling pathway is modulated in HSCs has not been fully elucidated.
Slug is a zinc-finger transcriptional repressor and belongs to the highly conserved Slug/Snail family of transcription factors found in a diverse number of species 2, 15. In mammals, this family consists of Snail1, Slug/Snail2, Snail3/Smuc, and Scratch. All the family members share an extreme N-terminal SNAG domain, which is required for transcriptional repression and nuclear localization. Slug/Snail family members also contain a DNA binding domain consisting of four to six C2H2-type zinc fingers in the C-terminal domain. The Slug/Snail family members are involved in many important biological regulation processes, such as epithelial-mesenchymal transition (EMT), mammary stem cell activity, cancer metastasis, and cellular reprogramming 16–19. Our previous studies show that Slug deficiency enhances self-renewal of HSCs during hematopoietic regeneration 2. Although it was suggested that Slug is a potential target of SCF/c-Kit signaling pathway in cancer cell lines overexpressing c-Kit 20, 21, the molecular mechanisms by which Slug regulates HSC self-renewal remains largely unknown.
In the present study, we dissected the molecular mechanisms by which Slug regulates the self-renewal capacity of HSCs. Using microarray analysis, we found that c-Kit expression is elevated in Slug-deficient HSCs during HSC regeneration. By qPCR analysis, luciferase reporter assay, and chromatin immunoprecipitation (ChIP) assays, we confirmed c-Kit as a direct target gene of Slug. Using bone marrow transplantation assays, we observed that the enhanced engraftment of Slug−/− HSCs at two weeks after transplantation was impaired by knockdown of c-Kit. On the other hand, deletion of Slug improved the impairment of HSC repopulating capacity caused by c-Kit inhibition by week eight after transplantation. To further investigate the functional interactions between the Slug and c-Kit signaling pathway in vivo, we overexpressed Slug, c-Kit, or both Slug and c-Kit in HSCs and performed serial BM transplantations. Our results indicated that overexpression of either Slug or c-Kit impairs the self-renewal ability of HSCs, suggesting that the balance between Slug and c-Kit controls HSC long-term repopulating potential. Furthermore, we investigated the reciprocal regulation of Slug and c-Kit signaling in HSCs. We found that Slug is largely induced by SCF/c-Kit signaling in HSCs via downstream MEK or PI3K/Akt pathways. Interestingly, SCF treatment induced c-Kit expression in Slug−/− but not Slug+/+ HSCs.
In addition, by qPCR and ChIP we identified c-Myc and FoxM1 as two downstream targets of SCF/c-Kit signaling pathway, and showed that knockdown of either c-Myc or FoxM1 inhibits the upregulation of endogenous Slug induced by SCF, suggesting that c-Myc and FoxM1 are key mediators required for the c-Kit-Slug negative feedback circuit. In summary, our findings uncover previously unrecognized molecular mechanisms of the SCF/c-Kit-Myc-FoxM1-Slug negative feedback circuit in HSCs.
MATERIALS AND METHODS
Mice
C57BL/6, C57BL/6.SJL, and Tie2-Cre mice 22 were purchased from The Jackson Laboratory. Slug knockout mice were generated as described previously 23 and backcrossed to C57BL/6 (CD45.2) background for more than 6 generations. Generation of FoxM1fl/fl mice was described previously 24 and backcrossed to C57BL/6 (CD45.2) background for more than 6 generations. For deletion of FoxM1 in BM, and FoxM1fl/fl mice were crossed to Tie2-Cre mice. Mice were used at 6–12 weeks old of age and all the animal studies were approved by the Animal Care and Use Committee at the University of Illinois at Chicago (Approval Number: 13–132).
Cell Culture
293T cells were cultured in DMEM (high glucose) medium containing 10% FBS. K562 cells were cultured in IMDM medium plus 10% FBS. Enriched fresh hematopoietic stem and progenitor cells (HSPCs) were cultured in vitro in Stemspan (STEMCELL Technologies) supplemented with 10 μg/ml heparin (Sigma), 10 ng/ml mouse SCF, 20 ng/ml mouse TPO, 20 ng/ml human IGF-II, 10 ng/ml mouse FGF-1, and 100 ng/ml human Angptl3 25. All recombinant proteins were purchased from PEPROTECH or PROSPEC companies. All cell culture products were purchased from Life Technologies unless otherwise mentioned. All small molecule inhibitors were purchased from Cayman Chemicals.
ChIP Assay
ChIP assay was performed with a ChIP-IT Express Enzymatic kit (Active Motif) according to the manufacturer’s instructions. Genomic DNA was pulled down by anti-Flag or IgG control antibodies (Thermo Fisher Scientific). Immunoprecipitated DNA fragments were amplified by using the two primers listed in Table S3.
Statistical Analyses
Unpaired Student’s t test was used to statistically analyze all the experiments. Microarray data was analyzed by ANOVA test. Differences with P < 0.05 were considered statistically significant and are denoted as *P < 0.05; **P < 0.01. All PCR results were repeated at least three times.
RESULTS
c-Kit is a Target Gene of Slug Transcription Factor in HSCs
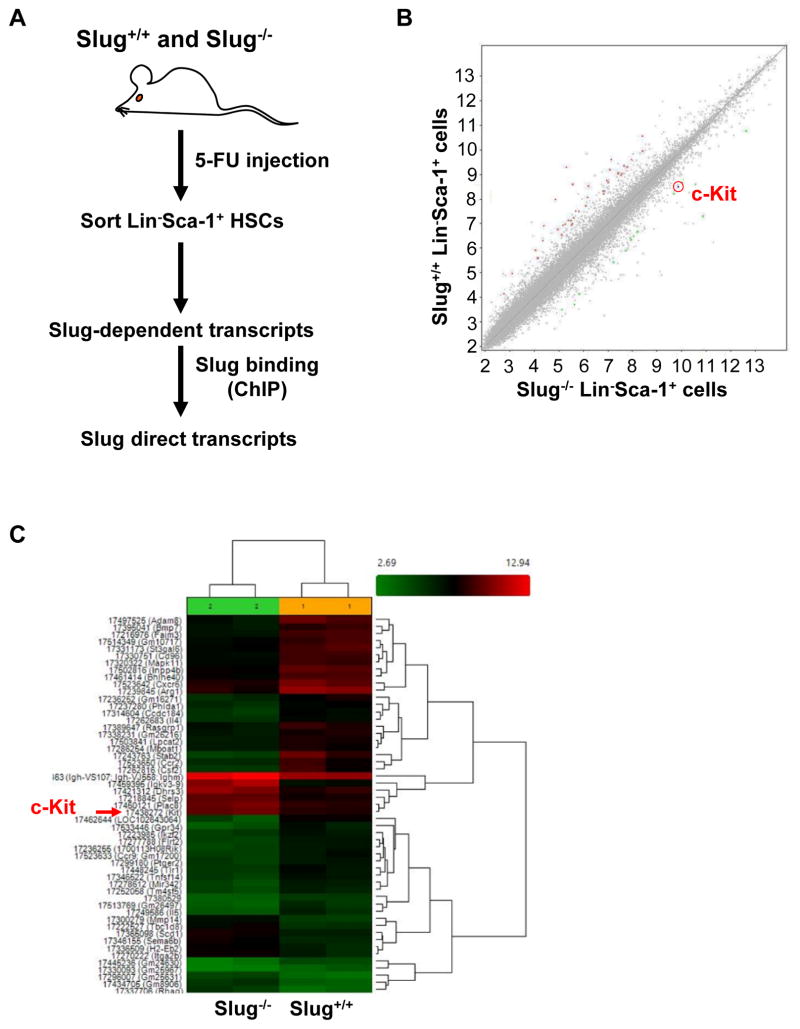
We previously showed that Slug deficiency enhances HSC self-renewal but not homing and differentiation 2. To gain insights into molecular pathways by which Slug regulates the self-renewal of HSCs, we sorted lineage-negative (Lin−) Sca-1+ HSPCs from the two groups of mice (Slug+/+ vs. Slug−/− mice) after 5-FU treatment 26. 5-FU treatment was shown to transiently alter HSC phenotype from Lin−Sca-1+c-Kit+ to Lin−Sca−1+c-Kit− 27, 28. Next, we compared gene expression profiles in the two groups of HSCs by performing microarray analysis (Figure 1A). We identified 14 genes that were upregulated, and 37 genes that were downregulated by more than 2.5 fold (p value < 0.05) in Slug−/− HSPCs compared to Slug+/+ HSPCs (Figure 1B, 1C, Table S1). Because Slug predominantly inhibits its gene targets29, it is likely that Slug is a direct transcriptional repressor of the 14 upregulated genes. Using online software INSECT2.0 30 to predict Slug binding sites in proximal promoter regions of these potential direct targets (~2.0 kb from transcription start site), three genes (c-Kit, Selp, and Sema6b) were selected (Table S2). Among the selected three genes, c-Kit has been shown to play critical roles in HSC self-renewal 6–9. Selp is associated with leukocyte cell-cell adhesion, migration and rolling, inflammatory response 31–33. Sema6b mostly involves retinal development and tumor progression in glioblastoma cells through its receptor Plexin-A4 34–36. Therefore, we decided to focus on c-Kit as a potential target gene of Slug.
