Summary
The commensal oral microbial flora has evolved with the human host to support colonization of the various intraoral sites without triggering a significant immune response. In exchange, the commensal microbes provide critical protection against invading pathogens. The intrinsic ability of the oral flora to create a symbiotic microbial community with the host can be disturbed, selecting for the overgrowth of a dysbiotic community that can result in dental diseases, such as caries and periodontitis. While the mechanisms of molecular pathogenesis in oral diseases are well characterized, much less is known about the molecular mechanisms used by the commensal flora to maintain oral health. Here we focus on the commensal species Streptococcus sanguinis, which is found in abundance in the early oral biofilm and is strongly correlated with oral health. S. sanguinis exhibits a variety of features that make it ideally suited as a model organism to explore the molecular basis for commensalism. As such, this review will describe our current mechanistic understanding of S. sanguinis commensalism and speculate upon its molecular traits that may be exploitable to maintain or restore oral health under conditions that would otherwise lead to disease.
Introduction
Our view of the etiology of the main oral diseases, caries and periodontitis, has been refined in recent years, largely due to rapid improvements in high throughput sequencing technologies. Numerous recent studies have provided a detailed picture of the connection between oral disease status and microbial dysbiosis among the oral flora (Hajishengallis and Lamont, 2012; Simon-Soro and Mira, 2015). The severity of disease is heavily influenced by the synergistic interactions of the individual members of the polymicrobial consortium, including metabolic cross-feeding and interspecies signaling. Thus, the etiology of caries and periodontitis (and other mucosal polymicrobial diseases) is largely a consequence of microbial ecology and poorly defined by Koch's Postulates (Hajishengallis and Lamont, 2016; Magalhaes et al., 2016; Stacy et al., 2016).
The polymicrobial nature of oral diseases challenges current treatment approaches and a refocus might be required, since the overall incidence of caries and periodontal disease in the population has not significantly improved in multiple decades. In fact, they continue to be among the most common human diseases worldwide (Kassebaum et al., 2015). While there is a wealth of detailed knowledge regarding the pathogenic mechanisms triggered by dysbiosis, much less is known about the mechanisms used by the commensal flora to prevent pathology triggered by the abundance of microbes found at most mucosal surfaces. This is one area of research that is going to require a much greater emphasis in the future if we expect to effectively prevent and/or cure diseases caused by dysbiosis. Recent advances in the various -omics technologies have provided unparalleled insights into the actual outcomes of polymicrobial interactions. The remaining challenge is to decipher the regulatory and mechanistic events leading to these interactions. Such studies still require detailed knowledge of key processes from individual organisms in order to test specific hypotheses derived from big data studies (McLean, 2014). S. sanguinis exhibits a variety of features that make it ideally suited as a model organism to explore the molecular basis for commensalism among the flora. As such, this review will describe our current mechanistic understanding of the commensal aspects of S. sanguinis biology and discuss potential avenues to exploit commensalism therapeutically.
Historical excursion
S. sanguinis was originally named due to its role in infective endocarditis. In a note to the Journal of Bacteriology, Niven and White described a new species isolated from about 100 cases of subacute bacterial endocarditis (Niven and White, 1946). About one third of those isolates failed characterization and were simply referred to as Streptococcus s.b.e. (for subacute bacterial endocarditis) (Niven and White, 1946). Overall the group was quite homogenous in its physiological and biochemical characteristics (White and Niven, 1946). Initial attempts to isolate Streptococcus s.b.e had largely failed at the time, as samples were mainly focused on throat isolations. However, a confirmed isolate was later obtained from an extracted tooth, which we now know coincides with its preferred colonization site. Since the only source of Streptococcus s.b.e. in the original publication came from the blood of endocarditis patients, it was named S. sanguis from the latin word for blood (Niven et al., 1946; White and Niven, 1946). This was later amended to S. sanguinis for grammatical reasons (Truper and De' Clari, 1997). Further serological characterization confirmed the isolation of a new species, since no cross reactivity was observed between rabbit serum from Streptococcus s.b.e. and other identified streptococci belonging to the various Lancefield groups (Washburn et al., 1946).
During the 1960s and 70s several publications coauthored by Jan Carlsson first identified the primary habitat of S. sanguinis. Culture-dependent detection of S. sanguinis in infants was strongly correlated to tooth eruption providing evidence for the tooth surface as the preferred colonization site (Carlsson et al., 1970). Carlsson also performed some of the earliest mixed culture studies of S. sanguinis and Streptococcus mutans to examine the ecological aspects of their interactions (Carlsson, 1971). In addition, he provided the first characterization of S. sanguinis pyruvate oxidase (SpxB, also PoxB), which is the main enzyme responsible for producing toxic quantities of hydrogen peroxide (H2O2) used to inhibit other species like S. mutans (Carlsson and Edlund, 1987; Carlsson et al., 1987; Kreth et al., 2008). The importance of H2O2 production for the promotion of homeostasis in the oral biofilm was first described in 1973 (Holmberg and Hallander, 1973) and is still the subject of ongoing research (see below).
Phenotypic characteristics
S. sanguinis is a Gram-positive, non-motile (although this has been challenged (Gurung et al., 2016); see below), non-spore forming, catalase-negative coccus. S. sanguinis is non-beta-hemolytic, but able to produce a green coloration on blood agar plates referred to as alpha-hemolysis (Facklam, 2002). S. sanguinis has been placed into the Streptococcus mitis group based upon 16S rRNA sequence analysis (Facklam, 2002). However, in a recent study based on the concatenated sequences of 50 ribosomal protein genes from 88 Streptococcus species, S. sanguinis is placed in it's own group together with S. gordonii (Teng et al., 2014). Thus far, S. sanguinis has only been isolated from humans.
Clinical evidence of S. sanguinis association with oral health
Several studies have demonstrated the importance of S. sanguinis during early colonization and its widespread distribution (Carlsson et al., 1970; Caufield et al., 2000; Aas et al., 2005). The cycle of early colonization by S. sanguinis likely occurs in every human after tooth eruption or extensive cleaning. Its contribution to oral biofilm homeostasis is inferred from its abundance at sites of oral health and its obvious decreased abundance at sites of active caries or periodontal disease (Becker et al., 2002; Colombo et al., 2009). Thus, S. sanguinis is not only one of the most ubiquitous and prolific inhabitants of the tooth surface, but it has evolved mechanisms to prevent obvious damage to the host as a consequence of its growth (i.e. a true commensal organism). Surprisingly little is known about the specific molecular mechanisms mediating this ability, with the exception of the few studies discussed later. Our limited knowledge in this area can be largely attributed to a historical emphasis upon mechanistic studies of pathogenesis, rather than commensalism.
In the seminal publication by Aas et al., the authors were able to define the normal bacterial flora of the adult oral cavity using culture independent 16S rRNA sequencing of bacterial samples from 9 different intra-oral sites (Aas et al., 2005). In this study, the teeth of all five subjects that were free of oral health-related problems were colonized by S. sanguinis. This was true for only one other identified species, S. mitis, which in contrast was found in all tested intraoral locations (Aas et al., 2005). In more recent studies, S. sanguinis was found to be among the shared taxa strongly associated with oral health (Corby et al., 2005; Bik et al., 2010; Belda-Ferre et al., 2012), which is consistent with the inverse correlation observed between S. sanguinis and S. mutans in adults (de Stoppelaar et al. 1970)(Giacaman et al., 2015). The impact of S. sanguinis relative abundance in the oral biofilm is also reflected in the overall functional expression profile of the dental plaque microbiome. Transcripts derived from just 9 species represented 71% of the total dental plaque transcriptome, with S. sanguinis providing the majority of detected transcripts (16%) (Peterson et al., 2014). In addition to adults, studies of children have also have reported a similar positive association between S. sanguinis abundance and oral health. In a defined study that compared 30 children with severe early childhood caries to healthy controls, S. sanguinis was only detected in healthy subjects or on teeth with sound enamel. In contrast, S. sanguinis was absent on white spot lesions, cavitated teeth, or on dentin (Becker et al., 2002). Later studies reported that S. sanguinis can still be detected in children with severe early childhood caries (Ge et al., 2008; Kanasi et al., 2010), but the relative levels of S. sanguinis and S. mutans in the oral cavity vary with oral health status (Ge et al., 2008). Similarly, the early colonization of S. sanguinis in infants has been shown to correlate with a significant delay in the colonization of S. mutans (Caufield et al., 2000). Overall, the vast majority of studies report an association of S. sanguinis with oral health. However, an over-representation of S. sanguinis in a caries active subject pool was recently reported, albeit with a very small cohort of 4 caries active subjects (Peterson et al., 2013). It is also worth noting that S. sanguinis seems to be among the species that are more prevalent in subjects with periodontal health as well (Stingu et al., 2008; Colombo et al., 2009; Mason et al., 2015). However, few specific details have been reported to explain why this is the case.
