Abstract
Background
Klebsiella pneumoniae can be a serious pathogen in nonhuman primates, particularly Neotropical monkeys.
Methods
During a K. pneumoniae outbreak in an owl monkey research colony, thirteen K. pneumoniae isolates were DNA fingerprinted by automated repetitive extragenic palindromic-polymerase chain reaction and the profiles compared to isolates obtained from other nonhuman primate species during the same time period and isolates from previous outbreaks.
Results
Eleven different types of K. pneumoniae were circulating in the owl monkey colony at the time of the outbreak. When comparing owl monkey isolates relatedness to previous colony outbreak isolates and squirrel monkey and capuchin monkey isolates, all were different.
Conclusions
These results agree with recent reports where K. pneumoniae nosocomial isolates in hospital settings can have high genetic diversity, and multiple strains can be circulating simultaneously. This potential genetic diversity should be considered when designing strategies for controlling K. pneumoniae outbreaks in captive nonhuman primate colonies.
Keywords: Aotus nancymai, automated repetitive extragenic palindromic-polymerase chain reaction, Cebidae, DNA fingerprinting, Klebsiellosis, owl monkey, rep-PCR
Introduction
Klebsiella pneumoniae is an emerging bacterial pathogen in humans and animals [11, 14]. Wide spread in the environment, K. pneumoniae, including antibiotic-resistant biotypes, can be found in agricultural foodstuff, farm produce, forest and botanical environments [3, 5, 8, 20, 21, 24]. Originally described as an occasional source of urinary tract infections and abscesses in humans, more recently, it has become a multidrug resistant life threatening infection requiring hospitalization [13, 22, 27]. In nonhuman primates, K. pneumoniae is an important pathogen known to cause pneumonia, peritonitis, and septicemia, threatening the viability of captive colonies [7, 9, 10, 19, 23, 26]. Numerous outbreaks have been reported, with neotropical nonhuman primates appearing to be particularly susceptible to fatal disease [2, 10, 17, 19]. Less obvious, but not less important, is that subclinical infections with some K. pneumoniae strains can alter immunological profiles in nonhuman primates, potentially affecting research studies [6].
In this report we describe a K. pneumoniae outbreak in a nonhuman primate research colony and the use of DNA fingerprinting to help determine isolates relatedness and help establish outbreak control and preventive measures.
Materials and Methods
Humane Care Guidelines
The owl monkey (Aotus nancymai) colony at the National Institute of Allergy and Infectious Diseases, National Institutes of Health, comprise an average daily census of approximately 200 animals, all adults of both sexes. Most animals were wild-caught in the Peruvian Amazon basin region by Center for Reproduction and Conservation of Nonhuman Primates (CRCP) personnel in Iquitos, Perú and transferred to the National Institute of Allergy and Infectious Diseases, National Institutes of Health through an agreement with the Pan American Health Organization. A few animals were transferred from other research institutions. Routine quarantine procedures comprised of physical examination, tuberculin testing, screening for intestinal bacterial pathogens by routine bacteriologic cultures, and wet mounts and fecal flotation for parasitological examination. The monkeys were housed and cared for according to the “Guide for the Care and Use of Laboratory Animals” and Animal Welfare Regulations [15]. All animals were enrolled in Institutional Animal Care and Use Committee-approved malaria candidate vaccine studies but not necessarily infected with malaria parasites at the time of the outbreak. Standard husbandry procedures included feeding Purina New World Primate Diet 5040 (Purina Mills, St. Louis, MO), Zupreem Primate Diet Canned (Zupreem, Shawnee, KS), diet supplements (a mash prepared in-house and bananas), and water ad-libitum. The animals were housed in stainless steel 6.0 square-foot cages (Primate Products Inc., Miami, FL) with PVC nesting boxes and wood perches. A 12:12 dark/light hour photoperiod cycle was observed, room temperature was maintained at 24° Celsius. Pair-housing (male/female) was done when possible.