Figure 1. Gene Profiling by microarray analysis Identifies c-Kit as a Target of Slug in HSCs During Hematopoietic Regeneration.
(A) Diagram for identifying HSC-enriched Slug target genes upon BM regeneration.
(B, C) Microarray analysis of gene expression profiles in HSPCs from Slug+/+ and Slug−/− mice after 5-FU treatment (n = 2). The mice were injected 5-FU at 150 mg/kg body weight. The BM cells were isolated at day 7 after 5-FU injection. Lin−Sca-1+ HSPC subpopulation was sorted by FACs and extracted RNA for microarray analysis. The red arrow indicates c-Kit (C). Microarray data was analyzed by ANOVA test.
See also Figure S1.
Using real-time PCR, we validated that c-Kit was upregulated ~5.3 fold in Slug−/− HSPCs when compared to Slug+/+ HSPCs (Figure S1A). Moreover, in the absence of extrinsic stress (i.e. 5-FU treatment), c-Kit mRNA expression was still 2.3-fold and 2.0-fold higher in Lin− and CD150+CD48− (hereafter called SLAM 3) cell populations, respectively, from Slug−/− than those from Slug+/+ mice (Figure S1B). We also evaluated c-Kit expression at protein level using flow cytometry. The results showed that mean fluorescence intensity (MFI) of c-Kit is significantly higher in distinct subsets of HSPCs from Slug−/− mice than counterparts from Slug+/+ mice (Figure S1C, S1D), indicating that endogenous Slug downregulates c-Kit expression under normal conditions.
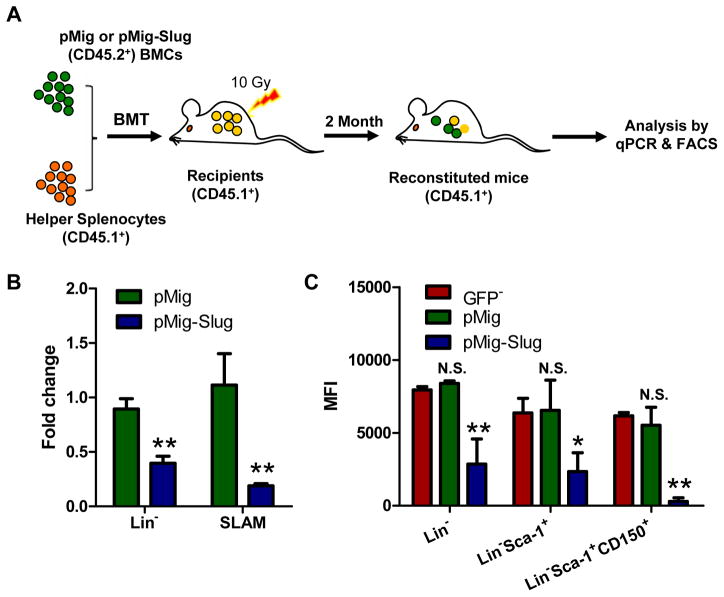
To rigorously assess the negative regulatory effects of Slug on the expression of c-Kit, we also overexpressed Slug (with a GFP marker and Flag tag) in HSPCs using Slug-overexpressing retroviruses and determined whether Slug inhibits the transcription of c-Kit. Our qPCR analysis showed that Slug overexpression downregulates the transcription of c-Kit by 400-fold in HSPCs (Figure S2). To assess the effects of Slug overexpression in HSCs in vivo, we infected wild-type (WT) HSPCs with retroviruses-expressing Slug and transplanted infected cells into lethally irradiated recipient mice. Eight weeks after BM transplantation, we isolated BM MNCs from reconstituted mice, and analyzed the expression of c-Kit in GPF+ HSPCs by qPCR and flow cytometry (Figure 2A). Our results showed that overexpression of Slug inhibited the expression of endogenous c-Kit by 2- and 4-folds in Lin− and SLAM subsets, respectively (Figure 2B). Furthermore, we examined the expression levels of c-Kit protein in various subsets of HSPCs by MFI analysis. We found that overexpression of Slug decreased the expression level of c-Kit protein by 2.8-fold, 2.7-fold, and 21-fold in Lin−, Lin−Sca-1+, and SLAM subsets, respectively (Figure 2C).
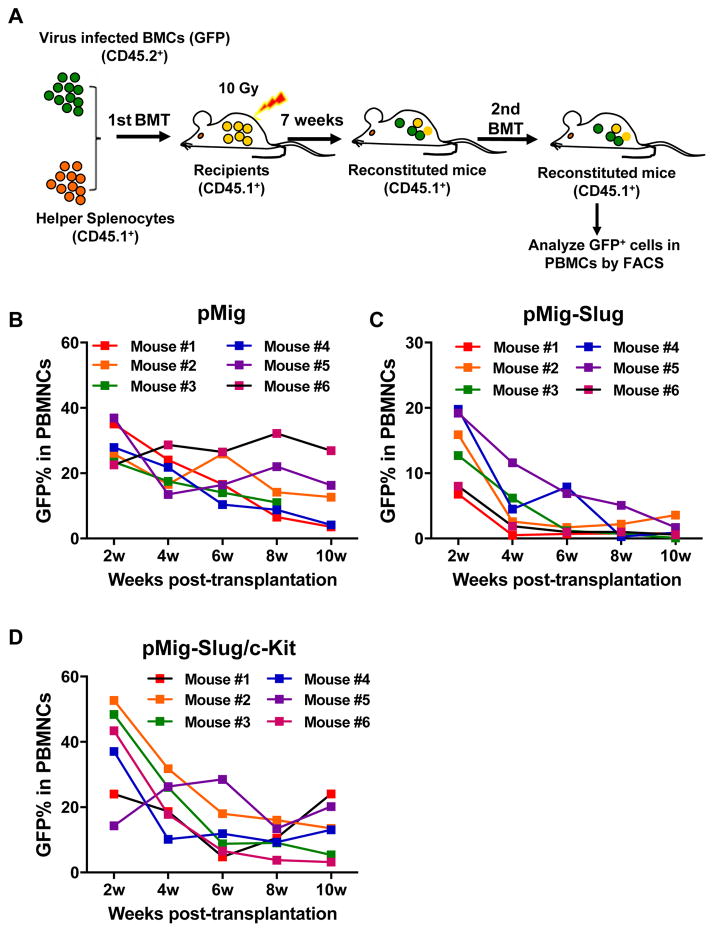
Figure 2. Overexpression of Slug Regulates c-Kit Expression in HSCs in vivo.
