ABSTRACT
Human breast adipose tissue is a heterogeneous cell population consisting of mature white adipocytes, multipotent mesenchymal stem cells, committed progenitor cells, fibroblasts, endothelial cells, and immune cells. Dependent on external stimulation, adipose-derived stem cells differentiate along diverse lineages into adipocytes, chondrocytes, osteoblasts, fibroblasts, and myofibroblasts. It is currently not fully understood how a high-fat diet reprograms adipose-derived stem cells into myofibroblasts. In our study, we used mouse models of a regular diet and of high-fat-diet-induced obesity to investigate the role of dietary fat on myofibroblast differentiation in the mammary stromal microenvironment. We found that a high-fat diet promotes myofibroblast differentiation by decreasing microRNA 140 (miR-140) expression in mammary adipose tissue through a novel negative-feedback loop. Increased transforming growth factor β1 (TGF-β1) in mammary adipose tissue in obese mice activates SMAD3 signaling, causing phospho-SMAD3 to bind to the miR-140 locus and inhibit miR-140 transcription. This prevents miR-140 from targeting SMAD3 for degradation, resulting in amplified TGF-β1/SMAD3 signaling and miR-140 downregulation-dependent myofibroblast differentiation. Using tissue and coculture models, we found that myofibroblasts and the fibrotic microenvironment created by myofibroblasts impact the stemness and proliferation of normal ductal epithelial cells and early-stage breast cancer invasion and stemness.
KEYWORDS: Obesity, breast cancer, fibrosis, miRNA, myofibroblasts
INTRODUCTION
The heterogeneous cell population found within the mammary stromal vascular fraction (SVF) is essential for extracellular matrix (ECM) remodeling and disease development. Despite the importance of the mammary adipose tissue, surprisingly little is known about normal tissue homeostasis. The SVF consists of multipotent mesenchymal stem cells, committed progenitor cells, fibroblasts, endothelial cells, and immune cells. Under the influence of a variety of hormones, growth factors, and external factors, mesenchymal stem cells (MSCs) in the stromal vascular fraction are able to differentiate into a multitude of cell types, including adipocytes, chondrocytes, osteoblasts, fibroblasts, and myofibroblasts. For example, to commit MSCs into the adipogenic lineage in vitro, the master regulators peroxisome proliferator-activated receptor gamma (PPARγ) and C/EBPα are induced using a combination of dexamethasone, indomethacin, insulin, and methylisobutylxanthine, whereas in osteoblast differentiation, RUNX2 is activated using a mixture of ascorbate, bone morphogenetic protein, dexamethasone, and 1,25-dihydroxy vitamin D3 (1). While these cocktails can be used to induce differentiation, the complex signaling pathways regulating adipose MSCs are not fully elucidated.
Fibroblasts are critical for the production and maintenance of the extracellular matrix through secretion of ECM proteins, including fibronectin and collagen as well as ECM-degrading matrix metalloproteinases. In addition to their role in remodeling the normal ECM, fibroblasts and their activated form, myofibroblasts, are prominent members of the wound healing response (2). This process is dictated by transforming growth factor β1 (TGF-β1) signaling. When TGF-β1 binds its receptor, it recruits and phosphorylates SMAD proteins, including SMAD3, which then enter the nucleus and act as transcription factors and cofactors to activate the expression of target genes. In fibroblasts and adipose stem cells, TGF-β1 signaling stimulates differentiation into myofibroblasts, a highly contractile and proliferative cell type that produces elevated levels of ECM proteins (3, 4).
Dysregulation of myofibroblasts and wound healing results in the development of fibrosis, a pathogenic accumulation of scar tissue. Fibrosis has been well characterized through the study of many tissue-specific fibrotic diseases, including those of the lung (pulmonary fibrosis [5], radiation-induced lung fibrosis [6, 7], and cystic fibrosis [8]), heart (endomyocardial fibrosis [9, 10]), and liver (cirrhosis [11]).
Obesity increases myofibroblast differentiation within mammary adipose tissue, promoting fibrotic remodeling of the microenvironment and breast cancer progression (12). However, the mechanisms of obesity-induced mammary adipose myofibroblast differentiation remain poorly understood.
Mammary adipose tissue is very sensitive to stimulation by environmental and dietary factors. When obesity occurs, increased fat storage triggers adipocytes to undergo hyperplasia and hypertrophy. This promotes adipose tissue hypoxia and inflammation (13, 14), leading to myofibroblast differentiation. Although the mechanisms are not fully known, fibrotic changes to the adipose tissue have been associated with increased breast cancer initiation and growth (12, 15). Fibrosis results in increased ECM stiffness, which has been shown to activate mechanosignaling through the Rho/ROCK signaling pathways. Rho/ROCK phosphorylate focal adhesion kinase, and the nuclear translocation of the oncogenic transcription factors YAP/TAZ promotes uncontrolled proliferation (16). Diet-induced obesity has also been shown to directly affect the structure of the mammary gland. Mammary glands from high-fat-diet mice exhibited dysregulated mammary duct development, reduced branching, incomplete myoepithelial lining, and increased ductal collagen deposition (17).