Two important questions remain: i) If S. sanguinis is associated with oral health and a delayed colonization of S. mutans, what causes its shift in abundance during disease development? Conventional wisdom posits this is simply a consequence of reduced viability due to acidification in dental plaque. However, it is also conceivable that initial impairments in S. sanguinis competitive abilities could precede and ultimately facilitate the sequence of events required for extensive plaque acidification. ii) Does S. sanguinis provide any protective function for periodontal health and perhaps more importantly, can this be exploited to prevent and/or mitigate periodontal disease (as well as caries)?
S. sanguinis presence in extra-oral diseases
Recent studies have suggested a potential mechanism for S. sanguinis to improve clinical outcomes in patients afflicted with cystic fibrosis, which is a systemic disease causing chronic respiratory infections due to defects in exocrine gland function (Elborn, 2016). S. sanguinis was demonstrated to inhibit the principal pathogen in cystic fibrosis lung infections, P. aeruginosa, due to its production of H2O2 (Whiley et al., 2015). The authors speculated that an early colonization of S. sanguinis and other H2O2-producing streptococcal species in the oral cavity and lung might influence the bacterial community structure of cystic fibrosis patients, potentially delaying disease onset and/or progression (Whiley et al., 2015). However, epidemiological evidence for such an effect is currently lacking. The authors also noted that synergistic effects were possible between other viridans group streptococci and P. aeruginosa, which actually led to enhanced pathogenicity (Whiley et al., 2015). While much is known about P. aeruginosa pathogenic mechanisms in cystic fibrosis, this is not the case for studies of commensal streptococci in this disease. Such studies may yield entirely new strategies to exploit bacterial antagonism as a therapeutic approach for cystic fibrosis. Although not the subject of this review, S. sanguinis is also known to be the etiological agent of several extra-oral diseases. The most prominent is its association with infective endocarditis, which is a relatively rare, but potentially fatal disease affecting the heart valves or endocardium of patients with predisposing heart defects. For further details, the interested reader is directed to several excellent reviews on this topic (Cahill and Prendergast, 2015, 2016). In even rarer instances, S. sanguinis has also been reported to cause both meningitis and severe bacteremia, sometimes as a result of surgical procedures or cancer (Kampe et al., 1995; Macaluso et al., 1998; Moon et al., 2010; Liu et al., 2013; Bijlsma et al., 2016).
Molecular mechanisms of S. sanguinis commensalism in the oral cavity
Initial attachment and biofilm development
As a pioneer colonizer of the tooth surface, S. sanguinis facilitates the subsequent colonization of other species in the oral biofilm (Kolenbrander et al., 2006). This is not a unique ability of S. sanguinis per se, as there are a variety of other pioneer colonizers involved as well. However, the overall abundance of S. sanguinis, especially in early biofilms, suggests a dominant role in this process. Its success as a pioneer colonizer is also reflected in its relatively large abundance of salivary pellicle adhesins compared to most other oral bacterial species (Kolenbrander et al., 2006; Peterson et al., 2014). The abundance of S. sanguinis in newly formed oral biofilms also suggests that it likely plays a central role in shaping biofilm ecology as these communities develop, although detailed studies in this area are still lacking. For additional information regarding oral streptococcal coadhesion and community assembly, the interested reader is referred to several excellent recent reviews (Nobbs et al., 2009; Nobbs et al., 2011; Wright et al., 2013; Jakubovics et al., 2014; Nobbs et al., 2015).
Oral biofilm formation begins with the attachment of S. sanguinis and other pioneer colonizers to macromolecular complexes formed on saliva-coated tooth surfaces (Diaz et al., 2006; Kreth and Herzberg, 2015). Negatively charged residues and electrostatic interactions with hydrophilic regions in salivary proteins facilitate their attachment to the tooth surface (Lamkin and Oppenheim, 1993; Lindh, 2002) forming what is referred to as the acquired enamel pellicle (AEP). Although S. sanguinis is able to directly adhere to saliva-free hydroxyapatite (Tanaka et al., 1996), the main mineral found in tooth enamel, the initial attachment process is most-likely driven by an interaction of the streptococcal surface with salivary components. Binding to salivary proteins is mediated via protein-protein or protein-carbohydrate interactions with receptors exposed on the bacterial surface. Amylase is the most abundant salivary protein and is present both in the AEP and in dental plaque (Orstavik and Kraus, 1973; Aguirre et al., 1987). S. sanguinis specifically binds to amylase via long filamentous pili (Okahashi et al., 2011). The genes encoding these pili are organized in an operon encoding three putative structural pilin subunits as well as a sortase involved in the surface anchoring of the pili proteins. Disruption of the pilus locus results in decreased single species biofilm formation on saliva coated glass slides, although it does not abolish amylase binding completely (Okahashi et al., 2011). Thus, additional surface proteins are likely to be involved in amylase binding. Besides anchoring to the AEP, another major advantage of adherence to salivary amylase is that the enzyme retains about 50% of its enzymatic function (Scannapieco et al., 1990). Since amylase can efficiently hydrolyze the alpha-1,4-glucosidic linkages in starch to glucose, maltose, and maltodextrins (Ramasubbu et al., 1996), they can provide a readily accessible source of easily metabolizable sugars that can be imported via high affinity carbohydrate transporters (Vadeboncoeur and Pelletier, 1997). This is presumably an important mechanism used to help newly attached cells of S. sanguinis rapidly spread over the tooth surface and develop into biofilm communities (Marsh et al., 1985).
SsaB is another surface exposed protein shown to mediate binding to saliva-coated hydroxyapatite, although the mechanism is unknown (Ganeshkumar et al., 1988). However, SsaB has been demonstrated to serve as the substrate-binding protein for an ATP-binding cassette (ABC) transporter for manganese (Mn2+) (Crump et al., 2014). Therefore, it is unlikely to be a classical adhesin. Since salivary proteins are largely negative charged (Gibbins et al., 2014) and might interact with the divalent cation Mn2+, it is possible that SsaB not only transports Mn2+, but also functions as an adhesin when Mn2+ is bound to salivary proteins.
Another major component of saliva and AEP are the mucins, the gel-forming components of mucus. Mucins are a diverse group of >20 glycoproteins that primarily serve as a hydrating and lubricating layer for mucosal epithelial cells (Frenkel and Ribbeck, 2015) and S. sanguinis is known to adhere specifically to salivary MUC7 via the surface receptor SrpA (Plummer and Douglas, 2006). In addition, S. sanguinis has an unusually high number of uncharacterized lipoproteins (LP) and surface exposed cell-wall anchored proteins (CWA). The genome of the common lab strain SK36 contains 60 LPs and 30 CWAs (Xu et al., 2007), any of which could conceivably promote attachment to the AEP and/or facilitate coadherence to other species. The number of such proteins found in S. sanguinis is considerable higher than in S. mutans and S. pneumoniae (Xu et al., 2007) further highlighting its role as a central player in early oral biofilm development.
A feature that is rather unusual for streptococci has been recently investigated in greater detail and might also aid in S. sanguinis biofilm development. First described in the mid-1970s, S. sanguinis is capable of surface-associated twitching motility (Henriksen and Henrichsen, 1975). This ability is mediated by a type IV secretion system comprised of surface associated pili (Gurung et al., 2016). Motility is achieved by retracting pilus-like structures, thus pulling the cell in one direction (Jarrell and McBride, 2008). These pili are organized in an operon located within a 22 kb pil locus separate from the aforementioned amylase binding pili. Mutagenesis of the pilT component of the complex indicates that its ATPase activity is likely the principal mediator of pilus retraction during twitching motility (Gurung et al., 2016). It is perhaps even more striking that this pilus locus is thus far absent from all other streptococcal genomes. Currently, it is unknown whether twitching motility has any relevance in vivo or if it provides any colonization advantage, but it is tempting to speculate that this conserved function enhances S. sanguinis development of biofilms on the tooth surface. Since S. sanguinis lacks an obvious chemotaxis system, its twitching motility seems unlikely to be utilized for carbon source acquisition. One important function unrelated to motility could be as a surface adhesin via pili tethering. The forces generated through pilus retraction are surprisingly strong (Gurung et al., 2016) and could potentially help to anchor the bacteria when encountering excessive shear forces.