(A) Scheme representing the strategy for overexpression of Slug in HSCs in vivo. Lin− BMCs (CD45.2+) were infected with pMig-Slug or pMig (vector control) and then mixed with splenocytes (1.5×106, CD45.1+ helper cells) before transplanted into lethally irradiated recipient mice (CD45.1+).
(B) qPCR analysis of the mRNA expression level of endogenous c-Kit in Lin− and SLAM populations.
(C) Flow cytometric analysis of the expression of endogenous c-Kit in the indicated hematopoietic subsets of pMig- and pMig-Slug reconstituted mice (n = 3). The levels of endogenous c-Kit were evaluated by flow cytometry. MFI, mean fluorescence intensity. Data are representative of two independent experiments. All data are represented as mean ± SD. Two-tailed Student’s t tests were used to assess statistical significance (*P < 0.05; **P, < 0.01).
See also Figure S2 and S3.
Next, we asked if the other Snail/Slug family members (Snail1, Snail3, Scratch) could also regulate c-Kit in HSCs. To this end, we examined expression of all the Snail/Slug family members in sorted hematopoietic cell subset by RT-PCR analysis 15. The results showed that Slug was selectively expressed in HSCs and myeloid lineages. By contrast, Snail3 was only highly expressed in lymphoid lineage. As a control, Vav-1 is evenly expressed in all cells of different lineages, and IL-7 receptor is only expressed in CLP, pro-B, and pro-T cells (Figure S3). All together, these findings support the concept that Slug acts as a negative regulator of c-Kit in HSC population.
Slug Functions as a Direct Transcription Repressor of c-Kit Promoter
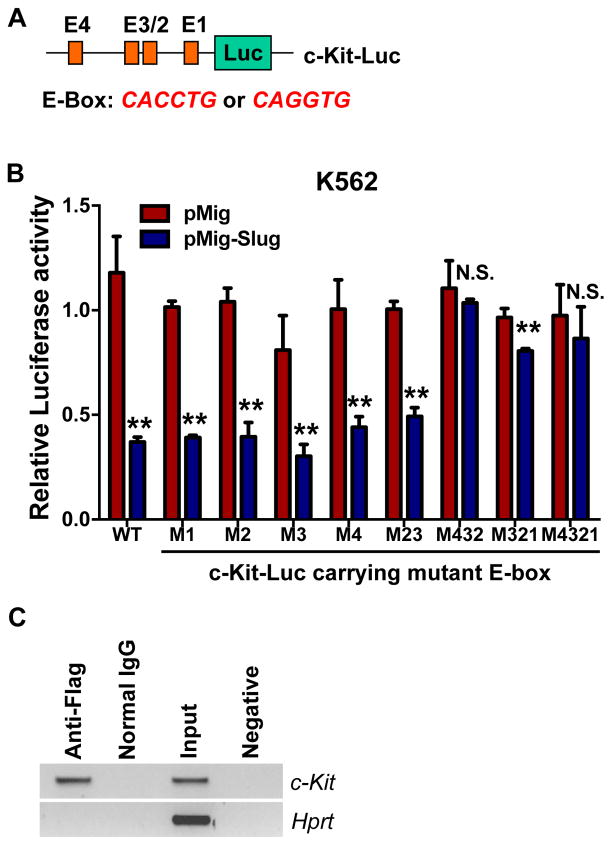
Given the negative regulation of c-Kit by Slug in HSCs, we hypothesized that Slug negatively modulates c-Kit by suppressing the transcriptional level of c-Kit. Thus, we analyzed the promoter sequence of c-Kit and identified four E-boxes (“CAGGTG” or “CACCTG”) for Slug binding sites (Figure 3A, S4A). To determine whether c-Kit is a true gene target of the Slug transcriptional repressor, we generated a luciferase reporter driven by the c-Kit promoter (c-Kit-Luc) and mutated each of the four E-boxes (Figure S4B). Next, we performed luciferase reporter assays to validate our bioinformatics prediction by transfecting each c-Kit promoter reporters together with a Slug expression plasmid in 293T (Figure S5) and K562 hematopoietic cell line. Our data revealed that the c-Kit luciferase reporters containing either WT or each mutant E-boxes were inhibited 50% by Slug overexpression (Figure 3B, S5A, S5B). Next we mutated different combinations of the four E-boxes in the c-Kit luciferase reporter and transfected them along with a Slug expression plasmid into K562 cells. Our results showed that Slug inhibited the c-Kit luciferase reporters containing both mutant E-box 2 and E-box 3. By contrast, the inhibitory effect of Slug on the luciferase activities of the c-Kit-Luc reporter was abolished if the three E-boxes (E-box 2, 3, 4) or all of the four E-boxes were simultaneously mutated (Figure 3B, S5C). In addition, Slug inhibited the c-Kit luciferase reporters harboring a combination of three mutation E-boxes (E-box 1, E-box 2, and E-box 3). Therefore, our data suggested that E-box 4, together with E-box 2 and 3, significantly contributed to the Slug-mediated negative regulation of c-Kit. Furthermore, we confirmed the inhibitory effects of Slug on c-Kit promoter in primary HSPCs. (Figure S5D).
Figure 3. Slug Directly Regulates c-Kit Promoter.
(A) Diagram of the luciferase reporters for the c-Kit promoter.
(B) c-Kit luciferase reporter assays. K562 cells were transfected with WT c-Kit-Luc or mutant c-Kit-Luc carrying mutations in different E-box together with pMig (vector control) or pMig-Slug, then cultured for 72 h before luciferase activity assay. pCMV-LacZ was included in each transfections as an internal control to normalized luciferase activity. Data are representative of three independent experiments. All data represent mean ± SD. Two-tailed Student’s t tests were used to assess statistical significance (*P < 0.05; **p < 0.01; N.S., not significant, P > 0.05).
(C) Analysis of Slug occupancy at the c-Kit promoter by ChIP analysis. Primary mouse HSPCs were transduced with retrovirus-expressing Flag-tagged Slug, expanded, and then performed ChIP assay. The genomic DNA was pulled down by anti-Flag and IgG control antibodies, respectively; and amplified by a pair of primers covering the E-box 1–3 (from −445 to −301 bp from ATG). Data are representative of two independent experiments. Hprt was amplified as a negative control.
See also Figure S4 and S5.
To determine whether Slug occupies the c-Kit promoter, we overexpressed a Flag-tagged Slug in HSPCs using retrovirus and then performed ChIP assay using anti-Flag antibody. A pair of primers was designed to amplify the DNA fragment covering the three E-boxes (E-box 1–3, from −445 to −301 bp from ATG) in c-Kit promoter (Figure 3A, S4). As shown in Figure 3C, a specific DNA fragment was amplified from the ChIP sample pulled down by anti-Flag antibody but not by normal IgG control, indicating that Slug specifically binds to the c-Kit promoter region in vivo. Taken together, our results demonstrated that Slug directly interacts with the c-Kit promoter in HSPCs.
