Abstract
A heparin oligosaccharide having a completely natural structure was successfully synthesized through a chemoenzymatic approach using an unnatural glycosyl acceptor, p-nitrophenyl glucuronide (GlcA-pNP). The use of an inexpensive and commercially available GlcA-pNP acceptor facilitates oligosaccharide recovery and purification on C-18 resin during chemoenzymatic synthesis. Oligosaccharide chain extension and modification afforded a heptasaccharide with gluconic acid residues at its reducing and non-reducing ends. Treatment with periodate oxidation followed by Smith degradation or alkaline elimination resulted in the selective cleavage of vicinal diol-containing glucronic acid residues affording highly sulfated heparin pentasaccharide having a completely natural structure. This methodology should facilitate the chemoenzymatic synthesis of a family of highly sulfated heparin oligosaccharides with unmodified structures for biological evaluation.
Heparin is the most highly sulfated member of the heparan sulfate (HS) family of linear, highly anionic polysaccharides1. HS consists of repeating, 1→4-linked disaccharide units of alternating uronic acid residue, either β-D-glucuronic acid (GlcA) or α-L-iduronic acid (IdoA) and α-D-glucosamine (GlcN) residue2–4. Both saccharide units in the HS disaccharide repeating units can be substituted with N-sulfo (S) or N-acetyl (Ac) groups and O-sulfo (S) groups (at the 2-O-position of the uronic acid residues and the 3-O- and 6-O-positions of the GlcN residue). Heparin is a unique form of HS, found within specialized granulated cells4, which has long highly sulfated domains5 consisting primarily of the trisulfated disaccharide repeating unit →4) IdoA2S (1→4) GlcNS6S (1→4.
Highly sulfated heparin chains display a number of prominent biological activities including growth factor6–8 and chemokine9 signaling and anticoagulant activity10. The pentasaccharide sequence containing a →4) IdoA2S (1→4) GlcNS6S (1→ at its reducing end with a central GlcNS3S6S residue, having the structure, →GlcNS6S→GlcA→GlcNS3S6S→IdoA2S→GlcNS6S→ (Fig. 1), is responsible for its specific binding to the serine protease inhibitor (Serpin) antithrombin III (AT). This AT pentasaccharide binding site has been chemically synthesized as the O-methylglycoside and is used clinically as an antithrombotic agent known as the ultra-low molecular weight heparin (ULMWH) fondaparinux (Arixtra®)11. Fondaparinux is a specific anti-factor Xa (FXa) agent and acts by binding to an allosteric site on AT making it a potent FXa inhibitor12, 13. Unfractionated heparins (UFHs), low molecular weight heparins (LMWHs) can contain a repeating trisulfated disaccharide sequence ([→4) IdoA2S (1→4) GlcNS6S (1→]6) flanking their AT pentasaccharide binding sites. This trisulfated disaccharide rich domain corresponds to heparins thrombin (FIIa) binding site and facilitates the assembly of the ternary heparin-AT-FIIa complex required for heparin’s global anticoagulant activity14.
Figure 1.
The antithrombin III-binding pentasaccharide found in heparin.
UFH and LMWH are polypharmacological agents, complex mixtures of molecules prepared from animal tissues15. The supply chain of pharmaceutical heparin has been poorly regulated and this presented safety concerns. A worldwide distribution of contaminated heparin in 2007, caused by its adulteration with a semi-synthetic oversulfated chondroitin sulfate contaminant, adversely affected the purity and safety of animal-sourced UFH and LMWH, and was associated with over 200 deaths in the US16, 17. Another potential threat is that other bioactive entities, such as viruses or prions that might remain associated with the HS chains in animal extracts. Thus, the cost-effective preparation of structurally defined heparin oligosaccharides (ULMWH) from non-animal sources is highly desirable.