Biofilm maturation
The biofilm developmental program includes the formation of extracellular polymeric substances (EPS), generating a matrix component that has several functions for the biofilm. The biofilm EPS or matrix provides a diffusion barrier that limits the entry of antimicrobial components either through size exclusion or binding and immobilization, effectively reducing the local concentration of antimicrobials (Stewart, 2003; Davenport et al., 2014). The biofilm matrix contains carbohydrates, proteins, lipids and extracellular DNA (eDNA) and also provides a cohesive mesh-like structure supporting biofilm integrity (Flemming et al., 2007; Flemming and Wingender, 2010). Among the best-investigated matrix components of the oral biofilm are the glucans. Treatment of those polymeric carbohydrates with glucan degrading enzymes results in significantly less biofilm biomass (Klein et al., 2015). S. sanguinis reference strain SK36 encodes two glucan forming glucosyltranferases (GTF), GtfB and GtfP respectively (Nobbs et al., 2009). However, this might not be entirely representative, since GTF activity was characterized in 10 strains with considerable variability in GTF activities (Herzberg et al., 1990) as well as inefficient glucan production and biofilm formation (Hamada et al., 1981; Kopec et al., 2001). Other studies have indicated that S. sanguinis GTF activity is influenced by a variety of environmental factors, such as sodium and potassium concentration or external pH (Keevil et al., 1984; Vacca Smith et al., 2000). Glucans synthesized by S. sanguinis GTFs have been shown to promote the adherence of diverse oral bacteria to saliva-coated hydroxyapatite and to increase biofilm formation (Yoshida et al., 2014). However, there is still scant evidence among the literature to indicate the overall contribution of glucans and GTFs to S. sanguinis biofilm formation. It has been suggested that S. sanguinis might also adhere to the extracellular glucan produced by other streptococci via putative glucan-binding proteins, such as GbpB, SspC, and SspD (Moraes et al., 2014). However, this has yet to be confirmed experimentally.
Extracellular DNA (eDNA) is another important component of the biofilm matrix for S. sanguinis as well as many other bacteria (Okshevsky and Meyer, 2015). For example, DNase treatment effectively disrupts S. sanguinis biofilms (Moraes et al., 2014), while mutant strains impaired in eDNA production also exhibit defects in biofilm formation (Zheng et al., 2011b; Ge et al., 2016). S. sanguinis produces high molecular weight chromosomally derived eDNA during aerobic growth in response to the H2O2 produced by the pyruvate oxidase enzyme SpxB (Kreth et al., 2008; Kreth et al., 2009). Presumably, it is the reduced eDNA production of the spxB mutant that is responsible for its deficient biofilms, which appear more sparsely populated than the wild type (Zheng et al., 2011b). A similar result could also be observed if H2O2 production is hindered by growth in environments with low oxygen tensions (Zheng et al., 2011a; Zheng et al., 2011c). The connection between oxygen availability and eDNA production correlates well with S. sanguinis role as a pioneer colonizer and early biofilm producer. Newly colonized sites on the tooth surface would be expected to be highly aerobic and conducive to the production of eDNA. However, once a biofilm has been established, the oxygen tension within the biofilm declines sharply (de Beer et al., 1994; Xu et al., 1998), which should result in a concomitant decrease in H2O2 and eDNA production. At this point, it is currently unclear whether eDNA release continues to occur through an H2O2-independent mechanism or if further eDNA production is simply not required once a biofilm is established. It should also be noted that oxygen is a readily available substrate to stimulate early supragingival biofilm formation, whereas the substrates for GTF activity (sucrose) are only available intermittently at best. In addition, since eDNA promotes S. sanguinis cell-cell aggregation (Kreth et al., 2009), this could be one of its key functions to stimulate biofilm formation, especially at newly colonized sites.
The social life of S. sanguinis
Both cell-cell and metabolic interactions between S. sanguinis and other members of the oral biofilm community are major determinants of oral ecology and ultimately oral health. Surface adhesin and/or lectin-mediated interactions with other species provide genetically encoded mechanisms to directly select the composition of organisms in developing mixed species biofilms. A variety of S. sanguinis coaggregation partners have been identified including species of Actinomyces, Prevotella and Porphyromonas (originally grouped in the genus Bacteroides), Capnocytophaga, Fusobacterium, and Candida (Jenkinson et al., 1990). Porphyromonas gingivalis was shown to adhere to S. sanguinis (Stinson et al., 1991; Lamont et al., 1992) via a surface exposed glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Maeda et al., 2004). Given the pathogenic potential of P. gingivalis, S. sanguinis might have measures in place to limit the overgrowth of P. gingivalis and other potentially damaging coaggregation partners via its aforementioned H2O2 production ability and perhaps other uncharacterized mechanisms as well. S. sanguinis was shown to antagonize a variety of periodontal pathogens (Herrero et al., 2016) in addition to its role in inhibiting the principal cariogenic species S. mutans (Kreth et al., 2008). It is worth mentioning that certain species antagonized by S. sanguinis also have mechanisms to inhibit S. sanguinis or weaken the effect of H2O2. For example, S. mutans is able to produce antimicrobial peptides called mutacins, which target closely related streptococci, including S. sanguinis (Merritt and Qi, 2012). Curiously, S. sanguinis seems to produce an extracellular (challisin-like) protease that can interfere with mutacin production by degrading the competence stimulating peptide CSP regulating mutacin gene expression (Wang and Kuramitsu, 2005). Furthermore, several oral species produce catalase, which might decrease the efficiency of H2O2 antimicrobial activity (Jakubovics et al., 2008). As shown for Aggregatibacter actinomycetemcomitans, H2O2 can also serve as a signaling molecule to regulate gene expression (Ramsey and Whiteley, 2009; Stacy et al., 2014). Thus, S. sanguinis may be able to utilize H2O2 production for interspecies signaling as well. It is also worth noting that saliva contains several H2O2 detoxifying enzymes (Ihalin et al., 2006; Ashby, 2008) that likely degrade the free H2O2 in the oral cavity. Therefore, the utility of H2O2 for interspecies interactions in the biofilm is presumably confined to microenvironments proximal to producers like S. sanguinis (Zhu and Kreth, 2012; Jakubovics et al., 2014). It is also worth noting that in vitro studies suggest H2O2 produced from streptococci can trigger cytotoxicity in endothelial cells (Okahashi et al., 2014). Thus, saliva exposure could be important for protecting against collateral damage from streptococcal H2O2 production during early biofilm development. Given the variety of functions of H2O2 for biofilm development and ecology, there may be potential therapeutic opportunities to exploit these pathways in commensal species for the prevention of dysbiosis or even for reshaping the species composition of a dysbiotic microbial community (Zhu and Kreth, 2012; Jakubovics et al., 2014).
pH modulating abilities of S. sanguinis in the oral biofilm
Plaque acidification not only triggers a net demineralization of tooth enamel, it also selects for the overgrowth of aciduric species like the lactobacilli and the mutans streptococci. As a countermeasure to prevent this, S. sanguinis and other commensals have a variety of mechanisms to raise the local plaque pH. S. sanguinis has two principal approaches to do this. i) The aforementioned pyruvate oxidase enzyme SpxB generates both CO2 and acetyl phosphate as end products in addition to H2O2 (Zhu and Kreth, 2012). ii) S. sanguinis catabolizes arginine via the arginine deiminase system (ADS) generating ornithine, ammonia, CO2, and ATP (Burne and Marquis, 2000). Since ammonia is a base, it can directly increase the local pH and S. sanguinis was recently demonstrated to be a substantial contributor to the alkali generation capacity of dental plaque (Huang et al., 2015). Huang et al. speculated that deficiencies in alkali production by the ADS might be equally important for caries development as the production of acid itself (Huang et al., 2015). Similarly, a variety of recent clinical studies have demonstrated net increases in plaque pH and lower caries scores when given arginine-supplemented dentifrices (Kraivaphan et al., 2013; Nascimento et al., 2014). In future studies, it will be interesting to determine if/how arginine supplementation influences plaque species composition in these subjects.