Slug Deficiency Partially Rescue Impaired HSC Repopulating Capacity Caused by c-Kit Knockdown
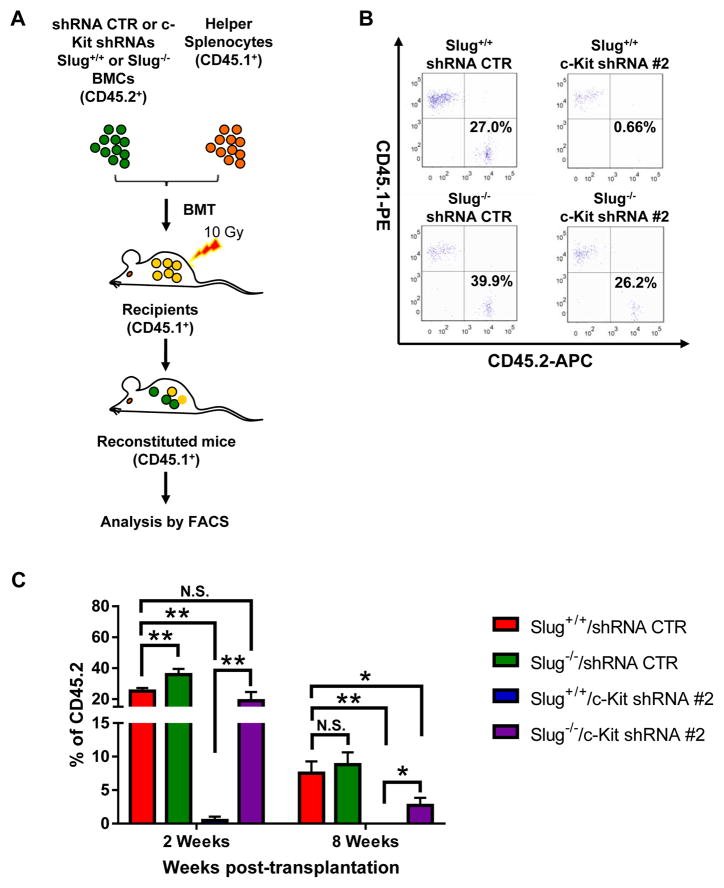
Although our robust evidence indicates that c-Kit is a direct transcriptional target of Slug in HSCs, a key question arises: does Slug regulate HSC self-renewal through c-Kit? To address this question, we generated lentiviral particles expressing scrambled shRNA (control) and three c-Kit shRNAs (#1, #2, #3). Our data showed that both c-Kit shRNA #2 and #3 dramatically knocked down the expression of endogenous c-Kit in primary HSPCs (Figure S6). Next, we transduced Slug+/+ HSPCs or Slug−/− HSPCs (CD45.2+) respectively, with lentiviruses expressing control shRNA or c-Kit-shRNA #2. After transduction, we transplanted the infected HSPCs into lethally irradiated recipient mice (CD45.1+) (Figure 4A). To assess the engraftments of four experimental groups (Slug+/+/shRNA control, Slug+/+/c-Kit shRNA#2, Slug−/−/shRNA control, and Slug−/−/c-Kit shRNA #2), we examined peripheral blood chimerism at 2 weeks, 4 weeks and 8 weeks after transplantation. As we expected, mice transplanted with Slug+/+/c-Kit shRNA #2 HSPCs exhibited much lower CD45.2 levels in peripheral blood up to 8 weeks after reconstitution, when compared to Slug+/+/shRNA control group (Figure 4B, 4C). By contrast, mice transplanted with Slug−/−/shRNA control showed accelerated donor-derived CD45.2 chimerism level in peripheral blood by week 2 after transplantation (Figure 4C). The Slug−/−/c-Kit shRNA #2 HSPCs-transplanted mice exhibited comparable CD45.2 chimerism level in peripheral blood at 2 weeks post-transplantation (Figure 4C). By week 8 after BM transplantation, CD45.2 chimerism level was slightly but not significantly higher in Slug−/−/shRNA control group than that in Slug+/+/shRNA control group. Mice transplanted with Slug−/−/c-Kit shRNA #2 exhibited significantly higher CD45.2 chimerism level than those with Slug+/+/c-Kit shRNA #2 transplantation (Figure 4C). A similar result was observed by week 4 after transplantation (Figure S7A). To confirm these results, we repeated the transplantation by using c-Kit shRNA #3. Similar results were observed by week 2 and 8 after BM transplantation (Figure S7B, S7C). In addition, we analyzed trilineage differentiation in these recipients and our data showed that Slug deficiency and/or knockdown of c-Kit did not notably affect multilineage differentiation of HSCs (Figure S8). These data indicated that Slug deficiency alleviates the impairment of HSC repopulating capacity due to knockdown of endogenous c-Kit.
Figure 4. Enhanced Repopulating Capacity of Slug−/− HSCs in Reconstituted Mice.
(A) Diagram of in vivo HSC repopulating assay. Donor Lin− BMCs from Slug+/+ or Slug−/− (CD45.2+) were infected with c-Kit-specific shRNA or control shRNA, and then injected together with helper splenocytes (CD45.1+) into recipient mice (CD45.1+) that received a prior irradiation (10 Gy). Ratio of donor-derived cells (CD45.2+) in peripheral blood was determined by flow cytometry.
(B) A representation of flow cytometric analysis of the ratio of donor-derived cells in peripheral blood was evaluated at 2 weeks after BM transplantation.
(C) Slug deficiency alleviated the impairment of in vivo HSC repopulating by knockdown of endogenous c-Kit. The percentage of donor-derived cells (CD45.2+) in peripheral blood was evaluated at 2 weeks (n = 4 mice) and 8 weeks (n = 3 mice) after BM transplantation, respectively. Data are representative of two independent experiments. All data are represented as mean ± SD. Two-tailed Student’s t tests were used to assess statistical significance (*P < 0.05, **p < 0.01, N.S., not significant, P > 0.05).
See also Figure S6, S7, and S8.
Slug and c-Kit Converge to Regulate HSC Self-Renewal
To further understand how Slug and c-Kit cooperate in regulating self-renewal of HSCs, we generated retroviruses that express Slug, c-Kit, or both Slug and c-Kit (fused by a 2A sequence 37, 38, thereafter called Slug/c-Kit) in pMIGR1 vector, which co-express a GFP marker via an internal ribosome entry site (IRES). HSPCs were infected with each retrovirus and followed by serial transplantation into lethally irradiated mice (Figure 5A). Our flow cytometric analysis showed that recipient mice transplanted with HSPCs infected with pMig-c-Kit exhibited very low chimerism (% of GFP+ cells) in peripheral blood by week 2 after transplantation (% of GFP+: 3.2–4.6). After 4 weeks of transplantation, GFP+ cells were not detectable in the recipient mice (Figure S9). By contrast, mice transplanted with HSPCs infected with control retroviruses and retroviruses expressing Slug or Slug/c-Kit showed comparable GFP levels in peripheral blood (data not shown). Therefore, we isolated BM MNCs from primary recipient mice at 7 weeks after first BM transplantation, and performed second BM transplantation. We analyzed percentage of GFP+ cells in peripheral blood in secondary recipient mice every two weeks after BM transplantation. Our data showed that the two groups of recipient mice transplanted with donor cells harboring retroviral vector only and donor cells overexpressing Slug/c-Kit, respectively, exhibited comparable percentage of GFP+ cells in peripheral blood at 10 weeks after transplantation (% of GFP+ cells in the control vector (pMig) group, 3.6~26.9%; % of GFP+ cells in the pMig-Slug/c-Kit group, 3.2~20.2%) (Figure 5B, 5D). By contrast, the percentage of GFP+ cells in the secondary recipient mice with donor cells overexpressing Slug decreased rapidly from 2 to 10 weeks (Figure 5C). We also analyzed lineage differentiation of donor HSCs in the recipients by flow cytometric analysis. Our data revealed that overexpression of Slug and/or c-Kit did not significantly affect HSC differentiation (Figure S10). Together, these results indicated that overexpression of either Slug or c-Kit impairs HSC self-renewal, suggesting that Slug and c-Kit converge to control HSC self-renewal.
Figure 5. Serial BM Transplantation Assay for HSCs Exhibits the Fine regulatory Interaction between Slug and c-Kit.
(A) Diagram of serial BM transplantation assay for in vivo HSC. CD45.2+ BM cells were transduced with retroviral particles containing pMig vector, pMig-Slug, or pMig-Slug/c-Kit, and then transplanted with helper splenocytes (CD45.1+) into lethally irradiated mice (CD45.1+). By 7 week of transplantation, primary BM cells were isolated from reconstituted mice and performed second transplantation. The percentage of GFP+ cells in peripheral blood was assessed every two weeks after second transplantation.
(B–D) Analysis of GFP percentage in peripheral blood by flow cytometry in pMig- (B), pMig-Slug (C), or pMig-Slug/c-Kit (D) reconstituted mice (n = 6). Data are representative of two independent experiments.
See also Figure S9.
