Abstract
Drought is one of the most important abiotic stresses that impair sesame (Sesamum indicum L.) productivity mainly when it occurs at flowering stage. However up to now, very few studies have attempted to investigate the molecular responses of sesame to drought stress. In this experiment, two genotypes having contrasting responses to drought (tolerant and sensitive) were submitted to progressive drought followed by recovering stage at flowering stage. RNAs were isolated from roots of plants before drought stress, at 3-time points during progressive drought, after rewatering, and sequenced using Illumina HiSeq 4000 platform. These RNA-Seq resources (BioSample IDs: SAMN06130606 and SAMN06130607) provided an opportunity to elucidate the molecular responses of sesame to drought and find out some candidate genes for drought tolerance improvement.
Keywords: Sesamum indicum, Drought stress, Illumina HiSeq, Transcriptome
1. Introduction
Sesame (Sesamum indicum L.) is an ancient oilseed crop mainly grown in drought-prone environments. It is highly valued because of its high oil yield (~ 55%), quality and stability [1]. Although sesame is considered as drought tolerant crop, its productivity is heavily affected by severe drought stress mainly when it occurs during anthesis [2]. To improve our understanding to sesame drought tolerance, several morphological, physiological and biochemical investigations have been conducted in the past years. However, data in regard with molecular responses to drought in sesame are very scarce. Recent gene expression profiling of some candidate gene families under moderate drought stress in sesame indicated that many genes are involved in drought responses especially transcription factors [3], [4], [5]. In this project, we took advantage of the availability of the sesame genome [6] to sequence for the first time the transcriptome of tolerant and sensitive genotypes at different time points under progressive drought stress and during recovering phase.
2. Direct link to deposited data
http://www.ncbi.nlm.nih.gov/bioproject/PRJNA356988
| Organism/cell line/tissue | Drought stressed root tissues of Sesamum indicum L./cultivated type |
| Sex | N/A |
| Sequencer or array type | Illumina HiSeq 4000 platform |
| Data format | Clean data |
| Experimental factors | Drought treatments and genotypes |
| Experimental features | Gene expression profiling under progressive drought stress and recovery phases (5 sampling dates). In total 30 samples from roots of drought sensitive and tolerant genotypes were collected and sequenced to elucidate the molecular mechanisms of response to drought |
| Consent | N/A |
| Sample source location | Key Laboratory of Biology and Genetic Improvement of Oil Crops, Ministry of Agriculture, Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences, No. 2 Xudong 2nd Road, 430062 Wuhan, Hubei, China |
3. Experimental design, materials and methods
3.1. Materials and stress treatment
Two sesame genotypes with different level of tolerance to drought [ZZM0635-drought tolerant (DT)] and [ZZM4782-drought sensitive (DS)] were obtained from the China National Genebank, Oil Crops Research Institute, Chinese Academy of Agricultural Sciences. Plants were sown in pots containing loam soil with known physicochemical properties mixed with 10% of added compound fertilizer. The experiment was carried out in a greenhouse under a completely randomized split-plot design with 6 replications and 3 plants per pot.
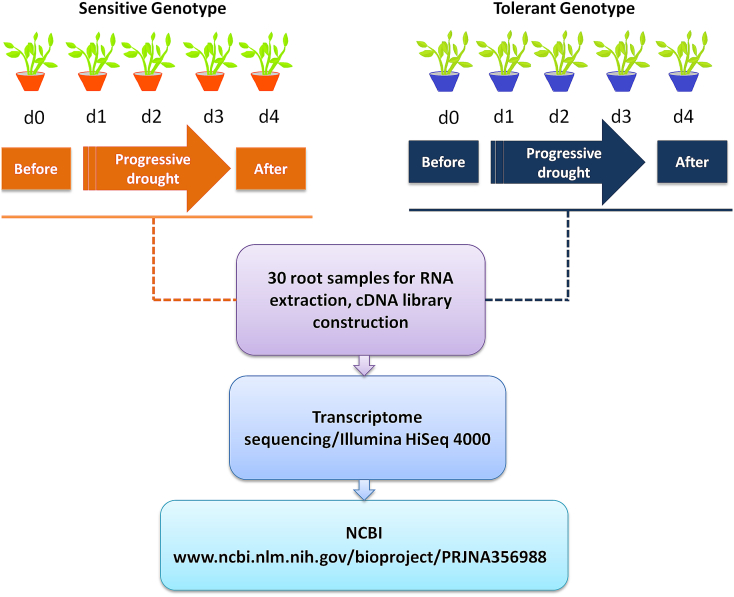
Seedlings were well watered to keep optimal soil moisture conditions (35% volumetric water content (vwc)). The water stress treatment was imposed at the early anthesis stage (47 days after sowing (DAS)) corresponding to the sampling date 1 (d0). The soil moisture gradually decreased by withholding water until it reached 15% vwc (d1), 9% vwc (d2), 6% vwc (d3) and plants showed mild-wilting to critical wilting signs. At 58 DAS, watering was resumed for 4 days to reach 35% vwc corresponding to the rewatering phase (sampling date 5 (d4)). At each of the above-indicated sampling dates, materials from three independent plants of the same pot (three biological replicates of root samples) were collected for RNA extraction (Fig. 1).
Fig. 1.
Schema representing the root transcriptome sequencing experiment under drought stress in two contrasting sesame genotypes.