Remaining under the radar – S. sanguinis interactions with the immune system
Commensal bacteria play crucial roles in the proper development of the immune system. For example, the development of intestinal immunity as well as the anatomical development of the gut-associated lymphoid tissue (GALT) is closely connected to the presence of the intestinal commensal microbiota (reviewed in (Palm et al., 2015). While those processes are well characterized for intestinal immunity, far less is known about analogous processes in the oral cavity (Lang et al., 2010). Current data suggests that commensal bacteria also influence immune system development during early colonization of the oral mucosa, while certain immune mechanisms are in place prior to colonization to provide initial protection (Lang et al., 2010). The gingival tissue must properly balance its response to bacterial colonization to benefit from the presence of commensal species, while still having the ability to mount a robust immune response when the ecology of the flora is disturbed. A comparative study of human immune cell responses to S. sanguinis and several other viridans streptococci illustrated the weak ability of S. sanguinis to activate CD45+, CD4+, and CD8+ cells, resulting in low levels of cytokine production and a poor anti–S. sanguinis humoral response (Salam et al., 2006). Likewise, incubating S. sanguinis with human oral keratinocytes results in no significant induction of antimicrobial peptide production, whereas both human beta defensin-3 and LL-37 are significantly induced by the closely related organism S. gordonii (Ji et al., 2007). While S. gordonii is also a pioneer colonizer of the human oral cavity, it is noteworthy that its abundance is typically maintained at much lower levels compared to S. sanguinis (Nyvad and Kilian, 1990; Tappuni and Challacombe, 1993; Mager et al., 2003; Li et al., 2004). Another study of gingival keratinocytes similarly observed no significant induction of matrix metalloproteinase 9 as well as a variety of beta defensins and cytokines in response to incubation with S. sanguinis cell wall extracts. Furthermore, the authors also observed that coincubation of F. nucleatum and S. sanguinis was able to reduce the inflammatory potential of F. nucleatum via a TLR2-dependent mechanism (Peyret-Lacombe et al., 2009). Similarly, when the commensal species S. sanguinis, S. mitis, or S. salivarius were coincubated with the periodontopathogen A. actinomycetemcomitans, HOK-18A oral keratinocytes produced significantly less IL-8 in response to A. actinomycetemcomitans (Sliepen et al., 2009). In this study, a diffusible factor was most likely responsible for the reduction in IL-8 production, as cell-free supernatants of these commensals reproduced this same anti-inflammatory effect (Sliepen et al., 2009). Interestingly, S. gordonii was not found to exhibit this same ability (Sliepen et al., 2009). In addition to oral keratinocytes, S. sanguinis also fails to elicit IL-8 and tumor necrosis factor- α production from human whole blood cells (Tietze et al., 2006), while S. sanguinis peptidoglycan was shown to reduce cytokine production in THP-1 monocytes stimulated with the purified lipopolysaccharide (LPS) of several periodontopathogens (Lee, 2015). The latter result is of particular interest because Gram positive bacteria constantly release large amounts of peptidoglycan fragments called muropeptides as a consequence of cell wall remodeling during normal growth and cell division (Dworkin, 2014). The next logical step would be to determine whether diffusible muropeptides from commensals like S. sanguinis might serve as global anti-inflammatory agents of the mucosae. It would be particularly intriguing if this effect occurred through the previously described TLR2-dependent pathway (Peyret-Lacombe et al., 2009), since lipoteichoic acids in peptidoglycan normally serve as potent TLR2 agonists to trigger inflammation (Moreillon and Majcherczyk, 2003; Draing et al., 2008). It is conceivable that the anti-inflammatory ability of the commensal flora could be one of the key mechanisms required to reduce the inflammatory potential of other lower abundance members of the flora in order to maintain host symbiosis.
Genomic comparison of key genes involved in S. sanguinis commensalism
The universal success of S. sanguinis as an early colonizing commensal species in the oral cavity suggests that key genes and pathways crucial for its commensalism should be conserved among most or all isolates (Fig. 1). By comparing 25 publically available S. sanguinis genomes, we screened for the presence of key genes and operons previously described in this review. In all 25 strains, we could detect the arginine deiminase system, the glucan binding protein PcsB, the dual function adhesin/Mn2+ transporter SsaB, and the pyruvate oxidase SpxB. These genes are all highly conserved and likely to be part of the core S. sanguinis genome. The glucosyltransferase GtfP and the glucan binding proteins, SspC and SspD, can be found in 24 of the 25 strains, indicating they too have a similar level of conservation. Twitching motility is conserved in most strains, but is not as highly conserved overall (Fig. 2 and Table 1). Since adhesion is a critical trait for S. sanguinis ability to colonize and establish biofilms, we also examined these genomes for several uncharacterized putative adhesins identified in the genome of SK36 (SSA_0227, SSA_0453, SSA_0805, SSA_1019, and SSA_1666) (Kitten et al., 2011). In nearly all of the genomes, obvious orthologs of these proteins were detectable, with each sharing over 80% identity to SK36 (Table 2). The predicted adhesins encoded by SSA_0453 and SSA_1019 can be found in 22 out of the 25 genomes, while the putative adhesins SSA_0227, SSA_0805 and SSA_1666 were found in 21 strains. It is worth noting that most publically available S. sanguinis genome sequences are draft genomes still in various stages of assembly. Adhesins in particular tend to be difficult to assemble due to the presence of repeat regions and are often found in the gaps between genome contigs. Therefore, it is possible that these aforementioned adhesins are actually present in all 25 strains, potentially even belonging to the core S. sanguinis genome. Overall, there seems to be a strong conservation for genes required for commensalism among S. sanguinis strains. In future studies, it will be of particular interest to determine whether the genetic pathways responsible for the anti-inflammatory qualities of S. sanguinis are also as highly conserved among strains. Such comparisons will have to wait until the genetic mechanisms of S. sanguinis immunomodulation are understood.
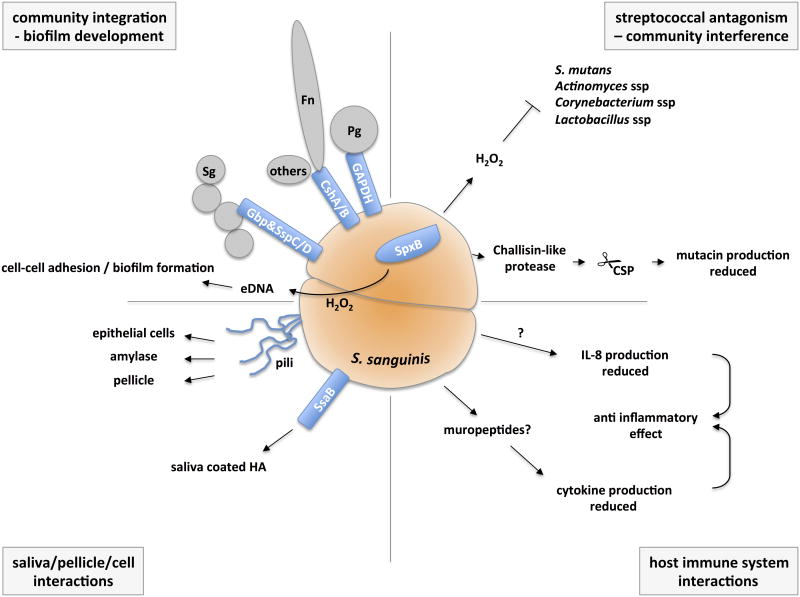
Fig. 1.
Summary of key S. sanguinis components important in commensalism. The schematic shows important components for S. sanguinis role as commensal organism, including community integration and biofilm development, community interference and streptococcal antagonism, and interactions with salivary proteins, host cells, and the immune system. Pg = P. gingivalis; Fn = F. nucleatum; Sg = S. gordonii; eDNA = extracellular DNA; CSP = competence stimulating peptide.
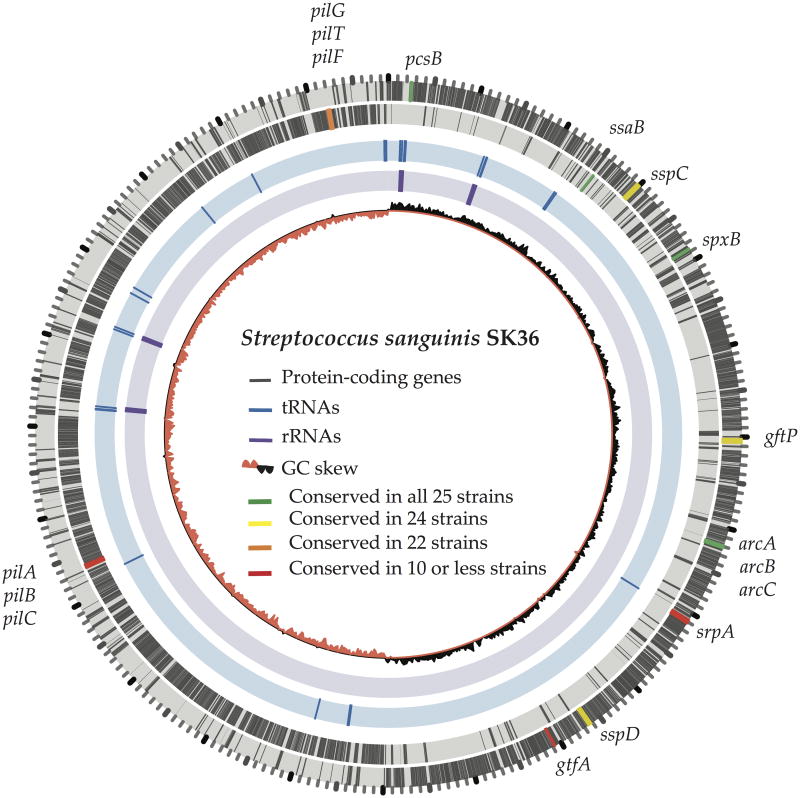
Fig. 2.