Slug Constitutes a Novel Negative Feedback Loop for Regulating the SCF/c-Kit Signaling Pathway
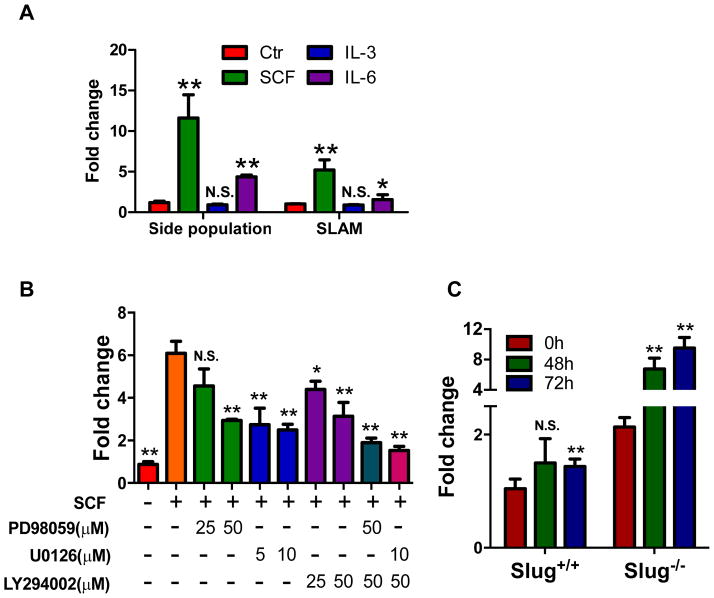
SCF/c-Kit is a critical signaling pathway that regulates the self-renewal capacity of HSCs 7–9. Previous studies reported that SCF enhances Slug expression in cell lines overexpressing c-Kit 20, 21. Because our data showed that Slug negatively regulates c-Kit expression, two related inevitable questions arise: as the ligand of c-Kit, does SCF increase the expression of endogenous Slug in HSCs? If so, does induction of endogenous Slug by SCF suppress the expression of c-Kit? To address these questions, we first tested whether exogenous SCF could stimulate the expression of endogenous Slug in HSCs. We sorted HSCs by flow cytometry using side population (SP) method or SLAM antibody combinations, and then treated both HSC subsets with individual cytokines (SCF, IL-3, or IL-6). The results showed that SCF significantly induced the transcriptional levels of endogenous Slug in both SP cells and SLAM cells by 11.5-fold and 5.2-fold, respectively. IL-6 induced transcriptional levels of Slug in SP and SLAM subsets by 4.4-fold and 1.6-fold, respectively. By contrast, IL-3 failed to induce the transcriptional level of Slug in both SP and SLAM populations (Figure 6A). Furthermore, we found that SCF induces expression of endogenous Slug in SP-HSCs in dose- and time-dependent manner (Figure S11A, S11B).
Figure 6. Role of Slug in Negative Feedback Regulation of SCF/c-Kit Signaling Pathway.
(A) qPCR analysis of endogenous Slug transcripts in indicated HSCs treated by various cytokines. HSCs were left without treatment as a control (Ctr) or treated with the indicated cytokines for 6 h, and then subjected to RNA extraction. Expression levels of target genes were normalized to Hprt levels. *P < 0.05; **p < 0.01; N.S., not significant.
(B) qPCR analysis of endogenous Slug transcripts in HSCs treated with a combination of SCF and signaling pathway inhibitors. HSCs were treated with the different combinations of SCF (100 ng/ml), PD98059, U0126, and LY294002 for 6 h before qPCR analysis. Data are representative of two independent experiments. *P < 0.05; **p < 0.01; N.S., not significant. There was a significant difference between the group treated with SCF alone and the groups treated with SCF+PD98059+LY294002 or SCF+U0126+LY294002.
(C) qPCR analysis of endogenous c-Kit transcripts in HSPCs. Slug+/+ and Slug−/− HSPCs were enriched by a single dose of 5-FU (300 mg/kg body weight), and then treated with or without SCF for 48 and 72 h. c-Kit expression was normalized to Hprt levels. *P < 0.05; **p < 0.01; N.S., not significant.
Data are representative of two independent experiments. Two-tailed Student’s t tests were used to assess statistical significance.
See also Figure S11.
In addition, we blocked the downstream of SCF/c-Kit signaling pathway by MEK pathway inhibitors (PD98059 and U0126) and PI3K/Akt pathway inhibitor (LY294002). Our data showed that either blocking MEK or PI3K/Akt pathway in part suppressed the induction of endogenous Slug by SCF, and blocking both the pathways (LY294002 plus PD98059 or U0126) almost abolished SCF-mediated induction of endogenous Slug, suggesting that both MEK and PI3K/Akt pathways are required for SCF to induce Slug expression in HSPCs (Figure 6B).
Because our data demonstrated that c-Kit is a direct target gene of Slug (Figure 3), we investigated whether deletion of Slug could lead to c-Kit upregulation by SCF. To address this, a higher dose of 5-FU (300 mg/kg body weight) was used to further enrich HSPCs from WT and Slug-deficient mice 2 (Figure S12). We harvested HSPCs 7 days after 5-FU injection and then treated the cells with SCF for different periods of time. Our qPCR analysis showed that SCF increased the expression of endogenous c-Kit in Slug-deficient HSPCs by 6.7-fold and 9.5-fold at 48 and 72 hours after treatment, respectively; whereas, SCF only slightly induced the expression of endogenous c-Kit in WT HSPCs, suggesting that SCF-mediated induction of Slug suppressed the induction of endogenous c-Kit at transcriptional level (Figure 6C).
Collectively, our data indicated that Slug functions as a novel negative-feedback regulator of the SCF/c-Kit signaling pathway (Figure 6).