There are two ways to prepare homogeneous heparin oligosaccharides, one uses a purely synthetic chemical method, based on repetitive steps of protection, activation, coupling and de-protection, and is very challenging for molecules larger than the pentasaccharide Arixtra®11. Chemoenzymatic synthesis, relies on combined chemical and enzymatic methods, mimicking the biosynthetic pathway of heparin, and represents a promising strategy to address many of these synthetic challenges. Our previous studies18, 19 have demonstrated that chemoenzymatic synthesis is capable of generating a series of heparin oligosaccharides in good overall yield and possessed excellent anticoagulant activity. One limitation of our current chemoenzymatic synthetic approach is that we begin with an unnautural inexpensive commercially available acceptor, p-nitrophenyl glucuronide (GlcA-pNP)18, 19, that is ultraviolet detectable, hydrophobic, easily binding to reversed phase chromatography resins, enabling the detection recovery, and purification of the resulting oligosaccharides using a C-18 resin. However, the resulting target molecules contain an unnatural structural feature. We have previously investigated the removal of pNP from chemoenzymatically synthesized HS oligosaccharides using ceric ammonium nitrate 20.
In the current study we faced the challenge of removing GlcA-pNP from chemoenzymatically synthesized highly sulfated oligosaccharide with domains consisting of repeating trisulfated disaccharide units. These heparin oligosaccharides represent potential precursors in the chemoenzymatic synthesis of Arixtra® and also display interesting biological activities18,21. We report the selective removal of GlcA-pNP moiety from a more structurally complex heparin-like oligosaccharide using both Smith degradation and alkaline elimination22–24. This approach has been previously used for micro scale analysis of polysaccharides relying on mass spectrometry (MS) and leaves unnatural moieties at the reducing end of the resulting degradation products25–28. We have modified this method to prepare a natural heparin pentasaccharide rich in trisulfated disaccharide units.
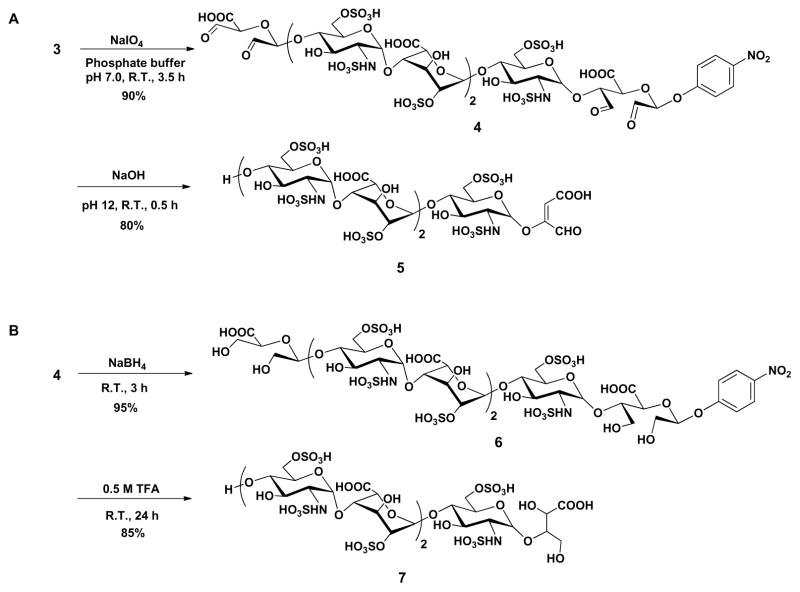
Based on our previous effort to synthetic homogeneous low-molecular-weight heparins18, the chemoenzymatic synthesis of heparin-like oligosaccharide containing IdoA2S-GlcNS6S (where S is sulfate) repeating unit was initiated from the commercially available p-nitrophenyl glucuronide (GlcA-pNP, 1) (Scheme 1). The monosaccharide acceptor 1 was repetitively elongated with PmHS2 (heparosan synthase 2 from Pasteurella multocida), UDP-N-trifluoroacetyl glucosamine (GlcNTFA) donor and UDP-glucuronic acid (GlcA), to form heptasaccharide 2. Selective removal of TFA group on nitrogen atom followed by N-sulfation, C5 epimerization /2-O-sulfation and 6-O-sulfation produced structure 3 for further modification. During this chemoenzymatic synthesis, the strong UV absorbance and high C-18 binding affinity of the p-NP group facilitate the detection and purification of the intermediates. The sulfated oligosaccharides were purified by Q-Sepharose. A total of 20 mg of 3 was prepared.
Scheme 1.