Bacterial Isolation
Rectal swabs taken from animals showing diarrhea, lethargy, or depression and, in fatal cases, from various organs during necropsy, were taken immediately to the bacteriology laboratory for routine culture, isolation, identification, and antimicrobial susceptibility testing. In addition, feed, supplies, and environmental swabs were collected to further assess the role environmental contamination could play in the transmission and persistence of infection. Briefly, specimens were placed in Cary-Blair transport medium and inoculated in Blood agar plate (BAP), MacConkey agar plate (MAC), Xylose-lysine-desoxycholate agar plate (XLD), Salmonella-Shigella agar plate (SS), Campylobacter agar plates (Campy & CVA), Anaerobic Reducible Blood Agar, GN Broth and/or Selenite Broth, and cold enriched in Thioglycollate broth (Remel Products, Lenexa, KS). BAP, MAC, XLD, & SS agar plates were incubated in ambient air at 37°C. Campylobacter selective agar plates incubated under micro-aerobic conditions at 42°C, enrichment broths (GN & Selenite) incubated for 24 hours at 37°C and sub-cultured to MAC and XLD agar plates, which were then incubated for 24 hours at 37°C. Cold Enrichment Broth (Thioglycollate) was incubated for 24 hours at 4°C and sub-cultured to a Yersinia agar plate (CIN). CIN plate was incubated for 48 hours at room temperature. Test plates were examined for colonies characteristic of bacterial agents which may be pathogenic, and suspect colonies isolated and evaluated for identification. Vitek II System Gram negative Card Panel and API 20 E System panel (BioMérieux, Durham, NC) was used for full identification. Sensitivity profiles were performed using the Biomic V3 Microbiology System for the disk diffusion method and the Trek Sensititre System for MIC Evaluation (Biomic V3 Microbiology System, Santa Barbara, CA). Pathogens screened for included Salmonella spp., Shigella spp., Campylobacter spp., pathogenic Escherichia coli, Yersinia spp., Klebsiella pneumoniae, Aeromonas spp., Staphylococcus aureus, and Pseudomonas aeruginosa.
Bacterial DNA Fingerprinting
Selected K. pneumoniae isolates were DNA fingerprinted by automated repetitive extragenic palindromic-polymerase chain reaction (rep-PCR, DiversiLab System, BioMérieux) of noncoding intergenic repetitive elements in the genomic DNA of the isolate. Briefly, genomic DNA was extracted with the UltraClean® microbial DNA isolation kit (MO Bio Laboratories, Inc., Carlsbad, CA). PCRs were performed with 90–140 ng of purified bacterial DNA and reagents in the DiversiLab™ DNA fingerprinting kit (BioMérieux). PCR was performed using an initial denaturation of 94°C for 120 s, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 70°C for 90 s, with a final extension at 70°C for 180 s. PCR products were separated in the Agilent 2100 BioAnalyzer® (Agilent Technologies, Santa Clara, CA) using a microfluidics DNA chip according to the protocol provided by Bacterial Barcodes, Inc. (BioMérieux). The rep-PCR fingerprint profiles were compared with DiversiLab™ v3.4 software (BioMérieux) using Pearson coefficients to determine distance matrices and the unweighted pair group method with arithmetic mean to create dendrograms. Reports were automatically generated and included in the dendrogram, virtual gel images, scatter plots, and demographic matrices to aid in interpretation of the data. Sample relationships for rep-PCR were designated as follows: indistinguishable (>97% similarity); similar (>95% similarity); and different (<95 % similarity) as recommended by the manufacturer (DiversiLab, BioMérieux). The obtained profiles were compared to samples obtained from a squirrel monkey (Saimiri spp.) and two capuchin monkeys (Cebus apella) housed in separate animal facilities and two strains isolated from owl monkeys during previous outbreaks in 1998 (A98) and 2001 (B01) respectively.
Pathology
Tissue samples from all major organs were collected at necropsy, fixed in neutral buffered 10% formalin for routine histopathological processing and light microscopy examination.
