Mosquito microbiota provide important physiological and ecological attributes to mosquitoes, including an impact on their susceptibility to pathogens, fitness, and sensitivity to mosquito control agents. Culex nigripalpus mosquito populations transmit various pathogens, including the Saint Louis and West Nile viruses, and proliferate in nutrient-rich environments, such as in wastewater treatment wetlands. Our study examined whether increases in nutrients within larval mosquito developmental habitats impact microbial communities associated with C. nigripalpus mosquitoes. We characterized the effects of organic enrichments on microbiomes associated with C. nigripalpus mosquitoes and identified potential bacterial microbiota that will be further investigated for whether they alter mosquito life history traits and for their potential role in the development of microbial-based control strategies.
KEYWORDS: aquatic chemistry, bacteria, disease vectors, food web, life stages, microbiome, mosquito, pollution
ABSTRACT
Pollution from nutrients in aquatic habitats has been linked to increases in disease vectors, including mosquitoes and other pestiferous insects. One possibility is that changes in mosquito microbiomes are impacted by nutrient enrichments and that these changes affect various traits, including larval development, susceptibility to larval control agents, and susceptibility of the adult mosquitoes to pathogens. We tested this hypothesis using field mesocosms supplemented with low- and high-organic-nutrient regimens and then sampled microbial communities associated with the naturally colonizing Culex nigripalpus mosquito vector. By high-throughput sequencing of 16S rRNA gene sequences, we found no significant differences in overall microbial communities associated with sampled mosquitoes, despite detecting discernible differences in environmental variables, including pH, dissolved oxygen, and nutrient amendments. Nevertheless, indicator species analysis revealed that members of the Clostridiales were significantly associated with mosquitoes that originated from high-nutrient enrichments. In contrast, members of the Burkholderiales were associated with mosquitoes from the low-nutrient enrichment. High bacterial variability associated with the life stages of the C. nigripalpus was largely unaffected by levels of nutrient enrichments that impacted larval microbial resources, including bacteria, ciliates, and flagellates in the larval environments.
IMPORTANCE Mosquito microbiota provide important physiological and ecological attributes to mosquitoes, including an impact on their susceptibility to pathogens, fitness, and sensitivity to mosquito control agents. Culex nigripalpus mosquito populations transmit various pathogens, including the Saint Louis and West Nile viruses, and proliferate in nutrient-rich environments, such as in wastewater treatment wetlands. Our study examined whether increases in nutrients within larval mosquito developmental habitats impact microbial communities associated with C. nigripalpus mosquitoes. We characterized the effects of organic enrichments on microbiomes associated with C. nigripalpus mosquitoes and identified potential bacterial microbiota that will be further investigated for whether they alter mosquito life history traits and for their potential role in the development of microbial-based control strategies.
INTRODUCTION
Nutrient pollution due to excess use of nitrogen and phosphorus can lead to an increased risk of vector-borne diseases (1–5). Previous field studies reported an increase in the abundance of mosquito vectors with an increase in nutrients in mosquito larval developmental sites (6–11). Moreover, increases in nutrient enrichments in mosquito larval developmental sites have been known to reduce the efficacy and persistence of larval control agents (12–15). Higher doses of mosquito larvicides are often required to have a significant reduction in mosquito larval population in organic-rich environments, suggesting a higher economic cost of mosquito control in polluted environments than in less polluted environments.
Nutrient enrichments are generally thought to cause changes in microbial communities, including bacteria, ciliates, flagellates, microalgae, and rotifers that are considered essential for larval mosquito development (9, 10, 16), and could alter aquatic food webs in an unpredictable manner. For example, elevation of nutrients in freshwater streams altered invertebrate predator-prey relationships from linear to curvilinear (17, 18). However, the underlying mechanisms of nutrients and mosquito vector interactions are not fully understood.
Culex nigripalpus Theobald is a major vector of Saint Louis encephalitis virus and is responsible for transmitting other pathogens in the southeastern United States, including West Nile virus (19, 20). Culex nigripalpus is among the dominant mosquito species found during early succession stages of newly developed aquatic habitats, including following rainfall and in polluted treatment wetlands (21–24). Significant genetic variations are known to exist among various populations of C. nigripalpus (25). Variations in abundance among larval developmental sites (21), susceptibility to infection and transmission of pathogens among geographic populations (26), and susceptibility to organophosphate-based pesticides (27) have also been documented. However, little is known about whether nutrient-mediated changes, including water quality variables and microbial consortia found in larval developmental habitats, can influence the mosquito-associated microbiomes for C. nigripalpus developing in different environments.
Bacteria associated with mosquitoes are found to be crucial sources of nutrition for successful larval development (28–31), affect mosquito susceptibility by various pathogens (32–35), impact resistance to pesticides (36, 37), and influence mosquito oviposition (38). As a result, understanding the effects of nutrients on microbial communities associated with mosquitoes is critical for disentangling the underlying causes of variability in disease transmission, variations in mosquito production among various aquatic habitats, and lack of susceptibility to pesticides. In addition, this knowledge is important for the development of novel microbial (e.g., Wolbachia) mosquito control strategies. We hypothesized that different nutrient regimens in larval habitats impact microbial communities associated with mosquitoes developing during the succession of these habitats. In order to test this hypothesis, we characterized microbiota associated with C. nigripalpus developing in two different resource (nutrient) regimens under natural field conditions. In addition, we characterized microbial communities in different life stages of C. nigripalpus to identify potential symbionts associated with all life stages.
RESULTS
Environmental variables in the water column.
The water quality indicators differed significantly between two contrasting larval environments (i.e., high and low nutrients) in outdoor experimental mesocosms (Fig. 1). Significantly lower pH values (Fig. 1A [F1, 4 = 55, P = 0.002]) and dissolved oxygen concentrations (Fig. 1B [F1, 4 = 25, P = 0.007]) were found in high-nutrient mesocosms. In contrast, higher chemical oxygen demand (COD) was found in the high-nutrient mesocosms (mean ± standard error [SE], 239 ± 45.8 mg/liter) compared to the low-nutrient mesocosms (143 ± 8 mg/liter) on day 7, although the difference was not statistically significant (P = 0.107). A similar trend but lower concentration was observed on day 9, with 195 ± 19.8 mg/liter and 150 ± 18.6 mg/liter in the high- and low-nutrient treatments, respectively.
FIG 1 .
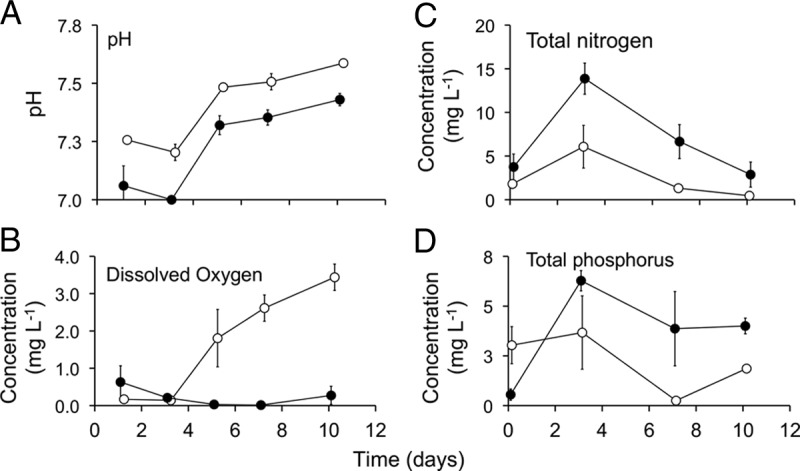
Water quality parameters. Mean ± SE (n = 3) pH, dissolved oxygen, total nitrogen, and total phosphorus in water of the low (○)- and high (●)-nutrient treatments of differentially treated larval habitats (mesocosms). The x axis represents time in days after mesocosms were uncovered and Culex mosquitoes laid egg rafts on water. The y axis denotes the concentration or values of water quality parameters. For example, the mesocosms were exposed to egg-laying female mosquitoes on 2 November 2015.