Construction of heparin-like heptasaccharide (3)
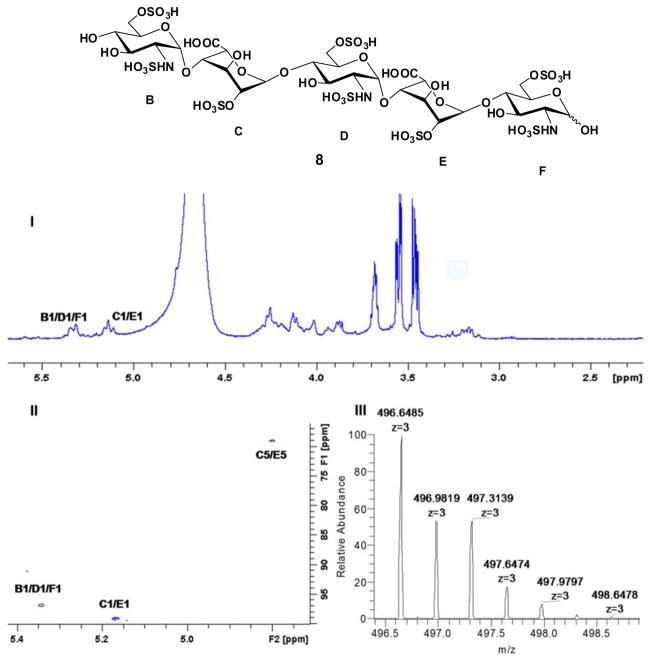
With substrate 3 in hand, a selective oxidative-cleavage of vicinal diol was carried out with sodium periodate (NaIO4) in neutral sodium phosphate buffer at 37 °C29 for 3.5 h, furnishing the corresponding aldehyde 4, which could be used to explore the cleavage of GlcA-pNP group using either alkaline elimination or Smith degradation method (Scheme 2). The periodate oxidation was terminated by the addition of ethylene glycol followed by dialysis (MWCO 100–500 Da) against distilled water and lyophilization. Alkaline elimination method was first investigated (Scheme 2A). A solution of oligosaccharide aldehyde 4 was adjusted to pH 12 with 0.5 M NaOH aqueous. After standing at room temperature for 30 min, the solution was neutralized with 0.5 M acetic acid. The resulting mixture was dialyzed (MWCO 100–500 Da) against distilled water and lyophilized to obtain the elimination compound 5 with an unnatural enol glycoside at the reducing end. Scheme 2B shows the Smith degradation of oligosaccharide aldehyde 4. Next, 4 was treated with sodium borohydride (NaBH4) in distilled water for 3 h at room temperature and then excess borohydride was quenched with 1 M acetic acid. After dialysis against distilled water and lyophilization, the oxidized-reduced oligosaccharide was dissolved in 0.5 M trifluoroacetic acid (TFA) solution at room temperature for 24 h. The reaction was terminated by the addition of 0.5 M NaOH to pH 7 and dialyzed to afford the degradation product 7. Gratifyingly, no glycosidic bond cleavage or de-sulfation was detected throughout this reaction scheme. Moreover, detection by LC-MS showed a yield of less than 5% of the desired target compound 8. This discovery suggested that the glycoside could be selectively cleaved while leaving the other glycosidic bonds within the oligosaccharide intact. Both the structures of 5 and 7 were clearly supported by LC-MS, 1D and 2D NMR spectrum (Fig. 2 and Supplementary data). In the 1H spectra of alkaline elimination product 5 and Smith degradation product 7, the disappearance of signals at 4.52 and 5.23 ppm show that two GlcA residues have been successfully oxidatively cleaved (Fig. 2 I–III). The anomeric signals appearing at 5.34 and 5.14 ppm confirmed the presence of GlcNS6S and IdoA2S residues, respectively (Fig. 2 II, III). In addition, according to H-H COSY spectrum of compound 5 and 7, the chemical shifts of H-2, H-4 and H-5 on IdoA residue appear at 4.26, 4.02 and 4.76 ppm, respectively, indicating that 2-O-sulfo groups were still present on the IdoA residues (Fig. 2 IV, V).
Scheme 2.
GlcA-pNP cleavage via alkaline elimination (A) and Smith degradation (B).