Results
Outbreak Report
On February 18th 2005, an owl monkey was found dead. At necropsy, the animal appeared dehydrated, and the perineal hair coat contained adherent dried fecal material. The distal colon contained fluid digesta with no formed feces. The cecum contained similar fluid digesta, and the cecal mucosa was diffusely thickened with multiple white to yellow plaques adhered to the mucosal surface. A sample of colon was taken and submitted for bacterial culture. Tissue samples from all major organs were collected and fixed in neutral buffered 10% formalin for routine histopathological examination. Microscopically, the small intestine showed diffuse congestion, the cecum and colon had mucosal erosions and fibrin exudation with bacterial colonization. The lesions were consistent with enteritis and typhlocolitis. On bacterial culture of the colon sample taken, Klebsiella pneumoniae was the only potential intestinal pathogen isolated. Rectal swabs were taken from all the monkeys on the same rack, and bacteriology cultures performed to detect subclinical infection with K. pneumoniae but all were found to be negative. Approximately two months later (April 21st), another animal was found dead and a third monkey four days later (April 25th). At necropsy, similar findings, enteritis and necrotic and hemorrhagic typhlocolitis, were observed with positive K. pneumoniae bacterial culture obtained from samples taken during necropsy. The room was placed under quarantine, and all animals were treated with oral antibiotics based on in-vitro sensitivity testing (trimethoprim-sulfamethoxazole 24mg/k PO, BID; Sulfatrim Pediatric, STI Pharma LLC, Langhome, PA). Despite antibiotic treatment, 17 additional owl monkeys died during the month of May with similar lesions at necropsy. K. pneumoniae was only isolated from 12 of the 17 cases, presumably because the animals were already on antibiotic treatment at the time of death. A sensitivity test performed on the isolates revealed that the isolates were resistant to trimethoprim-sulfamethoxazole. Treatment was switched to enrofloxacin (5mg/k IM, BID; Baytril, Bayer, Pittsburg, PA).
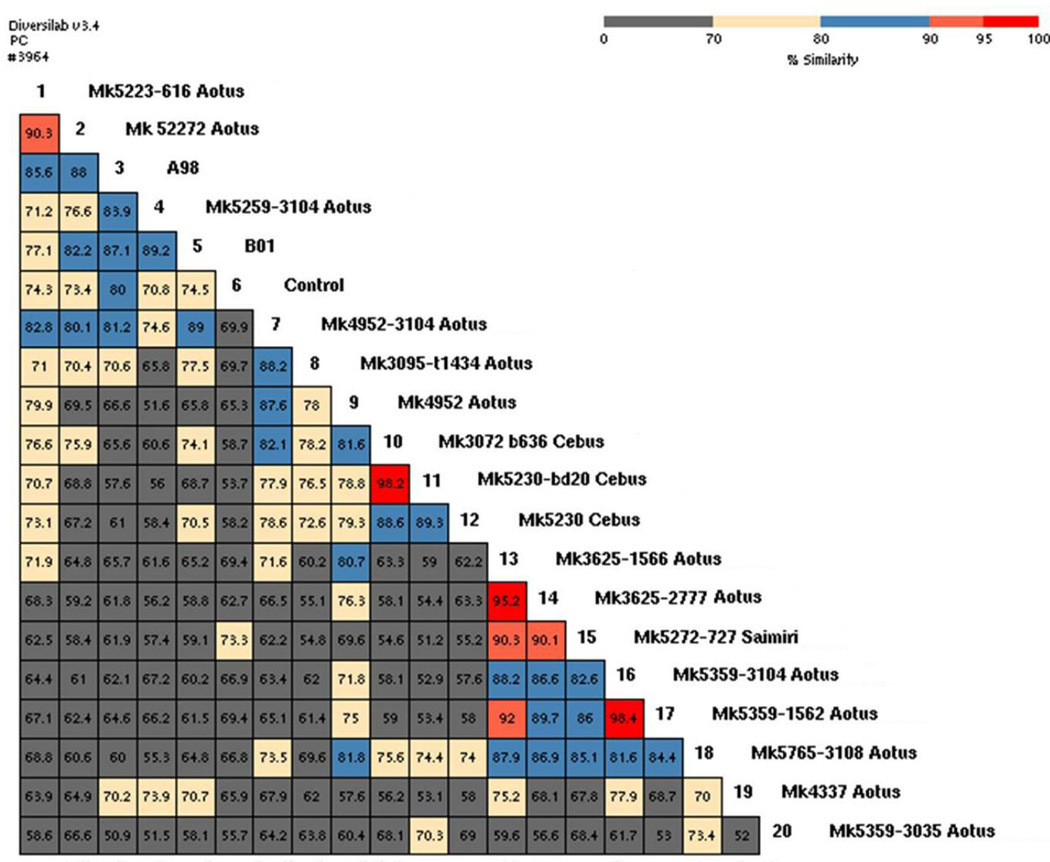