A higher concentration of total nitrogen (significance tested after Bonferroni correction) was also found in high-nutrient treatments than in the low-nutrient treatments and was variable across time (Fig. 1C [F1, 4 = 5.8, P = 0.07]). Similarly, a higher total phosphorus concentration was found in the high- than in the low-nutrient-treated mesocosms (Fig. 1D [F1, 4 = 4.7, P = 0.12]). Temperature and light intensity in the water column varied temporally but were relatively uniform among mesocosms and were not significantly affected by nutrient enrichments (data not shown).
The abundances of small (0.2- to 1.999-μm equivalent spherical diameter [ESD]) and large (2- to 60-μm ESD) organic particles differed significantly between the two treatments on the first day that the mesocosms were exposed to egg-laying female mosquitoes (Fig. 2). Mean total abundance of small particles was significantly greater in high-nutrient treatments than low-nutrient treatments (Fig. 2A and B [F1, 15 = 10.5, P = 0.005]). Similarly, the mean abundance of large particles was approximately 5-fold greater in the high-nutrient treatments than in the low-nutrient treatments (Fig. 2C and D [F1, 15 = 14.6, P = 0.002]).
FIG 2 .
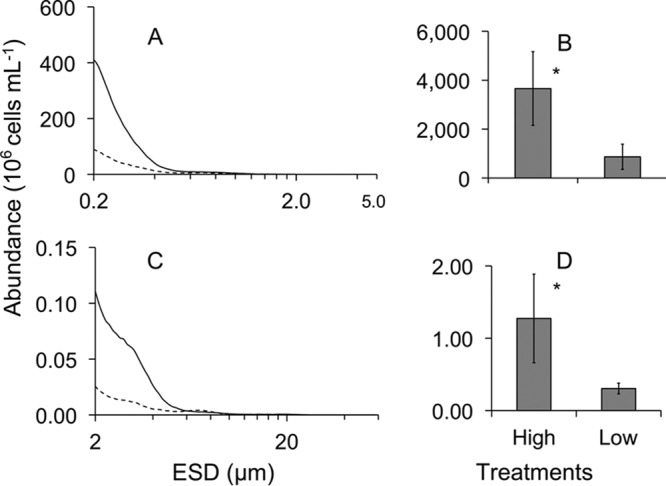
Heterotrophic and autotrophic particle (cell) abundance in water column. Small (0.2- to 1.999-μm equivalent spherical diameter [ESD]) (A) and large (2- to 60-μm ESD) (C) particle size distribution in high (solid lines)- and low (dashed lines)-organic-nutrient-enriched mesocosms, and mean ± SE total particle abundance (n = 3) of small (B) and large (D) particle sizes in water of low- and high-nutrient treatments on day 0.
Microeukaryote abundance.
Microscopic examination of water samples for microeukaryotes (i.e., ciliates, flagellates, and rotifers) revealed a significant difference in the combined abundance of these microorganisms between the two treatments (Fig. 3 [F1, 16 = 24.1, P = 0.0002]) and varied across time (F2, 15 = 4.3, P = 0.04). Significantly greater numbers of microeukaryotes were found in high-nutrient treatments on days 0 and 7, but that difference decreased by day 9.
FIG 3 .
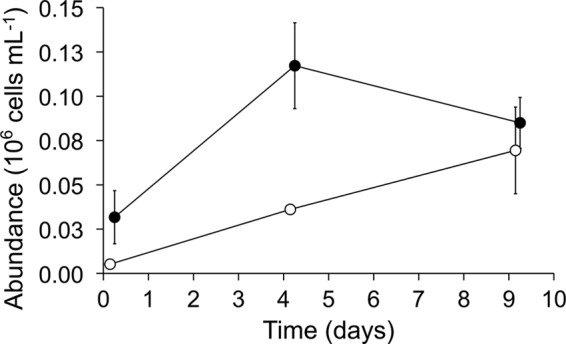
Microeukaryote abundance in water column. Mean ± SE (n = 3) abundance of ciliate protists, flagellates, and rotifers in the mesocosms with high (●)- and low (○)-nutrient treatments on days 0, 4, and 9 after egg laying by mosquitoes. Error bars not seen are contained within the symbols.
Mosquito larva abundance.
Microscopic examination of the dipper samples revealed no significant differences in total Culex larval abundance between the two treatments at either 5 days or 7 days after the mesocosms were exposed to naturally occurring mosquitoes (F1, 4 = 0.006, P = 0.9). The mean ± SE number of C. nigripalpus larvae found in the low-nutrient treatments was 41 ± 10, compared with the high-nutrient treatments, with 32 ± 27 larvae per dip sample. Very few individuals of the southern house mosquito Culex quinquefasciatus Say were observed on this sampling date in the low (average; <1 larvae)- and high (~4 larvae per dipper sample)-nutrient treatments. A similar trend was observed a week after uncovering the mesocosms (data not shown).
Diversity of bacterial communities in mosquitoes from different nutrient treatments.
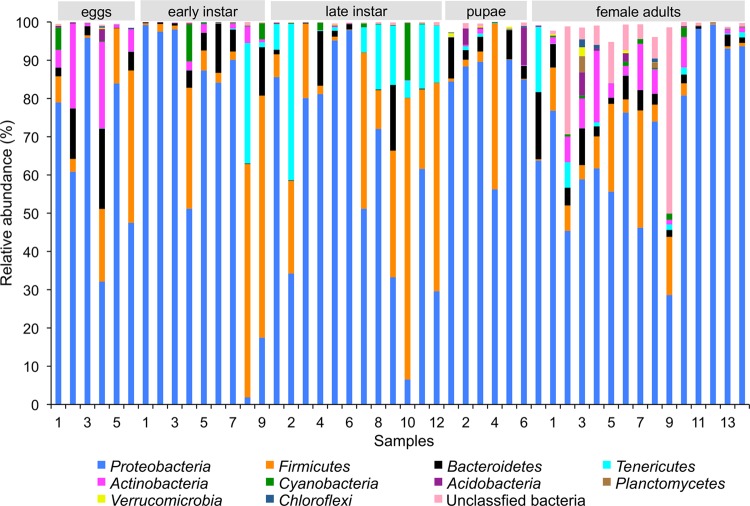
A total of 4,859,297 sequences in 1,751 operational taxonomic units (OTUs) were generated from 48 mosquito samples, including 6 half-egg rafts, 9 early and 12 late instar larvae, 7 pupae, and 14 newly (<1 day after eclosion) emerged non-blood-fed female adults of C. nigripalpus. Assembled and quality-checked sequences had a mean length of 416 bases with a mean overlap of 49.8 bases. On average, 83,408 sequences from egg rafts, 106,809 from early instar larvae, and 116,281 from late instar larvae, 89,430 from pupae, and 83,107 from female adults per sample were obtained. These were classified into 28 bacterial phyla with Proteobacteria, Firmicutes, Bacteroidetes, Tenericutes, Actinobacteria, and Acidobacteria dominating the bacterial phyla found associated with this mosquito species (Fig. 4). Proteobacteria accounted for nearly 70% of sequences, followed by Firmicutes, with 15% of all sequences. Approximately 56% of the sequences were identified as 240 genera, with Arcobacter (Epsilonproteobacteria: Campylobacteraceae) being the most abundant (29%) genus, followed by Thorsellia (7%). Few archaeal sequences (0.002%) were recovered from this mosquito species, and those were primarily from female adults.
FIG 4 .
