Abstract
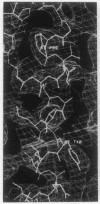
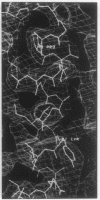
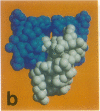
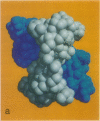
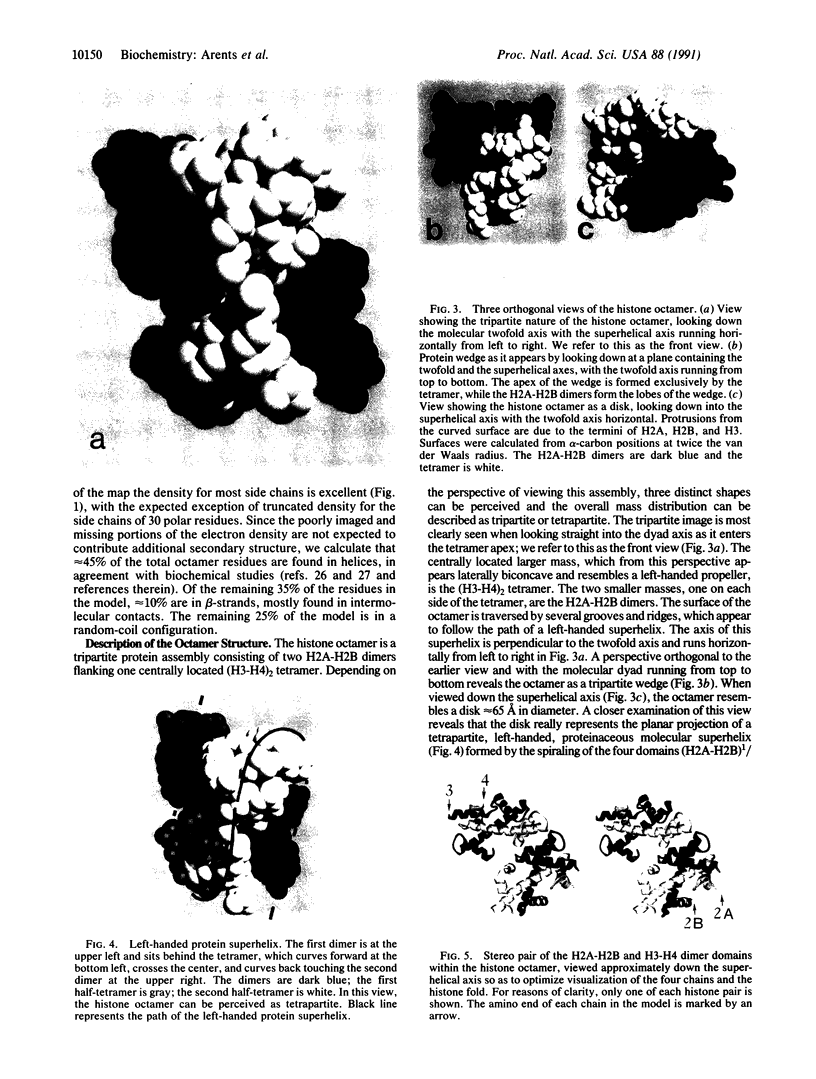
The structure of the octameric histone core of the nucleosome has been determined by x-ray crystallography to a resolution of 3.1 A. The histone octamer is a tripartite assembly in which a centrally located (H3-H4)2 tetramer is flanked by two H2A-H2B dimers. It has a complex outer surface; depending on the perspective, the structure appears as a wedge or as a flat disk. The disk represents the planar projection of a left-handed proteinaceous superhelix with approximately 28 A pitch. The diameter of the particle is 65 A and the length is 60 A at its maximum and approximately 10 A at its minimum extension; these dimensions are in agreement with those reported earlier by Klug et al. [Klug, A., Rhodes, D., Smith, J., Finch, J. T. & Thomas, J. O. (1980) Nature (London) 287, 509-516]. The folded histone chains are elongated rather than globular and are assembled in a characteristic "handshake" motif. The individual polypeptides share a common central structural element of the helix-loop-helix type, which we name the histone fold.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin J. P., Boseley P. G., Bradbury E. M., Ibel K. The subunit structure of the eukaryotic chromosome. Nature. 1975 Jan 24;253(5489):245–249. doi: 10.1038/253245a0. [DOI] [PubMed] [Google Scholar]
- Beaudette N. V., Fulmer A. W., Okabayashi H., Fasman G. D. Study of conformational states and reversibility of histone complexes. Biochemistry. 1981 Nov 10;20(23):6526–6535. doi: 10.1021/bi00526a003. [DOI] [PubMed] [Google Scholar]
- Bentley G. A., Lewit-Bentley A., Finch J. T., Podjarny A. D., Roth M. Crystal structure of the nucleosome core particle at 16 A resolution. J Mol Biol. 1984 Jun 15;176(1):55–75. doi: 10.1016/0022-2836(84)90382-6. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M., Cary P. D., Crane-Robinson C., Rattle H. W. Conformations and interactions of histones and their role in chromosome structure. Ann N Y Acad Sci. 1973 Dec 31;222:266–289. doi: 10.1111/j.1749-6632.1973.tb15268.x. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M., Rattle H. W. Simple computer-aided approach for the analyses of the nuclear-magnetic-resonance spectra of histones. Fractions F1, Fsa1, F2B, cleaved halves of F2B and F2B-DNA. Eur J Biochem. 1972 May 23;27(2):270–281. doi: 10.1111/j.1432-1033.1972.tb01836.x. [DOI] [PubMed] [Google Scholar]
- Brown A. P. Codon-level analysis of histone primary sequence: evidence of a repeat tetrapeptide origin and later inclusion of transcribed sequence. J Theor Biol. 1983 Oct 7;104(3):401–416. doi: 10.1016/0022-5193(83)90114-5. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Burlingame R. W., Love W. E., Moudrianakis E. N. Crystals of the octameric histone core of the nucleosome. Science. 1984 Jan 27;223(4634):413–414. doi: 10.1126/science.6691154. [DOI] [PubMed] [Google Scholar]
- Burlingame R. W., Love W. E., Wang B. C., Hamlin R., Nguyen H. X., Moudrianakis E. N. Crystallographic structure of the octameric histone core of the nucleosome at a resolution of 3.3 A. Science. 1985 May 3;228(4699):546–553. doi: 10.1126/science.3983639. [DOI] [PubMed] [Google Scholar]