c-Myc and FoxM1 Activates Slug Transcription in the SCF/c-Kit-Slug Negative-Feedback Loop
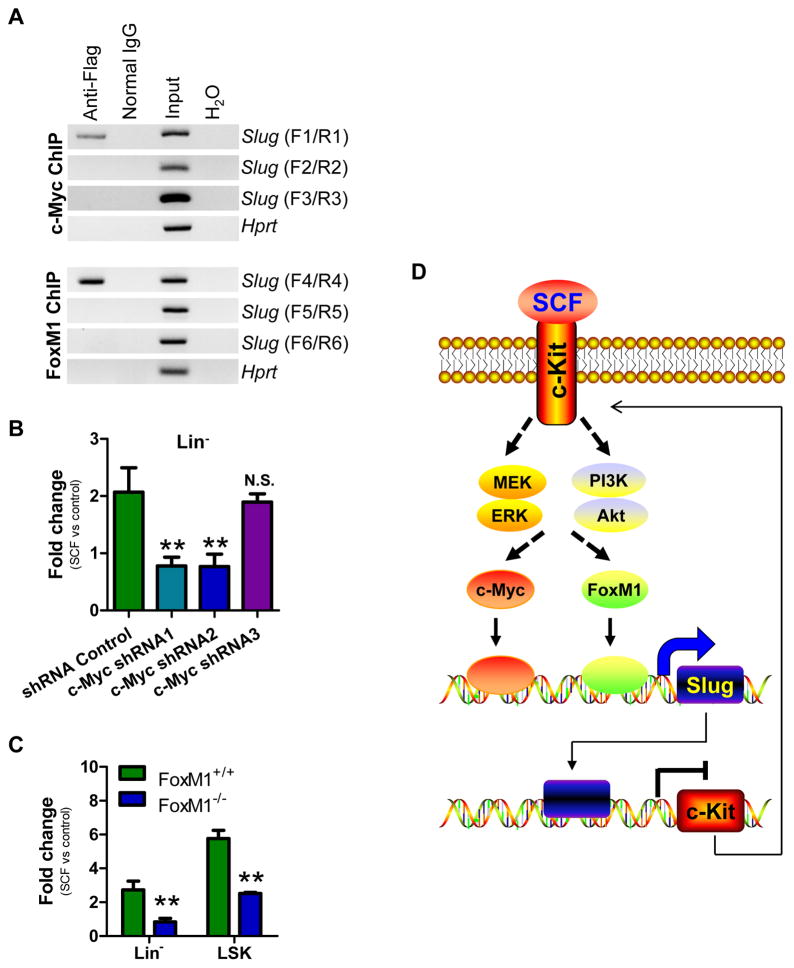
To better understand the molecular mechanism underlying the SCF/c-Kit-Slug negative-feedback loop, we analyzed the promoter region of Slug (~1.5 kb) and identified eight potential c-Myc recognition sites (“CANNTG”) and four FoxM1 putative sites [(C/T)AAA(C/T)A] (Figure S13). Both c-Myc and FoxM1 are transcription activators and can positively regulate various target genes. To test whether c-Myc and FoxM1 can indeed bind to the promoter of Slug, we first generated a luciferase reporter driven by Slug promoter (Slug-Luc) and then performed a reporter assay to validate this bioinformatics prediction by transfecting the reporter together with either c-Myc or FoxM1 plasmid in K562 cells. The results showed that both c-Myc and FoxM1 increased the transcriptional activity of the Slug-Luc reporter (Figure S14). In addition, we overexpressed a Flag-tagged c-Myc and FoxM1 in HSPCs using retrovirus and then determined whether c-Myc and FoxM1 directly bind to the promoter region of Slug by ChIP assay using anti-Flag antibody. The primers were designed to amplify the specific DNA fragments from the Slug promoter region (Figure 7A, S13). A specific DNA fragment was amplified from the c-Myc-associated complexes by primer pair F1/R1 and from the FoxM1-associated complexes by F4/R4 (Figure 7A), respectively. There was no DNA fragment amplified from DNA/protein complex pulled down by normal IgG (control antibody). These results indicated that both transcriptional factors directly bind to Slug promoter region. Although our data showed that both c-Myc and FoxM1 are capable of binding to the promoter region of Slug, the requirement of these two factors on induction of Slug by SCF still remains to be validated. To address this question, we knocked down endogenous c-Myc by shRNAs (Table S4). As shown in Figure S15A, four of five c-Myc specific shRNAs (#1, #2, #4, and #5) completely knocked down the expression of endogenous c-Myc in HSPCs. We selected three c-Myc shRNAs (#1, #2, and #3) to test their effects on SCF-mediated induction of Slug in HSPCs. Our data showed that c-Myc shRNAs #1 and #2 suppressed SCF-mediated induction of Slug in HSPCs by 70%, whereas Slug induction by SCF in the shRNA Control and c-Myc shRNA#3-expressing groups were comparable (Figure 7B). Furthermore, we found that SCF could upregulate the expression of endogenous c-Myc in HSPCs (Figure S15B), suggesting that c-Myc is a mediator for SCF to induce Slug in HSPCs.
Figure 7. Key Role of c-Myc and FoxM1 in SCF/c-Kit-Slug Feedback Loop.
(A) Analysis of c-Myc and FoxM1 occupancies at the Slug promoter by ChIP analysis. Primary BM cells were transduced with retroviral particles containing c-Myc and FoxM1, respectively. The genomic DNA was pulled down by anti-Flag and IgG control antibodies, and amplified with primer pairs F1/R1 (−856 to −691bp), F2/R2 (−1389 to −1234bp), F3/R3 (−1772 to −1605bp), F4/R4 (−597 to −410bp) F5/R5 (−1203 to −1012bp), F6/R6 (−1522 to −1312bp). Hprt was included as a loading control. Data are representative of two independent experiments.
(B) qPCR analysis of Slug transcripts in c-Myc-expressing HSPCs after treatment with SCF. HPSCs harboring c-Myc shRNA and control shRNA were left without treatment as a control or treated with SCF (100 ng/ml) for 12 hrs before qPCR analysis. Data are representative of two independent experiments. **p < 0.01; N.S., not significant.
(C) qPCR analysis of Slug transcripts in FoxM1−/− Lin− and LSK subpopulations following treatment with SCF. HSPCs were enriched from FoxM1+/+ and FoxM1−/− mice and treated with SCF (100 ng/ml) for 12 hrs or left without treatment (control). Slug transcripts was analyzed by qPCR and normalized to Gapdh level. Data are representative of two independent experiments. **p < 0.01.
(D) Action model of SCF/c-Kit-Myc/FoxM1-Slug negative-feedback circuit. SCF/c-Kit induces c-Myc and FoxM1 expression through the MEK and PI3K signaling, and leads to Slug upregulation. In turn, enhanced expression of Slug inhibits c-Kit expression, resulting in the blockage of SCF/c-Kit signaling pathway.
See also Figure S12, S13, S14, S15, and Table S1
FoxM1 was recently shown to upregulate Slug expression in human breast cancer cell lines 39. To determine whether or not FoxM1 is a positive regulator of Slug following SCF treatment, we enriched HSPCs from FoxM1+/+ and FoxM1−/− mice and treated them with or without SCF. Our qPCR analysis showed that Slug induction was decreased by 3-fold and 2.3-fold in FoxM1−/− Lin− cells and LSK cells, respectively, when compared to FoxM1+/+ Lin− cells and LSK cells (Figure 7C). A consistent result was observed by using a specific FoxM1-binding inhibitor FDI-6 (Figure S15C). Interestingly, FoxM1 expression increased by 8-fold at transcriptional level in HSPCs after treatment with SCF (Figure S15D). Thus, FoxM1 is involved in SCF/c-Kit signaling pathway and acts as a positive regulator of Slug following SCF treatment.
Collectively, our data supports a novel negative regulatory feedback mechanism for SCF/c-Kit signaling pathway (Figure 7D). In our action model, SCF binds to c-Kit and activates its downstream signaling MEK/ERK and PI3K/AKT, resulting in the upregulation of c-Myc and FoxM1, which induces Slug expression and subsequently reduces c-Kit expression, leading to the attenuation of SCF/c-Kit signaling.
DISCUSSION
The self-renewal capacity of HSCs is a critical feature that determines physiological functions of HSCs, such as homing, differentiation, and apoptosis. Considerable effort has been made to identify intrinsic and extrinsic regulators for HSC self-renewal. Here, we revealed that Slug negatively regulates c-Kit, a well-known regulator of HSC self-renewal; and conversely, SCF/c-Kit signaling positively regulates Slug expression in HSPCs. Moreover, we identified c-Myc and FoxM1 as important mediators for SCF/c-Kit to induce Slug expression. Together, SCF/c-Kit-Myc-FoxM1-Slug establishes a novel negative-feedback loop in the SCF/c-Kit signaling pathway.
The SCF/c-Kit signaling pathway plays many critical roles in HSC functions. For instance, upregulation of MMP-9 releases soluble SCF after BM ablation by 5-FU and causes the HSC status transition from quiescence to proliferation 40. Loss-of-function or high expression of c-Kit impairs the self-renewal of HSCs 7, 9.
However, it still remains to be elucidated how SCF/c-Kit regulates HSC functions and how the SCF/c-Kit pathway is regulated by other factors. Previous studies reported that Slug could be induced by SCF in cell lines overexpressing c-Kit and mediates the radioprotection 20, 21. Despite these descriptions of Slug in SCF/c-Kit pathway, it has not been shown that Slug is not only induced by SCF in HSPCs, but also negatively regulates SCF/c-Kit signaling through inhibition of c-Kit expression.