Dominant bacterial phyla found in different stages. Shown are bacterial phyla in eggs, early and late larval instars, pupae, and newly emerged female adults of Culex nigripalpus developed under the field conditions. Only phyla with an average abundance of >0.1% were included. Other unclassified sequences accounted for 0.3%. Archaea and an additional 16 phyla accounted for <0.1%. Egg samples 1, 4, and 5, early instar samples 7, 10, 11, and 18, late instar samples 14, 18, 20, 41, 42, 46, and 47, pupa samples 20 and 38, and female adult samples 32 to 36 were taken from high-nutrient regimens. The remaining 27 samples were derived from low-nutrient regimens.
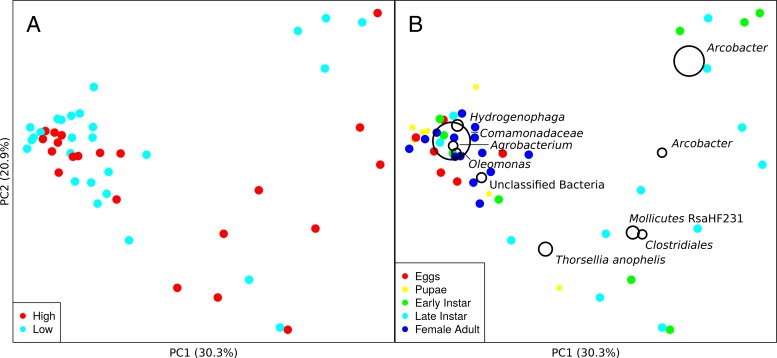
The principle coordinate (PCoA) ordinations based on weighted UniFrac measures revealed no significant differences in bacterial community composition found among mosquitoes developing in low- and high-nutrient treatments (Fig. 5A [multiresponse permutation procedure [MRPP; A = 0.005; P = 0.081]). The greatest variation (indicated by principal coordinate 1 [PC1]) detected among samples was attributed to the higher abundance of OTUs corresponding to Arcobacter (OTU 0) and an unidentified species of Comamonadaceae (OTU 2). Arcobacter (Epsilonproteobacteria) was mostly associated with larvae of C. nigripalpus, whereas the unidentified species in Comamonadaceae (Betaproteobacteria) was found associated with all life stages of C. nigripalpus, including egg rafts. Differences in bacterial communities among larval samples from low- and high-nutrient treatments were not significant (MRPP; A = 0.018; P = 0.084).
FIG 5 .
PCoA ordination based on the weighted UniFrac distance metric. Shown is ordination of microbial communities associated with Culex nigripalpus mosquitoes colored by either nutrient treatment level (A) or life stage (B). The effects of nutrients on microbial communities associated with the mosquitoes were not substantial (MRPP; A = 0.005, P = 0.08) between the two treatments. However, the microbiota in the different life stages differed weakly but significantly (MRPP; A = 0.08, P = 0.001). Large open circles indicate the 10 most abundant bacterial taxa associated with samples.
Indicator species analysis revealed that members of Clostridiales dominated bacterial communities associated with mosquitoes developing in high-nutrient regimens (indicator values = 0.5 to 0.8; P ≤ 0.01), whereas mosquitoes from low-nutrient treatments were enriched with Burkholderiales (see Table S1 in the supplemental material [indicator value = 0.48; P < 0.01]). Although differences among stages were apparent, indicator species analysis by life stages revealed that Comamonadaceae OTUs were strongly associated with all life stages, except pupal samples (see Table S1 and Fig. S1 in the supplemental material).
Indicator bacterial species (indicator value, ≥0.4; P < 0.05) associated with Culex nigripalpus Theobald developing in low- and high-organic-nutrient enrichments. Download TABLE S1, PDF file, 0.1 MB (25.7KB, pdf) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of the 16 most abundant (>1,000 sequences per sample) members of families recovered from different life stages of field-reared Culex nigripalpus mosquitoes. Others include 148 families (660,633 sequences) that had sequence abundance of <1,000 sequences per sample. The remaining 747 OTUs (692,057 sequences) were unidentified to families. Download FIG S1, EPS file, 0.3 MB (274.5KB, eps) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Nonmetric multidimensional scaling (NMDS) analysis based on Bray-Curtis distance measures revealed significant differences between microbiota samples that originated from different life stages of mosquitoes (Fig. 5B; see Fig. S2 in the supplemental material [MRPP; A = 0.08; P = 0.001]). Microbial samples from female adults were significantly separated from the immature stages and egg rafts of C. nigripalpus.
Nonmetric multidimensional scaling (NMDS) plot of bacterial communities from different life stages of field-reared Culex nigripalpus mosquitoes. Download FIG S2, PDF file, 0.1 MB (13.1KB, pdf) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Nine core taxa were found in all samples of all life stages of C. nigripalpus, including Thorsellia anophelis OTU 4, Oleomanas OTU 7, two unidentified species (OTUs 1 and 2) of Comamonadaceae, two Hydrogenophaga species (OTUs 18 and 1537), an unidentified species (OTU 71) of Cyanobacteria, an unidentified species (OTU 8) of Firmicutes, and an unidentified species (OTU 3) of Tenericutes (see Fig. S3 in the supplemental material).
Relative abundance of the 22 most abundant (average abundance of ≥1,000 sequences per sample) OTUs recovered from different life stages of field-reared Culex nigripalpus mosquitoes. Others include OTUs with average abundance of <1,000 sequences per sample and include 1,729 OTUs with a total of 1,968,535 sequences. The lowest taxonomic classifications of OTUs are presented. Download FIG S3, EPS file, 0.3 MB (281.2KB, eps) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial communities in egg rafts of Culex nigripalpus.
Bacterial sequences from egg rafts of C. nigripalpus grouped into 530 OTUs that were dominated by an unknown species (OTU 2) of Comamondaceae (35%), followed by Agrobacterium OTU 10 (12%) (see Table S2 and Fig. S3 in the supplemental material).
Indicator bacterial species (indicator value, ≥0.5; P < 0.05) associated with different life stages (egg to adults) of Culex nigripalpus. Download TABLE S2, PDF file, 0.1 MB (34.9KB, pdf) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial communities in immature stages of Culex nigripalpus.
An Epsilonproteobacteria member, Arcobacter (31%), and two OTUs corresponding to species in Betaproteobacteria (an unknown species in Comamondaceae [14%] and Vogesella [Neisseriaceae]) dominated bacterial communities associated with early larval instars. Thorsellia anophelis was also recovered from the early instar stage but in a much lower (<1%) proportion (see Table S3 in the supplemental material). Bacteria in late instar larvae were also dominated by Arcobacter (27%), Thorsellia anophelis (10.5%), and an unknown genus of Mollicutes (10%). Hydrogenophaga (14%), Thorsellia (11%), and an unknown species of Comamondaceae (10%) dominated bacterial communities in pupal samples.
Relative abundance (indicator value, ≥0.5; P ≥ 0.01) of bacterial taxa associated with developmental stages of Culex nigripalpus Theobald. Asterisks represent taxa that are either found in a lower proportion (i.e., <0.01 relative abundance per sample) or absent. Download TABLE S3, PDF file, 0.1 MB (32.2KB, pdf) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial communities in newly emerged Culex nigripalpus female adults.
Bacteria in newly emerged (<12 h after eclosion) non-blood-fed female adults reared from egg to adults outdoors were enriched with an unknown species (OTU 2) of Comamondaceae (20%), Oleomonas (7%), and Arcobacter (4.6%) (Table S3). Wolbachia was also found in 2 of the 14 adult samples, constituting 91% and <1% of their respective sequences but was absent in 12 other samples. These results suggest a likely inclusion of Culex quinquefasciatus, which is a known host of Wolbachia in those samples during DNA extraction. Other notable species were recovered at lower proportions (<1%) and include Thorsellia anophelis.
DISCUSSION
Effects of organic enrichments on microbial communities associated with Culex nigripalpus.