Figure 2.
NMR characterization of degradation oligosaccharides (R = pNP). Panel I, II, III show the 1D 1H NMR spectra of compound 3, 5 and 7, respectively. Peaks corresponding to the anomeric protons of these compounds can be clearly identified. Panel IV, V show the selected H-H correlation in 2D H-H COSY spectrum of compound 5 and 7, respectively. Three sets of peaks between 3.4–3.7 ppm in 1H NMR spectrum were from glycerol that came from the dialysis membrane.
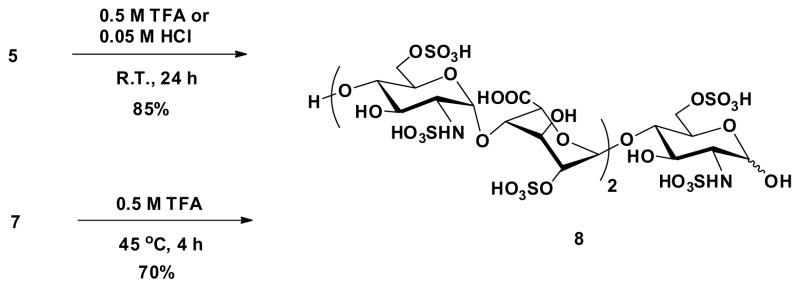
Acidic hydrolysis was used to remove the unnatural glycoside in oligosaccharides 5 and 7 to prepare natural heparin pentasaccharide (Scheme 3). Hydrolysis of compound 5 was easier since enol group was more acid labile. We found that either 0.5 M TFA or 0.05 M HCl aqueous could successfully produce target oligosaccharide 8 in good yield with <5% starting material left detected by LC-MS. However, removal of the unnatural glycoside from 7 turned out to be more challenging. Using the same conditions described above only recovered starting material. Finally, transformation of 7 to 8 was achieved in 0.5 M TFA solution at 45°C for 4 h. These reaction conditions need to be strictly controlled, as higher temperatures or longer reaction times result in significant decomposition. The NMR spectra and LC-MS of the target compound 8 clearly supports the success of this acid hydrolysis step (Figure 3). The anomeric signals at 5.34 and 5.14 ppm confirmed the presence of GlcNS6S and IdoA2S residues, respectively (Fig. 3 I). The selected anomeric and IdoA carbon region (64–104 ppm) in 2D 1H-13C HSQC spectrum also confirm the structure (Fig. 3 II). The structure was also characterized by LC-MS with m/z 496.6377 in negative mode (Fig. 3 III).
Scheme 3.
Acidic hydrolysis of glycoside on degradation oligosaccharides.
Figure 3.
NMR and HRMS of natural heparin oligosaccharide 8. Panel I shows the 1D 1H NMR spectrum. Panel II shows the selected anomeric and IdoA2S carbon region (64–104 ppm) in 2D 1H-13C HSQC spectrum. Panel III shows the HRMS (negative mode) of compound 8.
We next expanded the oligosaccharide substrates subjected to our alkaline degradation method to investigate the reaction scope (Table 1). These synthetic oligosaccharides contained both GlcA and IdoA residues and possessed different chain lengths, sulfation levels, and flanking residues. The first step, periodate oxidation forming the intermediate aldehyde, is critical to the entire degradation process. Although it was reported that the neighbouring environment might influence the reactivity of uronic acid residues27, no selectivity in periodate cleavage was observed for unsulfured GlcA or IdoA residues within the oligosaccharides tested. Oligosaccharides having different residues flanking GlcA (entries 1–3) were fully cleaved to monosaccharides at 37 °C and pH 7.0. Oligosaccharides containing both unsulfated GlcA and IdoA (entries 4–5) were also fully cleaved to monosaccharide products, thus, no selectivity between GlcA and IdoA was observed. Oligosaccharides containing GlcA residues and of different chain lengths and sulfation levels (entries 6–8) afforded complete cleavgae of GlcA residues and afforded oligosaccharides of chain lengths expected based on the spacing of these GlcA residues.
Table 1.