Thirteen owl monkey K. pneumoniae isolates were submitted for DNA fingerprinting. Pearson correlations between aligned rep-PCR profiles revealed at least eleven different isolate types (<95% similarity) circulating in the owl monkey colony between February and May. When comparing owl monkey K. pneumoniae isolate relatedness to previous outbreak isolates, all were different to the 1998 (A98) outbreak isolate (50.9% to 88% similarity) and the 2001 (B01) outbreak isolate (58.1% to 89.2% similarity). When comparing owl monkey K. pneumoniae isolates to a squirrel monkey isolate they were all different (54.8% to 90.3%). When comparing owl monkey to capuchin isolates, they were all different too (52.9% to 82.1% similarity). Lastly, when comparing the squirrel monkey isolate to the capuchin monkey isolates, these were also found to be different, showing only 51.2% to 55.2% similarity (Figures 1 and 2). One owl monkey (#3104), tested at different times during the outbreak, had three different K. pneumoniae isolates. A trimethoprim-sulfamethoxazole-resistant isolate was fingerprinted (sample number 18) and it was different (<95% similarity) to all other isolates. Bacterial culture of swabs taken from husbandry equipment and feed revealed K. pneumoniae was present on the leather gloves and nets used to catch the monkeys, the diet supplement prepared with ripe bananas, and the banana peels. Unfortunately, the K. pneumoniae isolates obtained from the bananas and equipment were not fingerprinted so we cannot tell if these were the same isolates found in the monkeys. Since no other potential source of infection was found, we decided to stop the use of bananas as a diet supplement and replace them with apples. All husbandry utensils were disinfected with diluted iodine after each use, and the cage wood perches replaced with PVC pipes to make sure they were effectively sanitized during cage washes. During the month of June, nine additional owl monkeys died, K. pneumoniae was isolated from five cases but it was not clear if the organism was the cause of death since, at necropsy, cardiomyopathy and renal disease was found in addition to typhlocolitis. At least two deaths were most likely due to cardiomyopathy, as indicated by the lesions observed at necropsy and the fact that these animals were on lifetime medication for cardiac insufficiency. By the middle of June, a trivalent bacterin prepared using K. pneumoniae isolates obtained from monkeys that died in the colony was tested in four healthy owl monkeys for potential side effects but not used in the whole population until the colony appeared to be stable a month later. One last case of K. pneumoniae associated death was observed in the monkey colony in August when one monkey died with typhlocolitis. This animal also had cardiomyopathy, nephritis, and glomerulosclerosis. No more Klebsiella associated morbidity or mortality has been observed since.
Figure 1.