Genome of Streptococcus sanguinis SK36. The two outer rings illustrate protein-encoding genes (in grey) on forward and reverse strands, respectively. The position and conservation of several functional genes (Tab. 1) are also highlighted. The innermost ring shows the GC skew. Interestingly, most genes are located on the leading strand of each replichore. Additionally, most tRNAs (blue, ring 3) and rRNAs (purple, ring) are clustered around the origin of replication, possibly to optimize the growth rate.
Table 1. Conservation of selected genes implicated in commensalism in 25 sequenced strains of S. sanguinis.
| Gene | Function | No. of strains present |
|---|---|---|
| arcA | Arginine deiminase system | 25 |
| arcB | Arginine deiminase system | 25 |
| arcC | Arginine deiminase system | 25 |
| pcsB | Glucan-binding | 25 |
| spxB | Pyruvate oxidase | 25 |
| ssaB | Adhesion, Mn2+ transport | 25 |
| gtfP | Glucosyl transferase | 24 |
| sspC | Glucan-binding | 24 |
| sspD | Glucan-binding | 24 |
| pilF | Twitching motility | 22 |
| pilG | Twitching motility | 22 |
| pilT | Twitching motility | 22 |
| gtfA | Glucosyl transferase | 10 |
| pilA | Pilus, attachment to amylase | 10 |
| pilB | Pilus, attachment to amylase | 10 |
| pilC | Pilus, attachment to amylase | 9 |
| srpA | Mucin binding | 1 |
Conservation of genes in 25 fully annotated genomes of S. sanguinis was determined by BLASTP. S. sanguinis SK36 proteins were used as queries, and an E-value of at least 1e-5, ≥70% coverage of the length of the query protein, and ≥70% amino acid identity was required to classify a protein as being conserved. The following strains were included in the analysis: SK36; VMC66; SK353; SK405; SK678; SK72; SK115; SK150; SK160; SK1; SK1057; SK330; SK408; SK1058; SK1087; SK1059; SK49; SK1056; SK355; SK340; ATCC 29667; CC94A; VT517; I141; 2908
Table 2. Conservation of predicted adhesins in 25 sequenced strains of S. sanguinis.
| SSA_designation | predicted function | No. of strains present |
|---|---|---|
| 1019 | collagen binding surface protein | 22 |
| 453 | pullanase/glycosidase | 22 |
| 227 | collagen-binding surface protein | 21 |
| 805 | collagen-binding surface protein | 21 |
| 1666 | collagen-binding surface protein | 21 |
Conservation of open reading frames listed as SSA_designations from strain SK36 which served as template in the search of 25 fully annotated genomes of S. sanguinis using BLASTP. An E-value of at least 1e-5, ≥70% coverage of the length of the query protein, and ≥70% amino acid identity was required to classify a protein as being conserved.
In the context of the oral biofilm, some of the traits described in this review are also conserved in other oral streptococcal species, such as the ability to produce competitive amounts of H2O2 via SpxB as well as alkali generation via the arginine deiminase system. The following oral streptococci encode spxB orthologs: Streptococcus gordonii, Streptococcus mitis, Streptococcus infantis, Streptococcus oralis, Streptococcus oligofermentans and Streptococcus cristatus (Zhu et al., 2014) and a recent investigation identified arginine deiminase activity in Streptococcus parasanguinis, Streptococcus intermedius, Streptococcus cristatus and Streptococcus gordonii (Huang et al., 2015). This observation is consistent with the notion that certain metabolic functions of the oral biofilm determine health and disease status. Given the abundance of S. sanguinis in the oral cavity, it presumably makes a substantial contribution to the total metabolic output of these pathways.
Conclusion
There has been tremendous progress made in our understanding of the mechanisms of molecular pathogenesis in both caries and periodontal disease. However, much less is known about the other side of the equation, which is what we refer to as molecular commensalism. It is clear that shifts in oral bacterial ecology are harbingers of disease, especially when it occurs at the expense of the commensal flora, S. sanguinis among them. This raises the interesting question of whether it is possible to restore and/or bolster the commensal flora as an alternative approach to improve oral health. While such a strategy seems logical, if not obvious, there is a dearth of in vivo evidence to support this approach. As we develop a more thorough understanding of the mechanisms of molecular commensalism, we can expect to discover new strategies to promote the competitiveness of the commensal flora during ecologically challenging conditions that might otherwise lead to disease. This could be through the exogenous management of critical genetic responses in the flora or simply via probiotic supplementation. A case in favor of the former approach could be illustrated by the improvements observed in plaque pH due to arginine supplemented dentifrice usage (Nascimento et al., 2014), while the latter approach is supported by the recent successes with probiotic treatments for Clostridium difficile induced colitis (CDI) [a summary of successful application of probiotics to treat or prevent CDI can be found in (Spinler et al., 2016)]. For an oral probiotic approach, S. sanguinis could be regarded as an ideal candidate (Pamer, 2016), as it is an efficient colonizer, does not express obvious virulence factors, and it can modulate the inflammatory response. Given the relative simplicity of S. sanguinis isolation, it may even be practical to create probiotic supplements using patient-specific isolates, thus providing S. sanguinis with its ideal ecological niche for colonization. Though, such an approach may not be appropriate for people with known heart valve defects, due to an elevated risk of infective endocarditis.
A surprisingly large number of polymicrobial diseases of the mucosae are the result of dysbiosis among the mucosal flora and most of these infections currently remain difficult to treat effectively. The oral system has proven to be an exceptional model system to characterize the key concepts driving pathogenesis in these types of diseases, since the organisms are easily accessible, well characterized, and many are genetically tractable. Indeed, the ecological basis for oral disease has been a driving force in the field for decades. For similar reasons, the oral system is ideally situated to serve as a preeminent model system of molecular commensalism as well. Presumably, the key mechanisms supporting symbiosis at the oral mucosa are also functioning analogously at other mucosal sites in the body, albeit with a different cast of characters. Thus, there is a prime opportunity for the oral microbiology and immunology community to further its leadership in our understanding of the interplay between the flora and host as well as potential strategies to exploit this knowledge for therapeutic benefit.
Acknowledgments
This work was supported by an NIH-NIDCR grant DE021726 to J.K. and NIH-NIDCR grants DE018893 and DE022083 to JM.
Footnotes
The authors declare no conflict of interest.