We tested the hypothesis that organic nutrient enrichment, a primary factor for eutrophication and pollution, would alter the microbial larval resources and thereby impact microbial communities associated with Culex disease vectors in replicated outdoor mesocosm experiments. Results of our study revealed that a significant increase in abundance of sestonic particles and planktonic microeukaryotes (i.e., ciliates, flagellates, and rotifers) in treatments with high-nutrient enrichments were in agreement with the bottom-up resource hypothesis: i.e., increasing nutrients will increase the abundance of both autotrophic and heterotrophic microorganisms (39, 40). This was further corroborated with the increase in chemical oxygen demand, a predictor of the amount of organic material available for oxidation, and microbial consumption in the high-nutrient treatments compared to low-nutrient treatments in this study. Total concentrations of nutrients increased immediately following the uncovering of the mesocosms, suggesting autotrophic and aerobic microbial colonization of the mesocosms.
Despite the apparent differences in microbiota and chemical variables in the water column (Fig. 1 to 3), microbial communities associated with mosquitoes developing in these two larval environments were not affected significantly. This could be due to a combination of factors, including food web-mediated factors, such as differences in the abundance of planktonic bacteriovores (i.e., flagellates, ciliates, and rotifers), and the remarkable variability in microbial communities among samples within each treatment group. Predation of bacteria by planktonic bacteriovores has been known to be intense (41), and these groups might have affected the bacterial diversity in the water column, a feeding zone of Culex mosquitoes. Alternatively, Culex larvae are considered omnivores, feeding on a variety of lower trophic microorganisms, including bacteria (16), and therefore the impact on bacterial diversity in the mosquitoes may not necessarily be affected by increases in bottom-up resources. A previous study of microbiota in other Culex mosquitoes sampled from different habitats with different nutrient concentrations (influenced by larval control treatments) also did not reveal significant differences in the bacterial communities in the larvae sampled from the different larval habitats (42–44).
Increases in nutrients such as nitrogen and phosphorus in aquatic habitats, as a result of runoff from agricultural practices and other nonpoint sources, have been reported to influence the abundance of disease vectors, particularly Culex mosquitoes that are more adapted to polluted environments (6, 8, 45, 46). In addition, organic enrichments have been shown to influence mosquito control strategies. For example, under high-organic-rich environments, the efficacy of a fungal biological control agent, Lagenidium giganteum, was significantly reduced (12). Other studies reported that increase in organic or inorganic mater in the water column negatively influenced the efficacy of the commonly used Bacillus-based larval control agents (13, 47). It is likely that the diverse microbial communities and sestonic particles ingested by mosquito larvae might provide immunity or protection of midgut epithelium that is considered a primary target of Bacillus thuringiensis serovar israelensis (Bti) toxins in polluted environments (13).
Mosquitoes, in general, and Culex mosquito vectors, in particular, are considered primary colonizers of newly created freshwater aquatic habitats and are well adapted to polluted environments (42, 46, 48). The difference in abundances of C. nigripalpus between treatments was not significant, suggesting that mosquito abundance was not influenced by water column nutrient concentrations, especially during the initial colonization of newly formed aquatic habitats. This study and others have shown that C. nigripalpus mosquitoes prefer to lay their eggs and develop in highly eutrophic habitats than their C. quinquefasciatus congeners (21, 23). Culex quinquefasciatus was considered to be the dominant species colonizer of eutrophic habitats (9, 24), but its abundance during the succession in our mesocosms was negligible compared to that of C. nigripalpus.
Bacterial communities associated with different life stages of Culex nigripalpus.
Microbial communities associated with C. nigripalpus sampled during the autumn varied significantly among life stages from the same cohort. Bacterial communities from female adults, eggs, and pupae were dominated by Alphabacteria and Betaproteobacteria, whereas bacteria from larvae were dominated by Arcobacter (Epsilonproteobacteria), Hydrogenophaga, and Agrobacterium (Alphaproteobacteria), Thorsellia (Gammaproteobacteria), and Clostridium (Firmicutes). Arcobacter is ubiquitous in aquatic environments and, as a member of Epsilonproteobacteria, might be associated with sulfur cycling (49, 50). The mesocosms used in this study were filled with well water, which contains a relatively high sulfur concentration (~100 mg sulfate/liter [D. Duguma, unpublished data]). Nearly 92% Arcobacter sequences were from larvae, whereas very few were found associated with eggs (0.4%), pupae (2.8%), and adults (4.6%), suggesting that Arcobacter found associated with the mosquitoes in this study might be waterborne and thus ingested by the mosquito larvae. Several Arcobacter spp. are known to be pathogenic to humans and animals (51) and associated with polluted environments (52). Although we have not ruled out experimentally that this bacterium can be transstadially transmitted across life stages or is a pathogen, Arcobacter found associated with pupae and adult C. nigripalpus mosquitoes was likely ingested by the larvae from the water and passed down to pupae and adults. Considering that members of this genus of bacteria are recognized as emerging pathogens to humans and animals (51), the recovery of a relatively small proportion (e.g., 4.6%) of Arcobacter sequences in female adults suggests further study, including a possibility that these bacteria might be harbored in salivary glands, with a direct implication for pathogen transmission (53). Salivary glands have been shown to harbor diverse bacterial communities (53).
Agrobacterium OTUs were found in all samples, from egg to adult stages of C. nigripalpus, with the highest abundance found associated with the eggs. Members of this genus are known to transfer genetic materials between themselves and other eukaryotes, such as plants (54), and this genus was among the dominant genera recovered from Aedes mosquitoes (55). Although we rinsed the egg rafts multiple times with distilled water, it is possible that some of the communities found associated with eggs might have been unintentionally cosampled from the water surface during the sampling of the egg rafts. Future studies will investigate whether some of the communities found associated with eggs are obligate symbionts.
Thorsellia anophelis was also found in C. nigripalpus, with the greatest abundance of this bacterium found in pupae (11.0%) and in late instar larvae (10.5%). The abundance of this species was considerably lower in eggs (0.02%), early instars (0.5%), and adults (0.14%), corroborating previous studies that this symbiont is likely ingested by larvae and transferred to the subsequent developmental stages (43, 56–58). The dominance and persistence of Thorsellia spp. in life stages of Culex mosquitoes in this study and in previous studies (43, 44) and Anopheles (56) mosquitoes suggest a strong consideration for the development of paratransgenic mosquito control (59).
In conclusion, differences in environmental habitat variations might not affect the internal bacterial communities associated with Culex mosquito vectors, which instead may be influenced by seasonal variations. For the first time, we identified microbial communities associated with C. nigripalpus across developmental stages and identified potential candidates that will be further investigated for their role in bionomics and control of this mosquito species.
MATERIALS AND METHODS
Mesocosm experiment.
Our experimental design involved two contrasting larval environments in outdoor experimental mesocosms during autumn 2015. Two different larval environmental conditions were created on 27 October 2015 by adding two nutrient regimens: 0.2 and 1% (wt/vol) (low and high, respectively) rabbit food (alfalfa pellets) to three replicated outdoor mesocosms filled with 378 liters of well water at the University of Florida, Florida Medical Entomology Laboratory (see Fig. S4 in the supplemental material). The surface area and depth of water were 0.85 m2 and 0.5 m, respectively. Outdoor mesocosms can be used to examine various ecological hypotheses, including the effects of nutrients and climate change on aquatic food webs (18, 42, 43, 60, 61). Alfalfa-based organic matter is commonly used to attract egg-laying female mosquitoes and supports the production of Culex mosquitoes for longer periods of time (9, 38, 43, 62). The organic matter was allowed to ferment in the mesocosms for ~1 week while covered with a tarp. Natural oviposition by Culex mosquitoes occurred in all mesocosms <24 h after uncovering the mesocosms (i.e., on 2 November 2015). Two egg rafts likely laid by two female Culex nigripalpus mosquitoes from each of the six mesocosms were sampled on day 1 (3 November 2015). One egg raft laid by an individual mosquito sampled from each of the mesocosms was placed in modified BioQuip mosquito-rearing chambers (see Fig. S5 in the supplemental material) and then submerged in each of the six mesocosms to allow access to larval microbial food resources and development of these mosquitoes under the field conditions. The submerged portion of the device has screen meshes (300 nylon) built into each mosquito breeder (BioQuip, Inc., Rancho Dominguez, CA, USA) to allow access to larval resources in the water column, whereas the above water portion of the device captures adults emerging from the same cohort of eggs. The second egg raft taken from each of the mesocosms was taken to the laboratory, triple rinsed with distilled water, and aseptically cut into two halves. One-half of the rafts from each of the containers were preserved in 95% ethanol for DNA extraction, while the remaining halves were placed in 200 ml of distilled water in sterile plastic cups and then allowed to hatch in an environmental chamber at a temperature of 27°C for positive morphological identification of the larvae to Culex nigripalpus.