Repetitive extragenic palindromic polymerase chain reaction (rep-PCR) of noncoding intergenic repetitive elements in the genomic DNA of Klebsiella pneumoniae isolate fingerprint profiles analyzed with Diversilab™ v3.4 strain typing software. Software generated dendrogram and virtual gel images show K. pneumoniae similarities between isolates obtained from owl monkeys and other nonhuman primates. Sample 6 is kit control. Samples A98 and B01 are strains isolated from owl monkeys during previous outbreaks (in 1998 and 2001, respectively). Only isolates 10 and 11 (isolated from capuchin monkeys) and isolates 16 and 17 (isolated from owl monkeys), were indistinguishable (>97% similarity). Isolates 13 and 14 (isolated from owl monkeys) were similar (>95% similarity), all other isolates were different (<95% similarity). Owl monkey number 3104, tested at different times during the outbreak, had three different K. pneumoniae isolates (isolates number 4, 7, and 16).
Figure 2.
Rep-PCR of noncoding intergenic repetitive elements in the genomic DNA of Klebsiella pneumoniae isolate fingerprint profiles analyzed with Diversilab™ v3.4 strain typing software. Software generated demographic matrix showing Klebsiella pneumoniae numerical similarity calculations between isolates obtained from owl monkeys and other nonhuman primates. Sample 6 is kit control. Samples A98 and B01 are strains isolated from owl monkeys during previous outbreaks (in 1998 and 2001, respectively). Isolates 10 and 11 (isolated from capuchin monkeys) and isolates 16 and 17 (isolated from owl monkeys), were indistinguishable (98.2% and 98.4% respectively). Isolates 13 and 14 (isolated from owl monkeys) were similar (95.2%), all other isolates were different (<95% similarity). Owl monkey number 3104 had three different K. pneumoniae isolates (4, 7, and 16) with similarity ranging between 74.6% and 63.4%.
Discussion
The wide genetic diversity observed in the K. pneumoniae isolates, during a relatively short period of time, suggested that not one but multiple strains were responsible for the outbreak. When trying to determine the source of the isolates, it became apparent that the animals were being infected while in captivity since on arrival, during quarantine, the animals were in most cases not infected. To asses this possibility, husbandry methods were scrutinized, feed, bedding, cages, and husbandry utensils, and environmental swabs taken and cultured to evaluate possible breaks in sanitation practices. As a result of this testing, K. pneumoniae was isolated from the leather gloves and nets used to catch the monkeys, from a diet supplement prepared with ripe bananas, and from banana peels. Unfortunately, the environmental K. pneumoniae isolates were not fingerprinted so we could not confirm the environmental isolates were the same found in the owl monkeys and responsible for the outbreak. However, since K. pneumoniae was not isolated from any other object in contact with the owl monkeys, it was concluded that the most likely source responsible for the introduction of K. pneumoniae isolates in the nonhuman primate colony were banana peels. Cross contamination most likely occurred while peeling the bananas to prepare the diet supplements. Further environmental contamination probably occurred through the use of husbandry utensils that had direct physical contact with infected monkeys and/or their feces. Based on antibiotic sensitivity testing, oral therapy with trimethoprim-sulfamethoxazole was initiated as soon as the outbreak started. However, on culture follow up, the bacterial isolates were resistant to the instituted therapy which prompted the current study. The trimethoprim-sulfamethoxazole-resistant isolate (sample number 18) that was fingerprinted was found to be different to all other isolates. We do not know if the strain was already drug-resistant when introduced in the colony or acquired resistance in the colony. The prolonged use of trimethoprim-sulfamethoxazole and the use of the contaminated blend, used as a diet supplement and as a vehicle for the administration of oral medications, may have contributed to the creation of the drug-resistant strains. Once bananas were no longer used as a diet supplement and all inanimate objects in contact with the monkeys disinfected, mortality decreased in the colony, the animals on treatment recovered, and no new cases observed, reducing K. pneumoniae infections to undetectable levels. Other changes instituted included sanitation of husbandry utensils with diluted bleach after each use and replacement of cage wood perches with PVC pipes to make sure they were effectively sanitized during cage wash. No K. pneumoniae outbreaks have occurred since these changes were instituted more than ten years ago. A Klebsiella bacterin was used shortly after the outbreak but vaccination was discontinued a year later without further cases noted.