References
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre A, Levine MJ, Cohen RE, Tabak LA. Immunochemical quantitation of alpha-amylase and secretory IgA in parotid saliva from people of various ages. Arch Oral Biol. 1987;32:297–301. doi: 10.1016/0003-9969(87)90024-0. [DOI] [PubMed] [Google Scholar]
- Ashby MT. Inorganic chemistry of defensive peroxidases in the human oral cavity. J Dent Res. 2008;87:900–914. doi: 10.1177/154405910808701003. [DOI] [PubMed] [Google Scholar]
- Becker MR, Paster BJ, Leys EJ, et al. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol. 2002;40:1001–1009. doi: 10.1128/JCM.40.3.1001-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, et al. The oral metagenome in health and disease. ISME J. 2012;6:46–56. doi: 10.1038/ismej.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma MW, Brouwer MC, Kasanmoentalib ES, et al. Community-acquired bacterial meningitis in adults in the Netherlands, 2006-14: a prospective cohort study. Lancet Infect Dis. 2016;16:339–347. doi: 10.1016/S1473-3099(15)00430-2. [DOI] [PubMed] [Google Scholar]
- Bik EM, Long CD, Armitage GC, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne RA, Marquis RE. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol Lett. 2000;193:1–6. doi: 10.1111/j.1574-6968.2000.tb09393.x. [DOI] [PubMed] [Google Scholar]
- Cahill TJ, Prendergast BD. Current controversies in infective endocarditis. F1000Res. 2015;4 doi: 10.12688/f1000research.6949.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill TJ, Prendergast BD. Infective endocarditis. Lancet. 2016;387:882–893. doi: 10.1016/S0140-6736(15)00067-7. [DOI] [PubMed] [Google Scholar]
- Carlsson J. Growth of Streptococcus mutans and Streptococcus sanguis in mixed culture. Arch Oral Biol. 1971;16:963–965. doi: 10.1016/0003-9969(71)90184-1. [DOI] [PubMed] [Google Scholar]
- Carlsson J, Edlund MB. Pyruvate oxidase in Streptococcus sanguis under various growth conditions. Oral Microbiol Immunol. 1987;2:10–14. doi: 10.1111/j.1399-302x.1987.tb00263.x. [DOI] [PubMed] [Google Scholar]
- Carlsson J, Edlund MB, Lundmark SK. Characteristics of a hydrogen peroxide-forming pyruvate oxidase from Streptococcus sanguis. Oral Microbiol Immunol. 1987;2:15–20. doi: 10.1111/j.1399-302x.1987.tb00264.x. [DOI] [PubMed] [Google Scholar]
- Carlsson J, Grahnen H, Jonsson G, Wikner S. Establishment of Streptococcus sanguis in the mouths of infants. Arch Oral Biol. 1970;15:1143–1148. doi: 10.1016/0003-9969(70)90005-1. [DOI] [PubMed] [Google Scholar]
- Caufield PW, Dasanayake AP, Li Y, Pan Y, Hsu J, Hardin JM. Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect Immun. 2000;68:4018–4023. doi: 10.1128/iai.68.7.4018-4023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo AP, Boches SK, Cotton SL, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corby PM, Lyons-Weiler J, Bretz WA, et al. Microbial risk indicators of early childhood caries. J Clin Microbiol. 2005;43:5753–5759. doi: 10.1128/JCM.43.11.5753-5759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump KE, Bainbridge B, Brusko S, et al. The relationship of the lipoprotein SsaB, manganese and superoxide dismutase in Streptococcus sanguinis virulence for endocarditis. Mol Microbiol. 2014;92:1243–1259. doi: 10.1111/mmi.12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport EK, Call DR, Beyenal H. Differential protection from tobramycin by extracellular polymeric substances from Acinetobacter baumannii and Staphylococcus aureus biofilms. Antimicrob Agents Chemother. 2014;58:4755–4761. doi: 10.1128/AAC.03071-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer D, Stoodley P, Roe F, Lewandowski Z. Effects of biofilm structures on oxygen distribution and mass transport. Biotechnol Bioeng. 1994;43:1131–1138. doi: 10.1002/bit.260431118. [DOI] [PubMed] [Google Scholar]
- Diaz PI, Chalmers NI, Rickard AH, et al. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl Environ Microbiol. 2006;72:2837–2848. doi: 10.1128/AEM.72.4.2837-2848.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draing C, Sigel S, Deininger S, et al. Cytokine induction by Gram-positive bacteria. Immunobiology. 2008;213:285–296. doi: 10.1016/j.imbio.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Dworkin J. The medium is the message: interspecies and interkingdom signaling by peptidoglycan and related bacterial glycans. Annu Rev Microbiol. 2014;68:137–154. doi: 10.1146/annurev-micro-091213-112844. [DOI] [PubMed] [Google Scholar]
- Elborn JS. Cystic fibrosis. Lancet. 2016 doi: 10.1016/S0140-6736(16)00576-6. [DOI] [PubMed] [Google Scholar]
- Facklam R. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin Microbiol Rev. 2002;15:613–630. doi: 10.1128/CMR.15.4.613-630.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Flemming HC, Neu TR, Wozniak DJ. The EPS matrix: the “house of biofilm cells”. J Bacteriol. 2007;189:7945–7947. doi: 10.1128/JB.00858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel ES, Ribbeck K. Salivary mucins in host defense and disease prevention. J Oral Microbiol. 2015;7:29759. doi: 10.3402/jom.v7.29759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeshkumar N, Song M, McBride BC. Cloning of a Streptococcus sanguis adhesin which mediates binding to saliva-coated hydroxyapatite. Infect Immun. 1988;56:1150–1157. doi: 10.1128/iai.56.5.1150-1157.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Shi X, Shi L, et al. Involvement of NADH Oxidase in Biofilm Formation in Streptococcus sanguinis. PLoS One. 2016;11:e0151142. doi: 10.1371/journal.pone.0151142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Caufield PW, Fisch GS, Li Y. Streptococcus mutans and Streptococcus sanguinis colonization correlated with caries experience in children. Caries Res. 2008;42:444–448. doi: 10.1159/000159608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacaman RA, Torres S, Gómez Y, Muñoz-Sandoval C, Kreth J. Correlation of Streptococcus mutans and Streptococcus sanguinis colonization and ex vivo hydrogen peroxide production in carious lesion-free and high caries adults. Arch Oral Biol. 2015;60:154–159. doi: 10.1016/j.archoralbio.2014.09.007. [DOI] [PubMed] [Google Scholar]
- Gibbins HL, Yakubov GE, Proctor GB, Wilson S, Carpenter GH. What interactions drive the salivary mucosal pellicle formation? Colloids Surf B Biointerfaces. 2014;120:184–192. doi: 10.1016/j.colsurfb.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung I, Spielman I, Davies MR, et al. Functional analysis of an unusual type IV pilus in the Gram-positive Streptococcus sanguinis. Mol Microbiol. 2016;99:380–392. doi: 10.1111/mmi.13237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. Dancing with the Stars: How Choreographed Bacterial Interactions Dictate Nososymbiocity and Give Rise to Keystone Pathogens, Accessory Pathogens, and Pathobionts. Trends Microbiol. 2016 doi: 10.1016/j.tim.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Torii M, Kotani S, Tsuchitani Y. Adherence of Streptococcus sanguis clinical isolates to smooth surfaces and interactions of the isolates with Streptococcus mutans glucosyltransferase. Infect Immun. 1981;32:364–372. doi: 10.1128/iai.32.1.364-372.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen SD, Henrichsen J. Twitching motility and possession of polar fimbriae in spreading Streptococcus sanguis isolates from the human throat. Acta Pathol Microbiol Scand B. 1975;83:133–140. doi: 10.1111/j.1699-0463.1975.tb00083.x. [DOI] [PubMed] [Google Scholar]
- Herrero ER, Slomka V, Bernaerts K, et al. Antimicrobial effects of commensal oral species are regulated by environmental factors. J Dent. 2016;47:23–33. doi: 10.1016/j.jdent.2016.02.007. [DOI] [PubMed] [Google Scholar]
- Herzberg MC, Gong K, MacFarlane GD, et al. Phenotypic characterization of Streptococcus sanguis virulence factors associated with bacterial endocarditis. Infect Immun. 1990;58:515–522. doi: 10.1128/iai.58.2.515-522.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg K, Hallander HO. Production of bactericidal concentrations of hydrogen peroxide by Streptococcus sanguis. Arch Oral Biol. 1973;18:423–434. doi: 10.1016/0003-9969(73)90167-2. [DOI] [PubMed] [Google Scholar]
- Huang X, Schulte RM, Burne RA, Nascimento MM. Characterization of the arginolytic microflora provides insights into pH homeostasis in human oral biofilms. Caries Res. 2015;49:165–176. doi: 10.1159/000365296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihalin R, Loimaranta V, Tenovuo J. Origin, structure, and biological activities of peroxidases in human saliva. Arch Biochem Biophys. 2006;445:261–268. doi: 10.1016/j.abb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Jakubovics NS, Yassin SA, Rickard AH. Community interactions of oral streptococci. Adv Appl Microbiol. 2014;87:43–110. doi: 10.1016/B978-0-12-800261-2.00002-5. [DOI] [PubMed] [Google Scholar]