Schematic sketch of outdoor mosquito-rearing mesocosms at the Florida Medical Entomology Laboratory. Mesocosms 1, 10, and 13 received high-nutrient enrichment, whereas 2, 9, and 14 received low-nutrient enrichment. Download FIG S4, PDF file, 0.2 MB (259.6KB, pdf) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Modified BioQuip environmental floating chamber for mosquito rearing under outdoor field conditions. Download FIG S5, PDF file, 0.2 MB (184.5KB, pdf) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mosquito and water sampling.
Samples of two (early and late instars) Culex nigripalpus larval stages, pupae, and adult mosquitoes developed from the same egg rafts in the BioQuip rearing chambers were taken on different days (see Table S4 in the supplemental material), preserved in 95% ethanol, and stored at −20°C until DNA extraction. In addition, mosquito larval samples were taken in five 350-ml standard dips from each of the mesocosms at days 7 and 9 after the mesocosms were exposed to egg-laying female mosquitoes to determine the identity and abundance of mosquitoes found in the mesocosms.
Sampling schedule of Culex nigripalpus mosquitoes developing in outdoor aquatic mesocosms for DNA extraction. Column heads represent mesocosm identification numbers, with mesocosms 1, 3, and 10 receiving high-nutrient treatments and 2, 9, and 14 receiving low-nutrient-treatment regimens. Numbers 1 through 3 in cells indicate the number of mosquitoes or the half of the six egg rafts used for DNA extraction. Asterisks indicate two groups of 3 larvae that were sampled on those dates. —, sampling was not carried out due to lack of appropriate life stages of mosquitoes. Download TABLE S4, PDF file, 0.1 MB (34.3KB, pdf) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Water samples were taken in 250-ml amber plastic bottles on days 2, 7, and 9 after mosquitoes colonized the mesocosms to determine total nitrogen, phosphorus, and chemical oxygen demand (COD) in the water column using a Hach DR3900 spectrophotometer (Hach Company, Loveland, CO). The water samples integrated both the surface water and ~10 cm below the surface of the water and were collected from the center of the mesocosm. Dissolved oxygen and pH in the water column were determined in situ using a YSI Professional Plus multiparameter instrument (YSI, Inc., Yellow Spring, OH). Temperature and light intensity in the mesocosms were monitored continuously using a Hobo Pendant temperature/light data logger (Onset Computer Corp., Bourne, MA).
Water samples were collected in duplicate 50-ml sterile centrifuge tubes on days 0, 4, and 9 after the mesocosms were opened and preserved with 1% Lugol’s iodine solution to quantify the abundance of microeukaryotes (i.e., ciliates, flagellates, and rotifers) found in the water column. The microeukaryotes were counted by direct microscopy on a hemocytometer using a Leica inverted microscope (Leica Microsystems, Inc., Buffalo Grove, IL). Particle size distributions for small (0.2- to 1.999-μm ESD) and large (2- to 60-μm ESD) particles that include both heterotrophic and autotrophic communities were determined using a Multisizer 4E Coulter Counter particle size analyzer (Beckman Coulter, Inc., Miami, FL) by a previously published procedure (42).
DNA extraction, PCR, and MiSeq Illumina library preparation.
The general scheme of this study followed procedures described in previous studies (43, 63). Briefly, pooled DNA samples from 1 to 3 individuals from each of the life stages of C. nigripalpus were extracted using the DNeasy blood and tissue kit following the manufacturer’s protocol (Qiagen, Valencia, CA) in a laminar flow hood. Prior to DNA extraction, mosquitoes were surface sterilized with 95% ethanol and rinsed three times using molecular biology-grade UltraPure water (Quality Biological, Inc., Gaithersburg, MD). The samples were gently vortexed for 10 s in between rinsing. The mosquitoes were left to air dry under a laminar flow hood before extraction. Pooling of individual insects for microbial analyses have been used routinely in characterization of community profiles in insects (43, 44, 64). Pooling of individuals may have several advantages, including maximizing the sequence yield per sample (above negative controls) to discern microbial community differences between treatment samples in insects (64). We also extracted DNA from egg rafts to determine if there were maternally transmitted symbionts (e.g., Wolbachia) or unknown symbionts were present in this species. DNA from adult males was not extracted in this study because males have no known significance in transmitting pathogens.
The PCR procedures, sequence assembly, and analyses followed previous procedures described in other studies (65–67). Briefly, ~460-bp amplicons were generated using PCR from the V3 and V4 regions of 16S rRNA genes using Pro341F and Pro805R, which target both bacteria and archaea (68). Amplicons from each of the samples and replicate no-template controls were tagged with unique 6-base barcodes, amplified using Illumina-specific primers, and sequenced according to a previously established protocol (67) with some modification. The modification included a second round of PCR with 15 cycles for samples with low amplification on the first round of PCR. In brief all PCR products were combined and subjected to 250-bp end sequencing (Reagent kit v2, 500) on a MiSeq (Illumina, San Diego, CA).
Data analysis.
Using AXIOME to manage sequences analysis (69), 16S rRNA gene reads were assembled by PANDAseq version 2.10 (70), with a quality threshold of 0.9 (which rejects sequences with low-quality scores), a minimum overlap of 10 bases, and a minimum assembled length of 100 bases, and sequences with ambiguous nucleotides were rejected. Operational taxonomic units (OTUs) were picked at 97% identity using the UPARSE algorithm USEARCH version 7.0.1090 (71) with de novo chimera checking. Taxonomic classification was performed on the representative sequence of each OTU using RDP version 2.2 (72) via QIIME (73), trained against the Greengenes (August 2013 revision) (74) reference set with a minimum posterior probability of 80%. Sequences were rarefied to the lowest number of sequences per sample (i.e., 43,611) for alpha and beta diversity analyses. To determine microbial community differences among mosquito samples originating from high- and low-nutrient treatments, principal coordinate analysis (PCoA) and nonmetric multidimensional scaling (NMDS) ordinations, based on the Bray-Curtis dissimilarity measures, were conducted using the vegan R package version 2.2-0 (75). In addition, a PCoA ordination based on UniFrac distance measures was carried out with QIIME to determine bacterial community differences among samples of multiple treatments and life stages. Multiresponse permutation procedures (MRPP) were used to test differences among sample groups based on distance measures. Core bacterial taxa were determined based on OTUs represented by at least one sequence per sample in all samples (76).
Repeated-measures analysis of variance (ANOVA) using JMP (77) was conducted to assess differences in environmental variables (e.g., nutrients, pH, dissolved oxygen, and microeukaryotes) and larval mosquito abundance in the water column between the two treatments. One-way ANOVA was performed to assess differences in mean abundance of total counts of small and large sestonic particles. Means were separated by Tukey’s test at P < 0.05, after performing Bonferroni correction on calculated P values.
Accession number(s).
All sequence data for this study were submitted to the European Bioinformatics Institute under accession no. PRJEB17885.
ACKNOWLEDGMENTS
Katja Engel, Daniel Velez, Sara Ortiz, and Arthur Simas-Domingos are thanked for their technical assistance. We thank James Newman for sketching schematic diagrams of the mesocosms, James McNealy for suggestions on the design of the modified BioQuip mosquito breeder, and Barry Alto for valuable comments on the manuscript.