Klebsiella pneumoniae is an important nonhuman primate pathogen, causing sporadic outbreaks with high mortality in captive populations, particularly New World monkeys [10, 17, 19]. Recently, it was found that even subclinical infections with some strains of K. pneumoniae can cause profound alterations in cytokine profiles in rhesus monkeys, making these animals unsuitable for immune function or infectious disease research [6]. Similar to our study, a recent report described an epidemiological investigation where different K. pneumoniae strains were present during a subclinical outbreak in a rhesus macaque colony [7, 12]. Clusters of these isolates are believed to be associated to specific animal vendors. This same diversity in K. pneumoniae isolates has been described in other animals [4, 11, 16, 18] and in humans in hospital settings were nosocomial isolates have been recently reported as having high genetic diversity [25].
K. pneumoniae outbreaks in nonhuman primate colonies have been associated with dietary supplements before [10]. A recent study demonstrated that bananas in particular have widely diverse enterobacteriacea associated microbial communities including, in addition to K. pneumoniae, E. coli, Salmonella spp., and Yersinia spp. [20]. Interestingly, these bacteria, including K. pneumoniae, appear to have a role protecting bananas from fungal infections similar to the role endogenous bacteria play in vertebrates [20]. Other studies have found vegetables and fruits carrying antibiotic resistant K. pneumoniae biotypes [3]. The blend used as a diet supplement and as a vehicle for the administration of oral medications in the current report was found to be contaminated with K. pneumoniae. This blend was stored up to seven days in the refrigerator increasing the chances of growth, spread, and possibly contributing to the creation of drug resistant strains. It was agreed that the blend must be used the same day it is prepared, and any leftover blend would be discarded on a daily basis.
DNA fingerprinting by automated repetitive extragenic palindromic-polymerase chain reaction has been reported as a highly sensitive alternative method for bacterial strain identification showing higher sensitivity than antibody-based methods and in a significantly shorter turnaround time compared to conventional methods [1]. In our case, rep-PCR allowed us to realize that multiple K. pneumoniae strains were circulating during the outbreak and take effective control measures. This potential genetic diversity should be considered when designing strategies for controlling and preventing K. pneumoniae outbreaks in captive nonhuman primate colonies. Preventive measures include, avoid potential cross contamination with K. pneumoniae when manipulating dietary supplements, particularly bananas and other agricultural foodstuff. Dietary supplements in the form of fruits and vegetables should be routinely disinfected with a diluted bleach solution and periodically evaluated by bacteriological cultures to verify they are free of K. pneumoniae and other potential pathogens. Periodically perform bacteriologic monitoring of husbandry equipment and enrichment devices in direct or indirect contact with colony nonhuman primates to verify sanitation practices are eliminating potential contamination with K. pneumoniae. Screen newly arrived nonhuman primates during quarantine by means of fecal or rectal swab cultures for potential asymptomatic carriers of K. pneumoniae and other potential pathogens. In case of a K. pneumoniae outbreak, fingerprint the isolates to find the extent of the biosecurity breach and potential source.
Acknowledgments
This study was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases, Comparative Medicine Branch; Laboratory of Malaria Immunology and Vaccinology; Department of Laboratory Medicine, Clinical Center, National Institutes of Health, and the Office of Research Services.
Footnotes
Abstract presented at the 64th AALAS National Meeting, Baltimore, MD, October 27–31, 2013.