- Callaway J. E., DeLange R. J., Martinson H. G. Contact site of histones 2A and 2B in chromatin and in solution. Biochemistry. 1985 May 21;24(11):2686–2692. doi: 10.1021/bi00332a014. [DOI] [PubMed] [Google Scholar]
- Eickbush T. H., Moudrianakis E. N. The compaction of DNA helices into either continuous supercoils or folded-fiber rods and toroids. Cell. 1978 Feb;13(2):295–306. doi: 10.1016/0092-8674(78)90198-8. [DOI] [PubMed] [Google Scholar]
- Eickbush T. H., Moudrianakis E. N. The histone core complex: an octamer assembled by two sets of protein-protein interactions. Biochemistry. 1978 Nov 14;17(23):4955–4964. doi: 10.1021/bi00616a016. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Brown R. S., Richmond T., Rushton B., Lutter L. C., Klug A. X-ray diffraction study of a new crystal form of the nucleosome core showing higher resolution. J Mol Biol. 1981 Feb 5;145(4):757–769. doi: 10.1016/0022-2836(81)90313-2. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Godfrey J. E., Baxevanis A. D., Moudrianakis E. N. Spectropolarimetric analysis of the core histone octamer and its subunits. Biochemistry. 1990 Jan 30;29(4):965–972. doi: 10.1021/bi00456a018. [DOI] [PubMed] [Google Scholar]
- Hendrickson W. A. Stereochemically restrained refinement of macromolecular structures. Methods Enzymol. 1985;115:252–270. doi: 10.1016/0076-6879(85)15021-4. [DOI] [PubMed] [Google Scholar]
- Klug A., Rhodes D., Smith J., Finch J. T., Thomas J. O. A low resolution structure for the histone core of the nucleosome. Nature. 1980 Oct 9;287(5782):509–516. doi: 10.1038/287509a0. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Martinson H. G., True R., Lau C. K., Mehrabian M. Histon-histone interactions within chromatin. Preliminary location of multiple contact sites between histones 2A, 2B, and 4. Biochemistry. 1979 Mar 20;18(6):1075–1082. doi: 10.1021/bi00573a022. [DOI] [PubMed] [Google Scholar]
- Mirzabekov A. D., Shick V. V., Belyavsky A. V., Bavykin S. G. Primary organization of nucleosome core particle of chromatin: sequence of histone arrangement along DNA. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4184–4188. doi: 10.1073/pnas.75.9.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T., Cary P. D., Abercrombie B. D., Crane-Robinson C., Bradbury E. M. A pH-dependent interaction between histones H2A and H2B involving secondary and tertiary folding. Eur J Biochem. 1976 Dec 11;71(2):337–350. doi: 10.1111/j.1432-1033.1976.tb11120.x. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Perez-Grau L., Bordas J., Koch M. H. Chromatin superstructure: synchrotron radiation X-ray scattering study on solutions and gels. Nucleic Acids Res. 1984 Mar 26;12(6):2987–2996. doi: 10.1093/nar/12.6.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAMACHANDRAN G. N., RAMAKRISHNAN C., SASISEKHARAN V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963 Jul;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Strynadka N. C., James M. N. Crystal structures of the helix-loop-helix calcium-binding proteins. Annu Rev Biochem. 1989;58:951–998. doi: 10.1146/annurev.bi.58.070189.004511. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]