D.D. conceived and designed the study. D.D. performed the experiments. D.D., M.W.H., and J.D.N. analyzed the data. D.D., J.D.N., and C.T.S. contributed reagents. D.D., M.W.H., C.T.S., and J.D.N. wrote the manuscript. All authors read and approved the final version of the manuscript.
D.D. was supported by funding from Florida Department of Agriculture and Consumer Services (project no. 00123786). J.D.N. was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC). Publication of this article was funded in part by the University of Florida Open Access Publishing Fund. The funders had no role in study design, data collection and interpretation, nor the decision to submit the work for publication.
REFERENCES
- 1.Johnson PT, Townsend AR, Cleveland CC, Glibert PM, Howarth RW, McKenzie VJ, Rejmankova E, Ward MH. 2010. Linking environmental nutrient enrichment and disease emergence in humans and wildlife. Ecol Appl 20:16–29. doi: 10.1890/08-0633.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camargo JA, Alonso A. 2006. Ecological and toxicological effects of inorganic nitrogen pollution in aquatic ecosystems: a global assessment. Environ Int 32:831–849. doi: 10.1016/j.envint.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Sanford MR, Chan K, Walton WE. 2005. Effects of inorganic nitrogen enrichment on mosquitoes (Diptera: Culicidae) and the associated aquatic community in constructed treatment wetlands. J Med Entomol 42:766–776. doi: 10.1093/jmedent/42.5.766. [DOI] [PubMed] [Google Scholar]
- 4.Sunish IP, Reuben R. 2001. Factors influencing the abundance of Japanese encephalitis vectors in ricefields in India. I. Abiotic. Med Vet Entomol 15:381–392. doi: 10.1046/j.0269-283x.2001.00324.x. [DOI] [PubMed] [Google Scholar]
- 5.Victor TJ, Reuben R. 2000. Effects of organic and inorganic fertilisers on mosquito populations in rice fields of southern India. Med Vet Entomol 14:361–368. doi: 10.1046/j.1365-2915.2000.00255.x. [DOI] [PubMed] [Google Scholar]
- 6.Duguma D, Walton WE. 2014. Effects of nutrients on mosquitoes and an emergent macrophyte, Schoenoplectus maritimus, for use in treatment wetlands. J Vector Ecol 39:1–13. doi: 10.1111/j.1948-7134.2014.12063.x. [DOI] [PubMed] [Google Scholar]
- 7.Awolola TS, Oduola AO, Obansa JB, Chukwurar NJ, Unyimadu JP. 2007. Anopheles gambiae s.s. breeding in polluted water bodies in urban Lagos, southwestern Nigeria. J Vector Borne Dis 44:241–244. [PubMed] [Google Scholar]
- 8.Lund A, McMillan J, Kelly R, Jabbarzadeh S, Mead DG, Burkot TR, Kitron U, Vazquez-Prokopec GM. 2014. Long term impacts of combined sewer overflow remediation on water quality and population dynamics of Culex quinquefasciatus, the main urban West Nile virus vector in Atlanta, GA. Environ Res 129:20–26. doi: 10.1016/j.envres.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Rodcharoen J, Mulla MS, Chaney JD. 1997. Organic enrichment of breeding sources for sustained productivity of mosquitoes (Diptera: Culicidae). J Vector Ecol 22:30–35. [PubMed] [Google Scholar]
- 10.Nguyen TT, Su T, Mulla MS. 1999. Bacteria and mosquito abundance in microcosms enriched with organic matter and treated with a Bacillus thuringiensis subsp. israelensis formulation. J Vector Ecol 24:191–201. [PubMed] [Google Scholar]
- 11.Chaves LF, Keogh CL, Vazquez-Prokopec GM, Kitron UD. 2009. Combined sewage overflow enhances oviposition of Culex quinquefasciatus (Diptera: Culicidae) in urban areas. J Med Entomol 46:220–226. doi: 10.1603/033.046.0206. [DOI] [PubMed] [Google Scholar]
- 12.Jaronski ST, Axtell RC. 1982. Effects of organic water pollution on the infectivity of the fungus Lagenidium giganteum (Oomycetes: Lagenidiales) for larvae of Culex quinquefasciatus (Diptera: Culicidae): field and laboratory evaluation. J Med Entomol 19:255–262. doi: 10.1093/jmedent/19.3.255. [DOI] [PubMed] [Google Scholar]
- 13.Ben-Dov E, Saxena D, Wang Q, Manasherob R, Boussiba S, Zaritsky A. 2003. Ingested particles reduce susceptibility of insect larvae to Bacillus thuringiensis. J Appl Entomol 127:146–152. doi: 10.1046/j.1439-0418.2003.00732.x. [DOI] [Google Scholar]
- 14.Mulla M, Darwazeh HA, Davidson EW, Dulmage HT. 1984. Efficacy and persistence of the microbial agent Bacillus sphaericus against mosquito larvae in organically enriched habitats. Mosq News 44:166–173. [Google Scholar]
- 15.Margalit J, Bobroglo H. 1984. The effect of organic materials and solids in water on the persistence of Bacillus thuringiensis var. israelensis serotype H-14. Z Angew Entomol 97:516–520. [Google Scholar]
- 16.Merritt RW, Dadd RH, Walker ED. 1992. Feeding behavior, natural food, and nutritional relationships of larval mosquitoes. Annu Rev Entomol 37:349–376. doi: 10.1146/annurev.en.37.010192.002025. [DOI] [PubMed] [Google Scholar]
- 17.Davis JM, Rosemond AD, Eggert SL, Cross WF, Wallace JB. 2010. Long-term nutrient enrichment decouples predator and prey production. Proc Natl Acad Sci U S A 107:121–126. doi: 10.1073/pnas.0908497107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hulot FD, Lacroix G, Lescher-Moutoué F, Loreau M. 2000. Functional diversity governs ecosystem response to nutrient enrichment. Nature 405:340–344. doi: 10.1038/35012591. [DOI] [PubMed] [Google Scholar]
- 19.Day JF, Stark LM. 2000. Frequency of Saint Louis encephalitis virus in humans from Florida, USA: 1990–1999. J Med Entomol 37:626–633. doi: 10.1603/0022-2585-37.4.626. [DOI] [PubMed] [Google Scholar]
- 20.Rutledge CR, Day JF, Lord CC, Stark LM, Tabachnick WJ. 2003. West Nile virus infection rates in Culex nigripalpus (Diptera: Culicidae) do not reflect transmission rates in Florida. J Med Entomol 40:253–258. doi: 10.1603/0022-2585-40.3.253. [DOI] [PubMed] [Google Scholar]
- 21.O’Meara GF, Cutwa-Francis M, Rey JR. 2010. Seasonal variation in the abundance of Culex nigripalpus and Culex quinquefasciatus in wastewater ponds at two Florida dairies. J Am Mosq Control Assoc 26:160–166. doi: 10.2987/09-5971.1. [DOI] [PubMed] [Google Scholar]
- 22.Day JF, Curtis GA. 1989. Influence of rainfall on Culex nigripalpus (Diptera: Culicidae) blood-feeding behavior in Indian River County, Florida. Ann Entomol Soc Am 82:32–37. doi: 10.1093/aesa/82.1.32. [DOI] [Google Scholar]
- 23.Rey JR, O’Meara GF, O’Connell SM, Cutwa-Francis MM. 2006. Factors affecting mosquito production from stormwater drains and catch basins in two Florida cities. J Vector Ecol 31:334–343. doi: 10.3376/1081-1710(2006)31[334:FAMPFS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 24.Wermelinger ED, Benigno CV, Machado RN, Cabello PH, Meira AM, Ferreira AP, Zanuncio JC. 2012. Mosquito population dynamic (Diptera: Culicidae) in a eutrophised dam. Braz J Biol 72:795–799. doi: 10.1590/S1519-69842012000500003. [DOI] [PubMed] [Google Scholar]
- 25.Nayar JK, Knight JW, Munstermann LE. 2002. Temporal and geographic genetic variation in Culex nigripalpus Theobald (Culicidae: Diptera), a vector of St. Louis encephalitis virus, from Florida. J Med Entomol 39:854–860. doi: 10.1603/0022-2585-39.6.854. [DOI] [PubMed] [Google Scholar]
- 26.Sardelis MR, Turell MJ, Dohm DJ, O’Guinn ML. 2001. Vector competence of selected North American Culex and Coquillettidia mosquitoes for West Nile virus. Emerg Infect Dis 7:1018–1022. doi: 10.3201/eid0706.010617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boike AH Jr, Rathburn CB Jr, Floore TG, Rodriguez HM, Coughlin JS. 1989. Insecticide tolerance of Culex nigripalpus in Florida. J Am Mosq Control Assoc 5:522–528. [PubMed] [Google Scholar]