References
- 1.Anderson PN, Hume ME, Byrd JA, Hernandez C, Stevens SM, Stringfellow K, Caldwell DJ. Evaluation of repetitive extragenic palindromic-polymerase chain reaction and denatured gradient gel electrophoresis in identifying Salmonella serotypes isolated from processed turkeys. Poultry Science. 2010;89:1293–1300. doi: 10.3382/ps.2009-00390. [DOI] [PubMed] [Google Scholar]
- 2.Berendt RF, Knutsen GL, Powanda MC. Nonhuman primate model for the study of respiratory Klebsiella pneumoniae infection. Infect Immun. 1978;22(1):275–281. doi: 10.1128/iai.22.1.275-281.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehme S, Werner G, Klare I, Reissbrodt R, Witte W. Occurrence of antibiotic-resistant enterobacteria in agricultural foodstuffs. Mol Nutr Food Res. 2004;48(7):522–531. doi: 10.1002/mnfr.200400030. [DOI] [PubMed] [Google Scholar]
- 4.Brisse S, Duijkeren Ev. Identification and antimicrobial susceptibility of 100 Klebsiella animal clinical isolates. Vet Microbiol. 2005;105(3–4):307–312. doi: 10.1016/j.vetmic.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Brown C, Seidler RJ. Potential Pathogens in the Environment: Klebsiella pneumoniae, a taxonomic and ecological enigma. Appl Microbiol. 1973;25(6):900–904. doi: 10.1128/am.25.6.900-904.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burke RL, West MW, Erwin-Cohen R, Selby EB, Fisher DE, Twenhafel NA. Alterations in cytokines and effects of dexamethasone immunosuppression during subclinical infections of invasive Klebsiella pneumoniae with hypermucoviscosity phenotype in rhesus (Macaca mulatta) and cynomolgus (Macaca fascicularis) macaques. Comp Med. 2010;60(1):62–70. [PMC free article] [PubMed] [Google Scholar]
- 7.Burke RL, Whitehouse CA, Taylor JK, Selby EB. Epidemiology of invasive Klebsiella pneumoniae with hypermucoviscosity phenotype in a research colony of nonhuman primates. Comp Med. 2009;59(6):589–597. [PMC free article] [PubMed] [Google Scholar]
- 8.Duncan DW, Razzell WE. Klebsiella biotypes among coliforms isolated from forest environments and farm produce. Appl Microbiol. 1972;24(6):933–938. doi: 10.1128/am.24.6.933-938.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Good RC, May BD. Respiratory pathogens in monkeys. Infect Immun. 1971;3(1):87–93. doi: 10.1128/iai.3.1.87-93.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gozalo A, Montoya E. Klebsiella pneumoniae infection in a New World nonhuman primate Center. Lab Primate Newsletter. 1991;30(2):13–15. [Google Scholar]
- 11.Haenni M, Ponsin C, Métayer V, Médaille C, Madec JY. Veterinary hospital-acquired infections in pets with a ciprofloxacin-resistant CTX-M-15-producing Klebsiella pneumoniae ST15 clone. J Antimicrob Chemother. 2012;67(3):770–771. doi: 10.1093/jac/dkr527. [DOI] [PubMed] [Google Scholar]
- 12.Hartman LJ, Selby EB, Whitehouse CA, Coyne SR, Jaissle JG, Twenhafel NA, Burke RL, Kulesh DA. Rapid real-time PCR assays for detection of Klebsiella pneumoniae with the rmpA or magA genes associated with the hypermucoviscosity phenotype: screening of nonhuman primates. J Mol Diagn. 2009;11(5):464–471. doi: 10.2353/jmoldx.2009.080136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hazen TH, Zhao L, Sahl JW, Robinson G, Harris AD, Rasko DA, Johnson JK. Characterization of Klebsiella sp. 10982, a colonizer of humans that contains novel antibiotic resistance alleles and exhibits genetic similarities to plant and clinical Klebsiella isolates. Antimicrob Agents Chemother. 2014;58(4):1879–1888. doi: 10.1128/AAC.01605-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Wang JY, Jiang W. An Increasing Prominent Disease of Klebsiella pneumoniae Liver Abscess: Etiology, Diagnosis, and Treatment. Gastroenterol Res Pract. 2013;2013:258514. doi: 10.1155/2013/258514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Research Council. Guide for the Care and Use of Laboratory Animals. 8th. Washington D.C.: The National Academies Press; 2011. [PubMed] [Google Scholar]