- Jakubovics NS, Gill SR, Vickerman MM, Kolenbrander PE. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiol Ecol. 2008;66:637–644. doi: 10.1111/j.1574-6941.2008.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell KF, McBride MJ. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol. 2008;6:466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- Jenkinson HF, Lala HC, Shepherd MG. Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect Immun. 1990;58:1429–1436. doi: 10.1128/iai.58.5.1429-1436.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S, Kim Y, Min BM, Han SH, Choi Y. Innate immune responses of gingival epithelial cells to nonperiodontopathic and periodontopathic bacteria. J Periodontal Res. 2007;42:503–510. doi: 10.1111/j.1600-0765.2007.00974.x. [DOI] [PubMed] [Google Scholar]
- Kampe CE, Vovan T, Alim A, Berenson J. Streptococcus sanguis bacteremia and colorectal cancer: a case report. Med Pediatr Oncol. 1995;24:67–68. doi: 10.1002/mpo.2950240116. [DOI] [PubMed] [Google Scholar]
- Kanasi E, Dewhirst FE, Chalmers NI, et al. Clonal analysis of the microbiota of severe early childhood caries. Caries Res. 2010;44:485–497. doi: 10.1159/000320158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassebaum NJ, Bernabe E, Dahiya M, Bhandari B, Murray CJ, Marcenes W. Global burden of untreated caries: a systematic review and metaregression. J Dent Res. 2015;94:650–658. doi: 10.1177/0022034515573272. [DOI] [PubMed] [Google Scholar]
- Keevil CW, West AA, Bourne N, Marsh PD. Inhibition of the synthesis and secretion of extracellular glucosyl- and fructosyltransferase in Streptococcus sanguis by sodium ions. J Gen Microbiol. 1984;130:77–82. doi: 10.1099/00221287-130-1-77. [DOI] [PubMed] [Google Scholar]
- Kitten T, Turner LS, Xu P. Biological implications of the Streptococcus sanguinis genome. In: Kolenbrander PE, editor. Oral microbial communities: genomic inquiries and interspecies communication. Washington: ASM Press; 2011. [Google Scholar]
- Klein MI, Hwang G, Santos PH, Campanella OH, Koo H. Streptococcus mutans-derived extracellular matrix in cariogenic oral biofilms. Front Cell Infect Microbiol. 2015;5:10. doi: 10.3389/fcimb.2015.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ, Jr, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- Kopec LK, Vacca Smith AM, Wunder D, Ng-Evans L, Bowen WH. Properties of Streptococcus sanguinis glucans formed under various conditions. Caries Res. 2001;35:67–74. doi: 10.1159/000047434. [DOI] [PubMed] [Google Scholar]
- Kraivaphan P, Amornchat C, Triratana T, et al. Two-year caries clinical study of the efficacy of novel dentifrices containing 1.5% arginine, an insoluble calcium compound and 1,450 ppm fluoride. Caries Res. 2013;47:582–590. doi: 10.1159/000353183. [DOI] [PubMed] [Google Scholar]
- Kreth J, Herzberg MC. Molecular Principles of Adhesion and Biofilm Formation. In: Chavez de Paz LE, Sedgley CM, Kishen A, editors. The root canal biofilm. Berlin, Heidelberg, New York: Springer; 2015. pp. 23–54. [Google Scholar]
- Kreth J, Zhang Y, Herzberg MC. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol. 2008;190:4632–4640. doi: 10.1128/JB.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Vu H, Zhang Y, Herzberg MC. Characterization of hydrogen peroxide-induced DNA release by Streptococcus sanguinis and Streptococcus gordonii. J Bacteriol. 2009;191:6281–6291. doi: 10.1128/JB.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkin MS, Oppenheim FG. Structural features of salivary function. Crit Rev Oral Biol Med. 1993;4:251–259. doi: 10.1177/10454411930040030101. [DOI] [PubMed] [Google Scholar]
- Lamont RJ, Hersey SG, Rosan B. Characterization of the adherence of Porphyromonas gingivalis to oral streptococci. Oral Microbiol Immunol. 1992;7:193–197. doi: 10.1111/j.1399-302x.1992.tb00024.x. [DOI] [PubMed] [Google Scholar]
- Lang ML, Zhu L, Kreth J. Keeping the bad bacteria in check: interactions of the host immune system with oral cavity biofilms. Endodontic Topics. 2010;22:17–32. [Google Scholar]
- Lee SH. Antagonistic effect of peptidoglycan of Streptococcus sanguinis on lipopolysaccharide of major periodontal pathogens. J Microbiol. 2015;53:553–560. doi: 10.1007/s12275-015-5319-6. [DOI] [PubMed] [Google Scholar]
- Li J, Helmerhorst EJ, Leone CW, Troxler RF, Yaskell T, Haffajee AD, et al. Identification of early microbial colonizers in human dental biofilm. J Appl Microbiol. 2004;97:1311–1318. doi: 10.1111/j.1365-2672.2004.02420.x. [DOI] [PubMed] [Google Scholar]
- Lindh L. On the adsorption behaviour of saliva and purified salivary proteins at solid/liquid interfaces. Swed Dent J Suppl. 2002:1–57. [PubMed] [Google Scholar]
- Liu YT, Lin CF, Lee YL. Streptococcus sanguinis meningitis following endoscopic ligation for oesophageal variceal haemorrhage. J Med Microbiol. 2013;62:794–796. doi: 10.1099/jmm.0.054213-0. [DOI] [PubMed] [Google Scholar]
- Macaluso A, Simmang C, Anthony T. Streptococcus sanguis bacteremia and colorectal cancer. South Med J. 1998;91:206–207. doi: 10.1097/00007611-199802000-00016. [DOI] [PubMed] [Google Scholar]
- Maeda K, Nagata H, Nonaka A, Kataoka K, Tanaka M, Shizukuishi S. Oral streptococcal glyceraldehyde-3-phosphate dehydrogenase mediates interaction with Porphyromonas gingivalis fimbriae. Microbes Infect. 2004;6:1163–1170. doi: 10.1016/j.micinf.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Magalhaes AP, Azevedo NF, Pereira MO, Lopes SP. The cystic fibrosis microbiome in an ecological perspective and its impact in antibiotic therapy. Appl Microbiol Biotechnol. 2016;100:1163–1181. doi: 10.1007/s00253-015-7177-x. [DOI] [PubMed] [Google Scholar]
- Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol. 2003;30:644–654. doi: 10.1034/j.1600-051x.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- Marsh PD, McDermid AS, Keevil CW, Ellwood DC. Environmental regulation of carbohydrate metabolism by Streptococcus sanguis NCTC 7865 grown in a chemostat. J Gen Microbiol. 1985;131:2505–2514. doi: 10.1099/00221287-131-10-2505. [DOI] [PubMed] [Google Scholar]
- Mason MR, Preshaw PM, Nagaraja HN, Dabdoub SM, Rahman A, Kumar PS. The subgingival microbiome of clinically healthy current and never smokers. ISME J. 2015;9:268–272. doi: 10.1038/ismej.2014.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean JS. Advancements toward a systems level understanding of the human oral microbiome. Front Cell Infect Microbiol. 2014;4:98. doi: 10.3389/fcimb.2014.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J, Qi F. The mutacins of Streptococcus mutans: regulation and ecology. Mol Oral Microbiol. 2012;27:57–69. doi: 10.1111/j.2041-1014.2011.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SY, Chung DR, Kim SW, et al. Changing etiology of community-acquired bacterial meningitis in adults: a nationwide multicenter study in Korea. Eur J Clin Microbiol Infect Dis. 2010;29:793–800. doi: 10.1007/s10096-010-0929-8. [DOI] [PubMed] [Google Scholar]
- Moraes JJ, Stipp RN, Harth-Chu EN, Camargo TM, Hofling JF, Mattos-Graner RO. Two-component system VicRK regulates functions associated with establishment of Streptococcus sanguinis in biofilms. Infect Immun. 2014;82:4941–4951. doi: 10.1128/IAI.01850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreillon P, Majcherczyk PA. Proinflammatory activity of cell-wall constituents from Gram-positive bacteria. Scand J Infect Dis. 2003;35:632–641. doi: 10.1080/00365540310016259. [DOI] [PubMed] [Google Scholar]
- Nascimento MM, Browngardt C, Xiaohui X, Klepac-Ceraj V, Paster BJ, Burne RA. The effect of arginine on oral biofilm communities. Mol Oral Microbiol. 2014;29:45–54. doi: 10.1111/omi.12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niven CF, Kiziuta Z, White JC. Synthesis of a Polysaccharide from Sucrose by Streptococcus S.B.E. J Bacteriol. 1946;51:711–716. doi: 10.1128/jb.51.6.711-716.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niven CF, Jr, White JC. A Study of Streptococci Associated with Subacute Bacterial Endocarditis. J Bacteriol. 1946;51:790. doi: 10.1128/jb.51.6.790-790.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus adherence and colonization. Microbiol Mol Biol Rev. 2009;73:407–450. doi: 10.1128/MMBR.00014-09. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Jenkinson HF, Jakubovics NS. Stick to your gums: mechanisms of oral microbial adherence. J Dent Res. 2011;90:1271–1278. doi: 10.1177/0022034511399096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Jenkinson HF, Everett DB. Generic determinants of Streptococcus colonization and infection. Infect Genet Evol. 2015;33:361–370. doi: 10.1016/j.meegid.2014.09.018. [DOI] [PubMed] [Google Scholar]
- Nyvad B, Kilian M. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 1990;24:267–272. doi: 10.1159/000261281. [DOI] [PubMed] [Google Scholar]
- Okahashi N, Sumitomo T, Nakata M, Sakurai A, Kuwata H, Kawabata S. Hydrogen peroxide contributes to the epithelial cell death induced by the oral mitis group of streptococci. PLoS One. 2014;9:e88136. doi: 10.1371/journal.pone.0088136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okahashi N, Nakata M, Terao Y, et al. Pili of oral Streptococcus sanguinis bind to salivary amylase and promote the biofilm formation. Microb Pathog. 2011;50:148–154. doi: 10.1016/j.micpath.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Okshevsky M, Meyer RL. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit Rev Microbiol. 2015;41:341–352. doi: 10.3109/1040841X.2013.841639. [DOI] [PubMed] [Google Scholar]