- 28.Coon KL, Vogel KJ, Brown MR, Strand MR. 2014. Mosquitoes rely on their gut microbiota for development. Mol Ecol 23:2727–2739. doi: 10.1111/mec.12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Díaz-Nieto LM, D’Alessio C, Perotti MA, Berón CM. 2016. Culex pipiens development is greatly influenced by native bacteria and exogenous yeast. PLoS One 11:e0153133. doi: 10.1371/journal.pone.0153133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitraka E, Stathopoulos S, Siden-Kiamos I, Christophides GK, Louis C. 2013. Asaia accelerates larval development of Anopheles gambiae. Pathog Glob Health 107:305–311. doi: 10.1179/2047773213Y.0000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coon KL, Brown MR, Strand MR. 2016. Gut bacteria differentially affect egg production in the anautogenous mosquito Aedes aegypti and facultatively autogenous mosquito Aedes atropalpus (Diptera: Culicidae). Parasit Vectors 9:375. doi: 10.1186/s13071-016-1660-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finney CA, Kamhawi S, Wasmuth JD. 2015. Does the arthropod microbiota impact the establishment of vector-borne diseases in mammalian hosts? PLoS Pathog 11:e1004646. doi: 10.1371/journal.ppat.1004646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glaser RL, Meola MA. 2010. The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS One 5:e11977. doi: 10.1371/journal.pone.0011977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zink SD, Van Slyke GA, Palumbo MJ, Kramer LD, Ciota AT. 2015. Exposure to West Nile virus increases bacterial diversity and immune gene expression in Culex pipiens. Viruses 7:5619–5631. doi: 10.3390/v7102886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dennison NJ, Jupatanakul N, Dimopoulos G. 2014. The mosquito microbiota influences vector competence for human pathogens. Curr Opin Insect Sci 3:6–13. doi: 10.1016/j.cois.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berticat C, Rousset F, Raymond M, Berthomieu A, Weill M. 2002. High Wolbachia density in insecticide-resistant mosquitoes. Proc Biol Sci 269:1413–1416. doi: 10.1098/rspb.2002.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duron O, Labbé P, Berticat C, Rousset F, Guillot S, Raymond M, Weill M. 2006. High Wolbachia density correlates with cost of infection for insecticide resistant Culex pipiens mosquitoes. Evolution 60:303–314. doi: 10.1111/j.0014-3820.2006.tb01108.x. [DOI] [PubMed] [Google Scholar]
- 38.Hazard EI, Mayer MS, Savage KE. 1967. Attraction and oviposition stimulation of gravid female mosquitoes by bacteria isolated from hay infusions. Mosq News 27:133–136. [Google Scholar]
- 39.Liess A, Diehl S. 2006. Effects of enrichment on protist abundances and bacterial composition in simple microbial communities. Oikos 114:15–26. doi: 10.1111/j.2006.0030-1299.14516.x. [DOI] [Google Scholar]
- 40.Hartman WH, Richardson CJ, Vilgalys R, Bruland GL. 2008. Environmental and anthropogenic controls over bacterial communities in wetland soils. Proc Natl Acad Sci U S A 105:17842–17847. doi: 10.1073/pnas.0808254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pernthaler J. 2005. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol 3:537–546. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- 42.Duguma D, Hall MW, Rugman-Jones P, Stouthamer R, Neufeld JD, Walton WE. 2015. Microbial communities and nutrient dynamics in experimental microcosms are altered after the application of a high dose of Bti. J Appl Ecol 52:763–773. doi: 10.1111/1365-2664.12422. [DOI] [Google Scholar]
- 43.Duguma D, Hall MW, Rugman-Jones P, Stouthamer R, Terenius O, Neufeld JD, Walton WE. 2015. Developmental succession of the microbiome of Culex mosquitoes. BMC Microbiol 15:140. doi: 10.1186/s12866-015-0475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duguma D, Rugman-Jones P, Kaufman MG, Hall MW, Neufeld JD, Stouthamer R, Walton WE. 2013. Bacterial communities associated with Culex mosquito larvae and two emergent aquatic plants of bioremediation importance. PLoS One 8:e72522. doi: 10.1371/journal.pone.0072522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wermelinger ED, Benigno CV, Machado RN, Nascimento TF, Ferreira AP, Meira AM, Souza MB, Zanuncio JC. 2010. Occurrence of Anopheles (Nyssorhynchus) rangeli (Gabaldon et al.) and Anopheles (Nyssorhynchus) evansae (Brethes) (Diptera: Culicidae) in an eutrophized dam. Neotrop Entomol 39:449–450. doi: 10.1590/S1519-566X2010000300022. [DOI] [PubMed] [Google Scholar]
- 46.Ishii T, Sohn SR. 1987. Highly polluted larval habitats of the Culex pipiens complex in central Sweden. J Am Mosq Control Assoc 3:276–281. [PubMed] [Google Scholar]
- 47.Aldemir A. 2009. Initial and residual activity of VectoBac 12 AS, VectoBac WDG, and VectoLex WDG for control of mosquitoes in Ararat Valley, Turkey. J Am Mosq Control Assoc 25:113–116. doi: 10.2987/08-5836.1. [DOI] [PubMed] [Google Scholar]
- 48.Batzer DP, Wissinger SA. 1996. Ecology of insect communities in nontidal wetlands. Annu Rev Entomol 41:75–100. doi: 10.1146/annurev.en.41.010196.000451. [DOI] [PubMed] [Google Scholar]
- 49.Talay F, Molva C, Atabay HI. 2016. Isolation and identification of Arcobacter species from environmental and drinking water samples. Folia Microbiol (Praha) 61:479–484 doi: 10.1007/s12223-016-0460-0. [DOI] [PubMed] [Google Scholar]
- 50.Hänel I, Tomaso H, Neubauer H. 2016. Arcobacter—an underestimated zoonotic pathogen? Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 59:789–794. doi: 10.1007/s00103-016-2350-7. [DOI] [PubMed] [Google Scholar]
- 51.Hsu TT, Lee J. 2015. Global distribution and prevalence of Arcobacter in food and water. Zoonoses Public Health 62:579–589. doi: 10.1111/zph.12215. [DOI] [PubMed] [Google Scholar]
- 52.Garren M, Raymundo L, Guest J, Harvell CD, Azam F. 2009. Resilience of coral-associated bacterial communities exposed to fish farm effluent. PLoS One 4:e7319. doi: 10.1371/journal.pone.0007319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma P, Sharma S, Maurya RK, Das De T, Thomas T, Lata S, Singh N, Pandey KC, Valecha N, Dixit R. 2014. Salivary glands harbor more diverse microbial communities than gut in Anopheles culicifacies. Parasit Vectors 7:235. doi: 10.1186/1756-3305-7-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunning Hotopp JC. 2011. Horizontal gene transfer between bacteria and animals. Trends Genet 27:157–163. doi: 10.1016/j.tig.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zouache K, Raharimalala FN, Raquin V, Tran-Van V, Raveloson LH, Ravelonandro P, Mavingui P. 2011. Bacterial diversity of field-caught mosquitoes, Aedes albopictus and Aedes aegypti, from different geographic regions of Madagascar. FEMS Microbiol Ecol 75:377–389. doi: 10.1111/j.1574-6941.2010.01012.x. [DOI] [PubMed] [Google Scholar]
- 56.Briones AM, Shililu J, Githure J, Novak R, Raskin L. 2008. Thorsellia anophelis is the dominant bacterium in a Kenyan population of adult Anopheles gambiae mosquitoes. ISME J 2:74–82. doi: 10.1038/ismej.2007.95. [DOI] [PubMed] [Google Scholar]