Alkaline Smith degradation of different synthetic oligosaccharides.
| Entry | Substrates | Productsa |
|---|---|---|
| 1 | GlcNAc-GlcA-(GlcNAc-GlcA)5-GlcNAc-GlcA-pNP | Monosaccharide-R |
| 2 | GlcNS-GlcA-(GlcNS-GlcA)5-GlcNS-GlcA-pNP | Monosaccharide-R |
| 3 | GlcNS6S-GlcA-(GlcNS6S-GlcA)5-GlcNS6S-GlcA-pNP | Monosaccharide-R |
| 4 | GlcNS-GlcA-GlcNS-IdoA-GlcNS-GlcA-pNP | Monosaccharide-R |
| 5 | GlcNS6S-GlcA-GlcNS6S3S-IdoA-GlcNS6S-GlcA-pNP | Monosaccharide-R |
| 6 | GlcNS6S-GlcA-(GlcNS6S-IdoA2S)5-GlcNS6S-GlcA-pNP | Undecassaccharide-Rb |
| 7 | GlcNS6S-GlcA-(GlcNS6S-IdoA2S)7-GlcNS6S-GlcA-pNP | Pentadecasaccharide-Rb |
| 8 | GlcNS-GlcA-(GlcNS-IdoA2S)7-GlcNS-GlcA-pNP | Pentadecassaccharide-Rb |
R=C4H3O3 (see 5 in Scheme 2 for structure of aglycone).
Confirmed by LC-MS (see Supplemental Information).
In conclusion, an unmodified heparin heptaasaccharide was successfully prepared through a chemoenzymatic synthesis using p-nitrophenyl glucuronide (GlcA-pNP) as a glycosylation acceptor, which greatly facilitates the synthesis. The selective removal of GlcA-pNP moiety from the reducing end of the molecule and GlcA from its non reducing end, using both Smith degradation and alkaline elimination, afforded the target pentasaccharide saccharide. To our knowledge, this is the first time to these two degradation methods has been applied to the preparative synthesis of an oligosaccharide. Most importantly, no de-sulfation was detected during the synthesis. Additional oligosaccharide substrates were investigated and demonstrated that this degradation method was specific for saccharide residues with vicinal diols, i.e., GlcA or IdoA or residues at the non-reducing end, could be applied to diverse oligosaccharides containing different chain lengths, sulfation levels, and flanking residues. This strategy should be useful in the construction of more structurally complex heparin oligosaccharides. Since these targets are all natural and with a GlcNS6S residue at their reducing end, they could be used as precursors in the chemoenzymatic synthesis of Arixtra® or other oligosaccharides with therapeutic anticoagulant and antithrombotic activities.
Supplementary Material
Acknowledgments
The authors are grateful for funding from the National Institutes of health in the form of HL125371, HL094463 and HL62244.
Notes and references
- 1.Linhardt RJ. J Med Chem. 2003;46:2551. doi: 10.1021/jm030176m. [DOI] [PubMed] [Google Scholar]
- 2.Pearson AG, Kiefel MJ, Ferro V, von Itzstein M. Org Biomol Chem. 2011;9:4614. doi: 10.1039/c1ob05165b. [DOI] [PubMed] [Google Scholar]
- 3.Solari V, Rudd TR, Guimond SE, Powell AK, Turnbull JE, Yates EA. Org Biomol Chem. 2015;13:6066. doi: 10.1039/c5ob00564g. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Linhardt RJ. Nat Prod Rep. 2014;31:1676. doi: 10.1039/c4np00076e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suflita M, Fu L, He W, Koffas M, Linhardt RJ. Appl Microbiol Biotechnol. 2015;99:7465. doi: 10.1007/s00253-015-6821-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop JR, Schuksz M, Esko JD. Nature. 2007;446:1030. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 7.Capila I, Linhardt RJ. Angew Chem Int Ed. 2002;41:390. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 8.Wu B, Wei N, Thon V, Wei M, Yu Z, Xu Y, Chen X, Liu J, Wang PG, Li T. Org Biomol Chem. 2015;13:5098. doi: 10.1039/c5ob00462d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Paz JL, Moseman EA, Noti C, Polito L, von Andrian UH, Seeberger PH. ACS Chem Biol. 2007;2:735. doi: 10.1021/cb700159m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Pedersen LC. Appl Microbiol Biotechnol. 2007;74:263. doi: 10.1007/s00253-006-0722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petitou M, van Boeckel CAA. Angew Chem Int Ed. 2004;43:3118. doi: 10.1002/anie.200300640. [DOI] [PubMed] [Google Scholar]