- Orstavik D, Kraus FW. The acquired pellicle: immunofluorescent demonstration of specific proteins. J Oral Pathol. 1973;2:68–76. doi: 10.1111/j.1600-0714.1973.tb01675.x. [DOI] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Flavell RA. Immune-microbiota interactions in health and disease. Clin Immunol. 2015;159:122–127. doi: 10.1016/j.clim.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamer EG. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science. 2016;352:535–538. doi: 10.1126/science.aad9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SN, Snesrud E, Liu J, et al. The dental plaque microbiome in health and disease. PLoS One. 2013;8:e58487. doi: 10.1371/journal.pone.0058487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SN, Meissner T, Su AI, et al. Functional expression of dental plaque microbiota. Front Cell Infect Microbiol. 2014;4:108. doi: 10.3389/fcimb.2014.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyret-Lacombe A, Brunel G, Watts M, Charveron M, Duplan H. TLR2 sensing of F. nucleatum and S. sanguinis distinctly triggered gingival innate response. Cytokine. 2009;46:201–210. doi: 10.1016/j.cyto.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Plummer C, Douglas CW. Relationship between the ability of oral streptococci to interact with platelet glycoprotein Ibalpha and with the salivary low-molecular-weight mucin, MG2. FEMS Immunol Med Microbiol. 2006;48:390–399. doi: 10.1111/j.1574-695X.2006.00161.x. [DOI] [PubMed] [Google Scholar]
- Ramasubbu N, Paloth V, Luo Y, Brayer GD, Levine MJ. Structure of human salivary alpha-amylase at 1.6 A resolution: implications for its role in the oral cavity. Acta Crystallogr D Biol Crystallogr. 1996;52:435–446. doi: 10.1107/S0907444995014119. [DOI] [PubMed] [Google Scholar]
- Ramsey MM, Whiteley M. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc Natl Acad Sci U S A. 2009;106:1578–1583. doi: 10.1073/pnas.0809533106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salam MA, Nakao R, Yonezawa H, Watanabe H, Senpuku H. Human T-cell responses to oral streptococci in human PBMC-NOD/SCID mice. Oral Microbiol Immunol. 2006;21:169–176. doi: 10.1111/j.1399-302X.2006.00272.x. [DOI] [PubMed] [Google Scholar]
- Scannapieco FA, Bhandary K, Ramasubbu N, Levine MJ. Structural relationship between the enzymatic and streptococcal binding sites of human salivary alpha-amylase. Biochem Biophys Res Commun. 1990;173:1109–1115. doi: 10.1016/s0006-291x(05)80900-3. [DOI] [PubMed] [Google Scholar]
- Simon-Soro A, Mira A. Solving the etiology of dental caries. Trends Microbiol. 2015;23:76–82. doi: 10.1016/j.tim.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Sliepen I, Van Damme J, Van Essche M, Loozen G, Quirynen M, Teughels W. Microbial interactions influence inflammatory host cell responses. J Dent Res. 2009;88:1026–1030. doi: 10.1177/0022034509347296. [DOI] [PubMed] [Google Scholar]
- Spinler JK, Ross CL, Savidge TC. Probiotics as adjunctive therapy for preventing Clostridium difficile infection - What are we waiting for? Anaerobe. 2016 doi: 10.1016/j.anaerobe.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacy A, McNally L, Darch SE, Brown SP, Whiteley M. The biogeography of polymicrobial infection. Nat Rev Microbiol. 2016;14:93–105. doi: 10.1038/nrmicro.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacy A, Everett J, Jorth P, Trivedi U, Rumbaugh KP, Whiteley M. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci U S A. 2014;111:7819–7824. doi: 10.1073/pnas.1400586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS. Diffusion in biofilms. J Bacteriol. 2003;185:1485–1491. doi: 10.1128/JB.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingu CS, Eschrich K, Rodloff AC, Schaumann R, Jentsch H. Periodontitis is associated with a loss of colonization by Streptococcus sanguinis. J Med Microbiol. 2008;57:495–499. doi: 10.1099/jmm.0.47649-0. [DOI] [PubMed] [Google Scholar]
- Stinson MW, Safulko K, Levine MJ. Adherence of Porphyromonas (Bacteroides) gingivalis to Streptococcus sanguis in vitro. Infect Immun. 1991;59:102–108. doi: 10.1128/iai.59.1.102-108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Ebara S, Otsuka K, Hayashi K. Adsorption of saliva-coated and plain streptococcal cells to the surfaces of hydroxyapatite beads. Arch Oral Biol. 1996;41:505–508. doi: 10.1016/0003-9969(95)00144-1. [DOI] [PubMed] [Google Scholar]
- Tappuni AR, Challacombe SJ. Distribution and isolation frequency of eight streptococcal species in saliva from predentate and dentate children and adults. J Dent Res. 1993;72:31–36. doi: 10.1177/00220345930720010401. [DOI] [PubMed] [Google Scholar]
- Teng JL, Huang Y, Tse H, et al. Phylogenomic and MALDI-TOF MS analysis of Streptococcus sinensis HKU4T reveals a distinct phylogenetic clade in the genus Streptococcus. Genome Biol Evol. 2014;6:2930–2943. doi: 10.1093/gbe/evu232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietze K, Dalpke A, Morath S, Mutters R, Heeg K, Nonnenmacher C. Differences in innate immune responses upon stimulation with Gram-positive and Gram-negative bacteria. J Periodontal Res. 2006;41:447–454. doi: 10.1111/j.1600-0765.2006.00890.x. [DOI] [PubMed] [Google Scholar]
- Truper HG, De' Clari L. Taxonomic Note: Necessary Correction of Specific Epithets Formed as Substantives (Nouns) “in Apposition”. Int J Syst Bacteriol. 1997;47:908–909. [Google Scholar]
- Vacca Smith AM, Ng-Evans L, Wunder D, Bowen WH. Studies concerning the glucosyltransferase of Streptococcus sanguis. Caries Res. 2000;34:295–302. doi: 10.1159/000016605. [DOI] [PubMed] [Google Scholar]
- Vadeboncoeur C, Pelletier M. The phosphoenolpyruvate:sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol Rev. 1997;19:187–207. doi: 10.1111/j.1574-6976.1997.tb00297.x. [DOI] [PubMed] [Google Scholar]
- Wang BY, Kuramitsu HK. Interactions between oral bacteria: inhibition of Streptococcus mutans bacteriocin production by Streptococcus gordonii. Appl Environ Microbiol. 2005;71:354–362. doi: 10.1128/AEM.71.1.354-362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MR, White JC, Niven CF., Jr Streptococcus S.B.E.: Immunological Characteristics. J Bacteriol. 1946;51:723–729. doi: 10.1128/jb.51.6.723-729.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley RA, Fleming EV, Makhija R, Waite RD. Environment and colonisation sequence are key parameters driving cooperation and competition between Pseudomonas aeruginosa cystic fibrosis strains and oral commensal streptococci. PLoS One. 2015;10:e0115513. doi: 10.1371/journal.pone.0115513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JC, Niven CF., Jr Streptococcus S.B.E.: A Streptococcus Associated with Subacute Bacterial Endocarditis. J Bacteriol. 1946;51:717–722. doi: 10.1128/jb.51.6.717-722.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CJ, Burns LH, Jack AA, et al. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28:83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu KD, Stewart PS, Xia F, Huang CT, McFeters GA. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl Environ Microbiol. 1998;64:4035–4039. doi: 10.1128/aem.64.10.4035-4039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Alves JM, Kitten T, et al. Genome of the opportunistic pathogen Streptococcus sanguinis. J Bacteriol. 2007;189:3166–3175. doi: 10.1128/JB.01808-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Konno H, Nagano K, et al. The influence of a glucosyltransferase, encoded by gtfP, on biofilm formation by Streptococcus sanguinis in a dual-species model. APMIS. 2014;122:951–960. doi: 10.1111/apm.12238. [DOI] [PubMed] [Google Scholar]
- Zheng L, Itzek A, Chen Z, Kreth J. Environmental influences on competitive hydrogen peroxide production in Streptococcus gordonii. Appl Environ Microbiol. 2011a;77:4318–4328. doi: 10.1128/AEM.00309-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Chen Z, Itzek A, Ashby M, Kreth J. Catabolite control protein A controls hydrogen peroxide production and cell death in Streptococcus sanguinis. J Bacteriol. 2011b;193:516–526. doi: 10.1128/JB.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng LY, Itzek A, Chen ZY, Kreth J. Oxygen dependent pyruvate oxidase expression and production in Streptococcus sanguinis. Int J Oral Sci. 2011c;3:82–89. doi: 10.4248/IJOS11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Kreth J. The role of hydrogen peroxide in environmental adaptation of oral microbial communities. Oxid Med Cell Longev. 2012;2012:717843. doi: 10.1155/2012/717843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Xu Y, Ferretti JJ, Kreth J. Probing oral microbial functionality--expression of spxB in plaque samples. PLoS One. 2014;9:e86685. doi: 10.1371/journal.pone.0086685. [DOI] [PMC free article] [PubMed] [Google Scholar]