Although our prior data showed that Slug deficiency enhances the self-renewal of HSCs 2, how Slug regulates HSC self-renewal is still unclear. In the current study, we found a novel feedback loop interconnecting Slug and SCF/c-Kit in HSCs: Slug is induced by SCF, and in turn downregulates the expression of endogenous c-Kit via the MEK and PI3K/Akt signaling pathways. Indeed, our data showed that reduced expression of c-Kit by overexpression of Slug or c-Kit shRNA is deleterious for HSCs and eventually causes the loss of HSCs, which are consistent with previous results 9. We also found that knockdown of endogenous c-Kit abolishes the enhanced HSC engraftment of Slug−/− HSCs, suggesting that c-Kit is required for Slug in regulating HSCs. Over a long period of time, Slug deficiency partially alleviated HSC loss caused by knockdown of c-Kit, suggesting that this negative feedback loop provides a regulatory mechanism for HSC self-renewal. This fine regulatory action model was further validated by serial BM transplantations using overexpression of Slug, c-Kit or Slug/c-Kit. Either overexpressing Slug or c-Kit individually impairs self-renewal of HSCs, whereas overexpression of both Slug and c-Kit maintains the self-renewal capacity of HSCs, suggesting that the expression level of c-Kit fine-tunes HSC self-renewal capacity. Thus, our findings clearly demonstrated that the regulatory negative feedback loop between SCF/c-Kit and Slug is critical for HSC self-renewal.
c-Myc, an unstable oncogenic nuclear factor, has been implicated in the regulations of many biological processes, including cell growth and division, apoptosis, angiogenesis and differentiation 13. c-Myc is essential for the self-renewal and differentiation of HSCs 13, 41, 42. Rodrigues et al. showed that the Myc-Slug/Twist regulatory circuit directs normal vessel formation in both the vascular and lymphatic systems 43. However, the regulatory model in SCF/c-Myc/Slug in HSCs still remains elusive. In our present study, c-Myc was required for the induction of endogenous Slug by SCF in HSCs. Meanwhile, SCF induced the expression of endogenous c-Myc, consistent with the previous study 11. Thus, our data suggest that c-Myc is a primary mediator of Slug in the SCF/c-Kit-Slug feedback loop. In addition, we found that FoxM1, a key transcription factor, is required for SCF in induction of Slug in HSPCs, which is in agreement with a recent report showing that overexpression of FoxM1 upregulates Slug expression in human breast cancer cell lines 39. Moreover, our data showed that FoxM1 is significantly induced by SCF in HSPCs, which may be due to SCF-activated PI3K and ERK signaling 14. Interestingly, it was showed that c-Myc and FoxM1 is a positive transcription activator for each other 44–46. Indeed, we demonstrated that SCF/c-Kit signaling positively regulates c-Myc and FoxM1, and then these transcription factors activate transcription of Slug in HSPCs using luciferase reporter and ChIP assays.
In summary, our current findings highlight how Slug functions as an important transcriptional repressor that finely regulates the SCF/c-Kit signaling pathway. Based on our findings, we proposed an action model for SCF/c-Kit-Myc/FoxM1-Slug in HSPCs (Figure 7E). In this model, SCF induces the expression of Slug in HSPCs through the primary mediators c-Myc and FoxM1, and sequentially elevated level of Slug suppresses c-Kit expression, thereby affecting HSC self-renewal. Our findings provide a novel insight into the molecular mechanism for how SCF/c-Kit governs HSC self-renewal.
Supplementary Material
Acknowledgments
This work was supported by NIH grants DK090478 and AG040182. We thank the Flow Cytometry Core and the Biological Resources Laboratory at University of Illinois at Chicago.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Z.Z. and W.S.W. designed and interpreted all of experiments. Z.Z. planed and conducted most of the experiments. Y.Z. conducted microarray analysis. P.Z., Y.S., Y.H., Z.Q., D.X., contribute to animal studies. Z.Z., J.M., and W.S.W wrote the manuscript.
References
- 1.Riether C, Schurch CM, Ochsenbein AF. Regulation of hematopoietic and leukemic stem cells by the immune system. Cell death and differentiation. 2015 Feb;22(2):187–198. doi: 10.1038/cdd.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun Y, Shao L, Bai H, Wang ZZ, Wu WS. Slug deficiency enhances self-renewal of hematopoietic stem cells during hematopoietic regeneration. Blood. 2010 Mar 4;115(9):1709–1717. doi: 10.1182/blood-2009-07-232934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell stem cell. 2013 Jul 3;13(1):102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nature reviews Molecular cell biology. 2007 Sep;8(9):703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- 5.Yang L, Bryder D, Adolfsson J, Nygren J, Mansson R, Sigvardsson M, et al. Identification of Lin(−)Sca1(+)kit(+)CD34(+)Flt3− short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. 2005 Apr 1;105(7):2717–2723. doi: 10.1182/blood-2004-06-2159. [DOI] [PubMed] [Google Scholar]
- 6.Lacombe J, Krosl G, Tremblay M, Gerby B, Martin R, Aplan PD, et al. Genetic interaction between Kit and Scl. Blood. 2013 Aug 15;122(7):1150–1161. doi: 10.1182/blood-2011-01-331819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller CL, Rebel VI, Helgason CD, Lansdorp PM, Eaves CJ. Impaired steel factor responsiveness differentially affects the detection and long-term maintenance of fetal liver hematopoietic stem cells in vivo. Blood. 1997 Feb 15;89(4):1214–1223. [PubMed] [Google Scholar]
- 8.Kimura Y, Ding B, Imai N, Nolan DJ, Butler JM, Rafii S. c-Kit-mediated functional positioning of stem cells to their niches is essential for maintenance and regeneration of adult hematopoiesis. PloS one. 2011;6(10):e26918. doi: 10.1371/journal.pone.0026918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin JY, Hu W, Naramura M, Park CY. High c-Kit expression identifies hematopoietic stem cells with impaired self-renewal and megakaryocytic bias. The Journal of experimental medicine. 2014 Feb 10;211(2):217–231. doi: 10.1084/jem.20131128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edling CE, Hallberg B. c-Kit--a hematopoietic cell essential receptor tyrosine kinase. The international journal of biochemistry & cell biology. 2007;39(11):1995–1998. doi: 10.1016/j.biocel.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Munugalavadla V, Dore LC, Tan BL, Hong L, Vishnu M, Weiss MJ, et al. Repression of c-kit and its downstream substrates by GATA-1 inhibits cell proliferation during erythroid maturation. Molecular and cellular biology. 2005 Aug;25(15):6747–6759. doi: 10.1128/MCB.25.15.6747-6759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie Y, Li Y, Kong Y. OPN induces FoxM1 expression and localization through ERK 1/2, AKT, and p38 signaling pathway in HEC-1A cells. International journal of molecular sciences. 2014;15(12):23345–23358. doi: 10.3390/ijms151223345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes & development. 2004 Nov 15;18(22):2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Ahmad A, Li Y, Banerjee S, Kong D, Sarkar FH. Forkhead box M1 transcription factor: a novel target for cancer therapy. Cancer treatment reviews. 2010 Apr;36(2):151–156. doi: 10.1016/j.ctrv.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue A, Seidel MG, Wu W, Kamizono S, Ferrando AA, Bronson RT, et al. Slug, a highly conserved zinc finger transcriptional repressor, protects hematopoietic progenitor cells from radiation-induced apoptosis in vivo. Cancer cell. 2002 Oct;2(4):279–288. doi: 10.1016/s1535-6108(02)00155-1. [DOI] [PubMed] [Google Scholar]