3.2. Transcriptome sequencing
After extraction, the total RNA was treated with DNase I and Oligo (dT) was used to isolate mRNA. The mRNA was mixed with the fragmentation buffer. Quantity and quality of mRNA were assessed by ND-1000 Nanodrop spectrometer (Nanodrop Technologies, USA) and on 2% denatured agarose gel. Then, the cDNA was synthesized using the mRNA fragments as templates. Short fragments were purified and resolved with EB buffer for end reparation and single nucleotide A (adenine) addition. The short fragments (200 ± 25 bp) were ligated with adapters and the suitable fragments were selected for the PCR amplification. During the QC steps, Agilent 2100 Bioanaylzer and ABI StepOnePlus Real-Time PCR System were used in quantification and quality check of the sample library. The libraries were sequenced using Illumina HiSeq 4000 [7].
3.3. Sequencing reads filtering and genome mapping
The sequencing reads containing low-quality, high content of unknown base (N) and adaptor sequences were removed before downstream analyses using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). After filtering, the remaining reads were called “clean reads” and stored in FASTQ. The clean reads were then mapped to the sesame reference genome [6] using HISAT. On average, 80.39% reads were mapped and the uniformity of the mapping result for each sample suggested that the samples are comparable.
3.4. Novel transcript prediction, gene expression analysis and differentially expressed gene detection
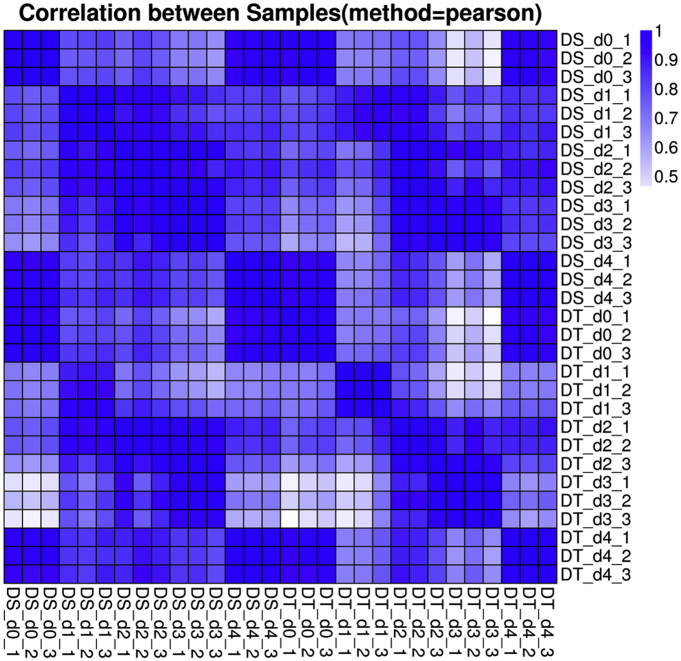
After genome mapping, the program StringTie was used for transcript assembly. Cuffcompare, a tool of cufflinks, was applied on the genome annotation information to identify novel transcripts in the samples. We further merged the novel coding transcripts with the reference transcripts to get the complete reference, and then we mapped clean reads to the complete reference using Bowtie2. RSEM package was used to calculate gene expression level for each sample. We calculated Pearson correlation between all samples using Cor, a function of R package (Fig. 2). The differentially expressed genes (DEG) were detected as described by Tarazona et al. [8] based on the parameters: Fold Change ≥ 2.00 and Probability ≥ 0.8.
Fig. 2.
Heatmap clustering analysis of the 30 transcriptome data.
Conflict of interest
The authors have no conflicts of interest.
Acknowledgement
Financial supports provided by the “Agricultural Science and Technology Innovation Project of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-OCRI)” and “China Agricultural Research System (no. CARS-15)” are gratefully acknowledged.
References
- 1.Pathak N., Rai A.K., Kumari R., Thapa A., Bhat K.V. Sesame crop: an underexploited oilseed holds tremendous potential for enhanced food value. Agric. Sci. 2014;5:519–529. [Google Scholar]
- 2.Kadkhodaie A., Zahedi M., Razmjoo J., Pessarakli M. Changes in some anti-oxidative enzymes and physiological indices among sesame genotypes (Sesamum indicum L.) in response to soil water deficits under field conditions. Acta Physiol. Plant. 2014;36(3):641–650. [Google Scholar]
- 3.Dossa K., Diouf D., Cissé N. Genome-wide investigation of Hsf genes in sesame reveals their segmental duplication expansion and their active role in drought stress response. Front. Plant Sci. 2016;7:1522. doi: 10.3389/fpls.2016.01522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dossa K., Wei X., Li D., Zhang Y., Wang L., Fonceka D., Yu J., Diouf D., Boshou L., Cissé N., Zhang X. Insight into the AP2/ERF transcription factor superfamily in sesame (Sesamum indicum) and expression profiling of the DREB subfamily under drought stress. BMC Plant Biol. 2016;16(1):171. doi: 10.1186/s12870-016-0859-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dossa K., Niang M., Assogbadjo A.E., Cissé N., Diouf D. Whole genome homology-based identification of candidate genes for drought resistance in (Sesamum indicum L.) Afr. J. Biotechnol. 2016;15(27):1464–1475. [Google Scholar]
- 6.Wang L., Yu S., Tong C., Zhao Y., Liu Y., Song C., Zhang Y., Zhang X., Wang Y., Hua W., Li D., Li D., Li F., Yu J., Xu C., Han X., Huang S., Tai S., Wang J., Xu X., Li Y., Liu S., Varshney R., Wang J., Zhang X. Genome sequencing of the high oil crop sesame. Genome Biol. 2014;15:R39. doi: 10.1186/gb-2014-15-2-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L., Li D., Zhang Y., Gao Y., Yu J., Wei X., Zhang X. Tolerant and susceptible sesame genotypes reveal waterlogging stress response patterns. PLoS One. 2016;11(3) doi: 10.1371/journal.pone.0149912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarazona S., Garcia-Alcade F., Dopazo J., Ferrer A., Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21(12):4436. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]