- 16.Nawaz M, Khan SA, Tran Q, Sung K, Khan AA, Adamu I, Steele RS. Isolation and characterization of multidrug-resistant Klebsiella spp. isolated from shrimp imported from Thailand. Int J Food Microbiol. 2012;155(3):179–184. doi: 10.1016/j.ijfoodmicro.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Obaldia N., 3rd Detection of Klebsiella pneumoniae antibodies in Aotus I. lemurinus (Panamanian owl monkey) using an enzyme linked immunosorbent assay (ELISA) test. Lab Anim. 1991;25(2):133–141. doi: 10.1258/002367791781082603. [DOI] [PubMed] [Google Scholar]
- 18.Paulin-Curlee GG, Singer RS, Sreevatsan S, Isaacson R, Reneau J, Foster D, Bey R. Genetic diversity of mastitis-associated Klebsiella pneumoniae in dairy cows. J Dairy Sci. 2007;90(8):3681–3689. doi: 10.3168/jds.2006-776. [DOI] [PubMed] [Google Scholar]
- 19.Pisharath HR, Cooper TK, Brice AK, Cianciolo RE, Pistorio AL, Wachtman LM, Mankowski JL, Newcomer CE. Septicemia and peritonitis in a colony of common marmosets (Callithrix jacchus) secondary to Klebsiella pneumoniae infection. Contemp Top Lab Anim Sci. 2005;44(1):35–37. [PubMed] [Google Scholar]
- 20.Rossmann B, Müller H, Smalla K, Mpiira S, Tumuhairwe JB, Staver C, Berg G. Banana-associated microbial communities in Uganda are highly diverse but dominated by Enterobacteriaceae. Appl Environ Microbiol. 2012;78(14):4933–4941. doi: 10.1128/AEM.00772-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shahid M, Malik A, Adil M, Jahan N, Malik R. Comparison of beta-lactamase genes in clinical and food bacterial isolates in India. J Infect Dev Ctries. 2009;3(8):593–598. doi: 10.3855/jidc.550. [DOI] [PubMed] [Google Scholar]
- 22.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, NISC Comparative Sequencing Program Group. Henderson DK, Palmore TN, Segre JA. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 2012;4(148):148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soto E, LaMon V, Griffin M, Keirstead N, Beierschmitt A, Palmour R. Phenotypic and genotypic characterization of Klebsiella pneumoniae isolates recovered from nonhuman primates. J Wildl Dis. 2012;48(3):603–611. doi: 10.7589/0090-3558-48.3.603. [DOI] [PubMed] [Google Scholar]
- 24.Talbot HW, Jr, Yamamoto DK, Smith MW, Seidler RJ. Antibiotic resistance and its transfer among clinical and nonclinical Klebsiella strains in botanical environments. Appl Environ Microbiol. 1980;39(1):97–104. doi: 10.1128/aem.39.1.97-104.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Li B, Tsang AK, Yi Y, Woo PC, Liu CH. Genotypic analysis of Klebsiella pneumoniae isolates in a Beijing Hospital reveals high genetic diversity and clonal population structure of drug-resistant isolates. PLoS One. 2013;8(2):e57091. doi: 10.1371/journal.pone.0057091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitehouse CA, Keirstead N, Taylor J, Reinhardt JL, Beierschmitt A. Prevalence of hypermucoid Klebsiella pneumoniae among wild-caught and captive vervet monkeys (Chlorocebus aethiops sabaeus) on the island of St. Kitts. J Wildl Dis. 2010;46(3):971–976. doi: 10.7589/0090-3558-46.3.971. [DOI] [PubMed] [Google Scholar]
- 27.Yoon da H, Jeon YJ, Bae EY, Jeong DC, Kang JH. Liver abscess due to Klebsiella pneumoniae in a healthy 12-year-old boy. Korean J Pediatr. 2013;56(11):496–499. doi: 10.3345/kjp.2013.56.11.496. [DOI] [PMC free article] [PubMed] [Google Scholar]