- 16.Liu X, Sun H, Qi J, Wang L, He S, Liu J, et al. Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT-MET mechanism for optimal reprogramming. Nature cell biology. 2013 Jul;15(7):829–838. doi: 10.1038/ncb2765. [DOI] [PubMed] [Google Scholar]
- 17.Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nature cell biology. 2009 Jun;11(6):694–704. doi: 10.1038/ncb1875. [DOI] [PubMed] [Google Scholar]
- 18.Shih JY, Yang PC. The EMT regulator slug and lung carcinogenesis. Carcinogenesis. 2011 Sep;32(9):1299–1304. doi: 10.1093/carcin/bgr110. [DOI] [PubMed] [Google Scholar]
- 19.Phillips S, Prat A, Sedic M, Proia T, Wronski A, Mazumdar S, et al. Cell-state transitions regulated by SLUG are critical for tissue regeneration and tumor initiation. Stem cell reports. 2014 May 6;2(5):633–647. doi: 10.1016/j.stemcr.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Losada J, Sanchez-Martin M, Rodriguez-Garcia A, Sanchez ML, Orfao A, Flores T, et al. Zinc-finger transcription factor Slug contributes to the function of the stem cell factor c-kit signaling pathway. Blood. 2002 Aug 15;100(4):1274–1286. [PubMed] [Google Scholar]
- 21.Catalano A, Rodilossi S, Rippo MR, Caprari P, Procopio A. Induction of stem cell factor/c-Kit/slug signal transduction in multidrug-resistant malignant mesothelioma cells. The Journal of biological chemistry. 2004 Nov 5;279(45):46706–46714. doi: 10.1074/jbc.M406696200. [DOI] [PubMed] [Google Scholar]
- 22.Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Developmental biology. 2001 Feb 15;230(2):230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 23.Inukai T, Inoue A, Kurosawa H, Goi K, Shinjyo T, Ozawa K, et al. SLUG, a ces-1-related zinc finger transcription factor gene with antiapoptotic activity, is a downstream target of the E2A-HLF oncoprotein. Molecular cell. 1999 Sep;4(3):343–352. doi: 10.1016/s1097-2765(00)80336-6. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Kiyokawa H, Dennewitz MB, Costa RH. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proceedings of the National Academy of Sciences of the United States of America. 2002 Dec 24;99(26):16881–16886. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang CC, Kaba M, Ge G, Xie K, Tong W, Hug C, et al. Angiopoietin-like proteins stimulate ex vivo expansion of hematopoietic stem cells. Nature medicine. 2006 Feb;12( 2):240–245. doi: 10.1038/nm1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szilvassy SJ, Weller KP, Chen B, Juttner CA, Tsukamoto A, Hoffman R. Partially differentiated ex vivo expanded cells accelerate hematologic recovery in myeloablated mice transplanted with highly enriched long-term repopulating stem cells. Blood. 1996 Nov 1;88(9):3642–3653. [PubMed] [Google Scholar]
- 27.Venezia TA, Merchant AA, Ramos CA, Whitehouse NL, Young AS, Shaw CA, et al. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol. 2004 Oct;2(10):e301. doi: 10.1371/journal.pbio.0020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Randall TD, Weissman IL. Phenotypic and functional changes induced at the clonal level in hematopoietic stem cells after 5-fluorouracil treatment. Blood. 1997 May 15;89(10):3596–3606. [PubMed] [Google Scholar]
- 29.Hemavathy K, Ashraf SI, Ip YT. Snail/slug family of repressors: slowly going into the fast lane of development and cancer. Gene. 2000 Oct 17;257(1):1–12. doi: 10.1016/s0378-1119(00)00371-1. [DOI] [PubMed] [Google Scholar]
- 30.Parra RG, Rohr CO, Koile D, Perez-Castro C, Yankilevich P. INSECT 2.0: a web-server for genome-wide cis-regulatory modules prediction. Bioinformatics. 2015 Dec 12; doi: 10.1093/bioinformatics/btv726. [DOI] [PubMed] [Google Scholar]
- 31.Aigner S, Ruppert M, Hubbe M, Sammar M, Sthoeger Z, Butcher EC, et al. Heat stable antigen (mouse CD24) supports myeloid cell binding to endothelial and platelet P-selectin. Int Immunol. 1995 Oct;7(10):1557–1565. doi: 10.1093/intimm/7.10.1557. [DOI] [PubMed] [Google Scholar]
- 32.Dole VS, Bergmeier W, Mitchell HA, Eichenberger SC, Wagner DD. Activated platelets induce Weibel-Palade-body secretion and leukocyte rolling in vivo: role of P-selectin. Blood. 2005 Oct 1;106(7):2334–2339. doi: 10.1182/blood-2005-04-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayadas TN, Johnson RC, Rayburn H, Hynes RO, Wagner DD. Leukocyte rolling and extravasation are severely compromised in P selectin-deficient mice. Cell. 1993 Aug 13;74( 3):541–554. doi: 10.1016/0092-8674(93)80055-j. [DOI] [PubMed] [Google Scholar]
- 34.Matsuoka RL, Sun LO, Katayama K, Yoshida Y, Kolodkin AL. Sema6B, Sema6C, and Sema6D expression and function during mammalian retinal development. PloS one. 2013;8(4):e63207. doi: 10.1371/journal.pone.0063207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tawarayama H, Yoshida Y, Suto F, Mitchell KJ, Fujisawa H. Roles of semaphorin-6B and plexin-A2 in lamina-restricted projection of hippocampal mossy fibers. J Neurosci. 2010 May 19;30(20):7049–7060. doi: 10.1523/JNEUROSCI.0073-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kigel B, Rabinowicz N, Varshavsky A, Kessler O, Neufeld G. Plexin-A4 promotes tumor progression and tumor angiogenesis by enhancement of VEGF and bFGF signaling. Blood. 2011 Oct 13;118(15):4285–4296. doi: 10.1182/blood-2011-03-341388. [DOI] [PubMed] [Google Scholar]
- 37.Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004 May;22(5):589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Z, Gao Y, Gordon A, Wang ZZ, Qian Z, Wu WS. Efficient generation of fully reprogrammed human iPS cells via polycistronic retroviral vector and a new cocktail of chemical compounds. PloS one. 2011;6(10):e26592. doi: 10.1371/journal.pone.0026592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang C, Chen H, Tan G, Gao W, Cheng L, Jiang X, et al. FOXM1 promotes the epithelial to mesenchymal transition by stimulating the transcription of Slug in human breast cancer. Cancer letters. 2013 Oct 28;340(1):104–112. doi: 10.1016/j.canlet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Heissig B, Hattori K, Dias S, Friedrich M, Ferris B, Hackett NR, et al. Recruitment of stem and progenitor cells from the bone marrow niche requires MMP-9 mediated release of kit-ligand. Cell. 2002 May 31;109(5):625–637. doi: 10.1016/s0092-8674(02)00754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satoh Y, Matsumura I, Tanaka H, Ezoe S, Sugahara H, Mizuki M, et al. Roles for c-Myc in self-renewal of hematopoietic stem cells. The Journal of biological chemistry. 2004 Jun 11;279(24):24986–24993. doi: 10.1074/jbc.M400407200. [DOI] [PubMed] [Google Scholar]
- 42.Baena E, Ortiz M, Martinez AC, de Alboran IM. c-Myc is essential for hematopoietic stem cell differentiation and regulates Lin(−)Sca-1(+)c-Kit(−) cell generation through p21. Experimental hematology. 2007 Sep;35(9):1333–1343. doi: 10.1016/j.exphem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 43.Rodrigues CO, Nerlick ST, White EL, Cleveland JL, King ML. A Myc-Slug (Snail2)/Twist regulatory circuit directs vascular development. Development. 2008 Jun;135(11):1903–1911. doi: 10.1242/dev.011296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wierstra I, Alves J. FOXM1c transactivates the human c-myc promoter directly via the two TATA boxes P1 and P2. The FEBS journal. 2006 Oct;273(20):4645–4667. doi: 10.1111/j.1742-4658.2006.05468.x. [DOI] [PubMed] [Google Scholar]
- 45.Wierstra I, Alves J. FOXM1c and Sp1 transactivate the P1 and P2 promoters of human c-myc synergistically. Biochemical and biophysical research communications. 2007 Jan 5;352(1):61–68. doi: 10.1016/j.bbrc.2006.10.151. [DOI] [PubMed] [Google Scholar]
- 46.Blanco-Bose WE, Murphy MJ, Ehninger A, Offner S, Dubey C, Huang W, et al. C-Myc and its target FoxM1 are critical downstream effectors of constitutive androstane receptor (CAR) mediated direct liver hyperplasia. Hepatology. 2008 Oct;48(4):1302–1311. doi: 10.1002/hep.22475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.