- 57.Kämpfer P, Lindh JM, Terenius O, Haghdoost S, Falsen E, Busse HJ, Faye I. 2006. Thorsellia anophelis gen. nov., sp. nov., a new member of the Gammaproteobacteria. Int J Syst Evol Microbiol 56:335–338. doi: 10.1099/ijs.0.63999-0. [DOI] [PubMed] [Google Scholar]
- 58.Lindh JM, Terenius O, Faye I. 2005. 16S rRNA gene-based identification of midgut bacteria from field-caught Anopheles gambiae sensu lato and A. funestus mosquitoes reveals new species related to known insect symbionts. Appl Environ Microbiol 71:7217–7223. doi: 10.1128/AEM.71.11.7217-7223.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilke AB, Marrelli MT. 2015. Paratransgenesis: a promising new strategy for mosquito vector control. Parasit Vectors 8:342. doi: 10.1186/s13071-015-0959-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stewart RIA, Dossena M, Bohan DA, Jeppesen E, Kordas RL, Ledger ME, Meerhoff M, Moss B, Mulder C, Shurin JB, Suttle B, Thompson R, Trimmer M, Woodward G. 2013. Mesocosm experiments as a tool for ecological climate-change research. Adv Ecol Res 48:71–181. doi: 10.1016/B978-0-12-417199-2.00002-1. [DOI] [Google Scholar]
- 61.Kent AD, Yannarell AC, Rusak JA, Triplett EW, McMahon KD. 2007. Synchrony in aquatic microbial community dynamics. ISME J 1:38–47. doi: 10.1038/ismej.2007.6. [DOI] [PubMed] [Google Scholar]
- 62.Leal WS, Barbosa RM, Xu W, Ishida Y, Syed Z, Latte N, Chen AM, Morgan TI, Cornel AJ, Furtado A. 2008. Reverse and conventional chemical ecology approaches for the development of oviposition attractants for Culex mosquitoes. PLoS One 3:e3045. doi: 10.1371/journal.pone.0003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dada N, Jumas-Bilak E, Manguin S, Seidu R, Stenström TA, Overgaard HJ. 2014. Comparative assessment of the bacterial communities associated with Aedes aegypti larvae and water from domestic water storage containers. Parasit Vectors 7:391. doi: 10.1186/1756-3305-7-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rubin BER, Sanders JG, Hampton-Marcell J, Owens SM, Gilbert JA, Moreau CS. 2014. DNA extraction protocols cause differences in 16S rRNA amplicon sequencing efficiency but not in community profile composition or structure. Microbiologyopen 3:910–921. doi: 10.1002/mbo3.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bartram AK, Lynch MDJ, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. 2011. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl Environ Microbiol 77:3846–3852. doi: 10.1128/AEM.02772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kennedy K, Hall MW, Lynch MD, Moreno-Hagelsieb G, Neufeld JD. 2014. Evaluating bias of Illumina-based bacterial 16S rRNA gene profiles. Appl Environ Microbiol 80:5717–5722. doi: 10.1128/AEM.01451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ross AA, Neufeld JD. 2015. Microbial biogeography of a university campus. Microbiome 3:66. doi: 10.1186/s40168-015-0135-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takahashi S, Tomita J, Nishioka K, Hisada T, Nishijima M. 2014. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS One 9:e105592. doi: 10.1371/journal.pone.0105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lynch MDj, Masella AP, Hall MW, Bartram AK, Neufeld JD. 2013. AXIOME: automated exploration of microbial diversity. Gigascience 2:3. doi: 10.1186/2047-217X-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. 2012. PANDAseq: paired-end assembler for Illumina sequences. BMC Bioinformatics 13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 72.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ, Suggests M. 2007. The vegan package. Community Ecol package 10. https://CRAN.R-project.org/package=vegan. [Google Scholar]
- 76.Rivière D, Desvignes V, Pelletier E, Chaussonnerie S, Guermazi S, Weissenbach J, Li T, Camacho P, Sghir A. 2009. Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J 3:700–714. doi: 10.1038/ismej.2009.2. [DOI] [PubMed] [Google Scholar]
- 77.SAS, Inc 2013. JMP release 11.0. SAS Institute, Inc, Cary, NC. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Indicator bacterial species (indicator value, ≥0.4; P < 0.05) associated with Culex nigripalpus Theobald developing in low- and high-organic-nutrient enrichments. Download TABLE S1, PDF file, 0.1 MB (25.7KB, pdf) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of the 16 most abundant (>1,000 sequences per sample) members of families recovered from different life stages of field-reared Culex nigripalpus mosquitoes. Others include 148 families (660,633 sequences) that had sequence abundance of <1,000 sequences per sample. The remaining 747 OTUs (692,057 sequences) were unidentified to families. Download FIG S1, EPS file, 0.3 MB (274.5KB, eps) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Nonmetric multidimensional scaling (NMDS) plot of bacterial communities from different life stages of field-reared Culex nigripalpus mosquitoes. Download FIG S2, PDF file, 0.1 MB (13.1KB, pdf) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of the 22 most abundant (average abundance of ≥1,000 sequences per sample) OTUs recovered from different life stages of field-reared Culex nigripalpus mosquitoes. Others include OTUs with average abundance of <1,000 sequences per sample and include 1,729 OTUs with a total of 1,968,535 sequences. The lowest taxonomic classifications of OTUs are presented. Download FIG S3, EPS file, 0.3 MB (281.2KB, eps) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Indicator bacterial species (indicator value, ≥0.5; P < 0.05) associated with different life stages (egg to adults) of Culex nigripalpus. Download TABLE S2, PDF file, 0.1 MB (34.9KB, pdf) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance (indicator value, ≥0.5; P ≥ 0.01) of bacterial taxa associated with developmental stages of Culex nigripalpus Theobald. Asterisks represent taxa that are either found in a lower proportion (i.e., <0.01 relative abundance per sample) or absent. Download TABLE S3, PDF file, 0.1 MB (32.2KB, pdf) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Schematic sketch of outdoor mosquito-rearing mesocosms at the Florida Medical Entomology Laboratory. Mesocosms 1, 10, and 13 received high-nutrient enrichment, whereas 2, 9, and 14 received low-nutrient enrichment. Download FIG S4, PDF file, 0.2 MB (259.6KB, pdf) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Modified BioQuip environmental floating chamber for mosquito rearing under outdoor field conditions. Download FIG S5, PDF file, 0.2 MB (184.5KB, pdf) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sampling schedule of Culex nigripalpus mosquitoes developing in outdoor aquatic mesocosms for DNA extraction. Column heads represent mesocosm identification numbers, with mesocosms 1, 3, and 10 receiving high-nutrient treatments and 2, 9, and 14 receiving low-nutrient-treatment regimens. Numbers 1 through 3 in cells indicate the number of mosquitoes or the half of the six egg rafts used for DNA extraction. Asterisks indicate two groups of 3 larvae that were sampled on those dates. —, sampling was not carried out due to lack of appropriate life stages of mosquitoes. Download TABLE S4, PDF file, 0.1 MB (34.3KB, pdf) .
Copyright © 2017 Duguma et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.