- 12.Bauer KA, Hawkins DW, Peters PC, Petitou M, Herbert JM, van Boeckel CAA, Meuleman DG. Cardiovasc Drug Rev. 2002;20:37. doi: 10.1111/j.1527-3466.2002.tb00081.x. [DOI] [PubMed] [Google Scholar]
- 13.Nagler M, Haslauer M, Wuillemin WA. Thromb Res. 2012;129:407. doi: 10.1016/j.thromres.2011.10.037. [DOI] [PubMed] [Google Scholar]
- 14.Onishi KSAA, Dordick JS, Linhardt RJ. Front Biosci. 2016;21:1372. doi: 10.2741/4462. [DOI] [PubMed] [Google Scholar]
- 15.Bhaskar U, Sterner E, Hickey AM, Onishi A, Zhang F, Dordick JS, Linhardt RJ. Appl Microbiol Biotechnol. 2012;93:1. doi: 10.1007/s00253-011-3641-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerrini M, Beccati D, Shriver Z, Naggi A, Viswanathan K, Bisio A, Capila I, Lansing JC, Guglieri S, Fraser B, Al-Hakim A, Gunay NS, Zhang Z, Robinson L, Buhse L, Nasr M, Woodcock J, Langer R, Venkataraman G, Linhardt RJ, Casu B, Torri G, Sasisekharan R. Nat Biotech. 2008;26:669. doi: 10.1038/nbt1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szajek AY, Chess E, Johansen K, Gratzl G, Gray E, Keire D, Linhardt RJ, Liu J, Morris T, Mulloy B, Nasr M, Shriver Z, Torralba P, Viskov C, Williams R, Woodcock J, Workman W, Al-Hakim A. Nat Biotech. 2016;34:625. doi: 10.1038/nbt.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Cai C, Chandarajoti K, Hsieh PH, Li L, Pham TQ, Sparkenbaugh EM, Sheng J, Key NS, Pawlinski R, Harris EN, Linhardt RJ, Liu J. Nat Chem Biol. 2014;10:248. doi: 10.1038/nchembio.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y, Masuko S, Takieddin M, Xu H, Liu R, Jing J, Mousa SA, Linhardt RJ, Liu J. Science. 2011;334:498. doi: 10.1126/science.1207478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai C, Li L, Harvey C, Liu J, Linhardt RJ. Tetrahedron Lett. 2013;54:4471. doi: 10.1016/j.tetlet.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, Pempe EH, Liu J. J Biol Chem. 2012;287:29054. doi: 10.1074/jbc.M112.358523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fransson L-Å, Carlstedt I. Carbohyd Res. 1974;36:349. doi: 10.1016/s0008-6215(00)83056-6. [DOI] [PubMed] [Google Scholar]
- 23.Lindberg B, Lönngren J, Thompson JL. Carbohyd Res. 1973;28:351. doi: 10.1016/s0008-6215(00)82321-6. [DOI] [PubMed] [Google Scholar]
- 24.Whistler RL, Chang PK, Richards GN. J Am Chem Soc. 1959;81:4058. [Google Scholar]
- 25.Arbatsky NP, Kondakova AN, Shashkov AS, Drutskaya MS, Belousov PV, Nedospasov SA, Petrova MA, Knirel YA. Org Biomol Chem. 2010;8:3571. doi: 10.1039/c004090h. [DOI] [PubMed] [Google Scholar]
- 26.Baird JK, Holroyde MJ, Ellwood DC. Carbohyd Res. 1973;27:464. [Google Scholar]
- 27.Fransson L-Å, Malmström A, Sjöberg I, Huckerby TN. Carbohyd Res. 1980;80:131. [Google Scholar]
- 28.Fransson L-Å, Sjöberg I, Havsmark B. Eur J Biochem. 1980;106:59. [PubMed] [Google Scholar]
- 29.Elevated temperature (37 °C) and pH ≥7.0 is required to fully cleave all the vicinal diols.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.