MicroRNAs (miRNAs) are approximately 22-nucleotide-long single-stranded RNAs which are key regulators of virtually all aspects of cellular function through the RNA interference pathway. miRNAs associate with the RNA-induced silencing complex (RISC) in the cytoplasm, and through specific binding to the 3′ untranslated region (UTR) guide the RISC to target mRNAs. The RISC degrades the mRNA before it can be translated, therefore posttranscriptionally regulating gene expression. miRNA 140 (miR-140) was first identified as a regulator of embryonic cartilage development (18, 19) and is also a tumor suppressor in breast cancer. We have shown that miR-140 inhibits expression of stem cell factors SOX2, SOX9, and ALDH1, suppressing breast cancer stem cells and preventing breast cancer initiation and progression. In estrogen receptor-positive breast cancer, miR-140 is downregulated due to estrogen-mediated transcriptional inhibition, whereas in basal-like breast cancers, epigenetic mechanisms of inhibition have been demonstrated (20–22). We have shown that miR-140 is highly expressed in preadipocytes (23) and is necessary for adipose-derived stem cell adipogenesis under normal physiological conditions (24). Although several studies have observed that miR-140 targets TGF-β1 signaling through SMAD3 and TGF-β1 receptor 1 (TGFBR1) mRNA degradation via binding to their 3′ UTRs and that upregulation of miR-140 inhibits TGF-β1 signaling in lung tissue (25), the mechanism for the loss of the miR-140 protection pathway in obesity and fibrosis is unknown.
In this paper, we address the critical question of how a high-fat diet dysregulates miR-140 and TGF-β1 signaling in the mammary adipose stromal vascular fraction. Using in vivo mouse models of high-fat-diet-induced obesity, we find that miR-140 is downregulated in SVF cells isolated from obese mice. We identify a new SMAD3 binding site that inhibits miR-140 expression and a TGF-β1/SMAD3/miR-140 negative-feedback loop that is critical for myofibroblast differentiation in the mammary glands of obese mice. Finally, through coculture models, we show that high-fat-diet dysregulation of miR-140 impacts both normal and malignant ductal epithelial cells.
RESULTS
A high-fat diet downregulates miR-140 in mammary stromal cells.
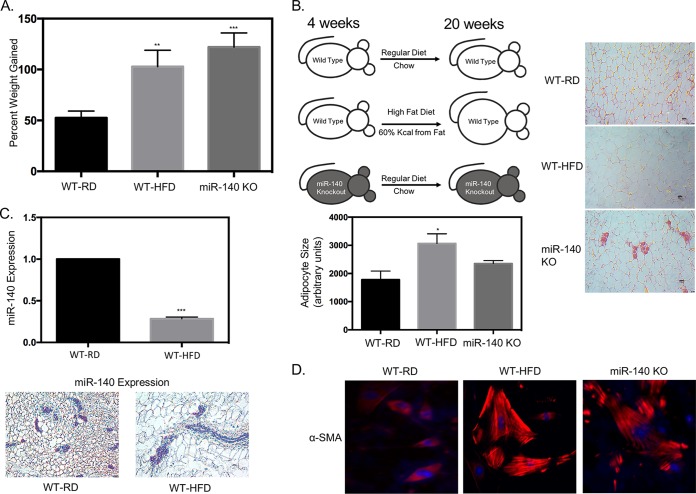
Our previous study (24) demonstrated that miR-140 expression in SVF cells is necessary to maintain normal adipogenesis under regular-diet conditions. In this study, we wanted to investigate the impact of a high-fat diet on miR-140 expression. We predicted that a long-term high-fat diet and obesity would dysregulate miR-140 and that miR-140 has a role in the adipocyte hypertrophy and hyperplasia that result from obesity. Starting at 4 weeks of age, we fed female C57BL/6 mice a high-fat diet (WT-HFD mice; 60% kcal from fat) and compared them to age-matched control female mice fed a normal chow diet (WT-RD mice). We observed that WT-RD mice had a percent weight gain (mean ± SD) of 52.55% ± 3.3% and WT-HFD mice of 102.8% ± 8.04% after 16 weeks of a high-fat diet (Fig. 1A). The mice were sacrificed, and the mammary fat pads were resected for examination. Histological assessment of hematoxylin-and-eosin-stained mammary fat pad sections showed global increases in adipocyte size in the WT-HFD mouse mammary fat pad, one of the characteristics of obesity (26) (Fig. 1B). To determine whether miR-140 was dysregulated in adipose tissue from obese mice, we performed quantitative real-time PCR (qRT-PCR). We found that miR-140 was significantly downregulated in SVF cells from WT-HFD mouse mammary adipose tissue (Fig. 1C, upper panel). As the SVF is a heterogeneous cell population, we performed RNA in situ staining to examine the expression of miR-140 within the context of the mammary fat pad. We used 5′ digoxigenin-labeled miR-140 RNA probes to stain paraffin-embedded tissue and found significant downregulation of miR-140 specific to the stromal cells of the mammary fat pad (Fig. 1C, lower panel). Obesity has been shown to be associated with an increase in tissue fibrosis. Using immunofluorescence analysis of stromal cells 10 days after adipogenic induction, we observed a significant increase in staining for the myofibroblast marker αSMA in the stromal cells from obese mice (Fig. 1D), suggesting an increase in myofibroblast differentiation in obese-mouse SVF cells. To investigate whether the downregulation of miR-140 seen in SVF cells from obese mice promoted myofibroblast differentiation, we isolated SVF cells from the mammary adipose tissue of age-matched chow-fed miR-140 knockout (miR-140 KO) mice (27). SVF cells from miR-140 KO mice exhibited high expression of αSMA after adipogenic differentiation, similar to that of SVF cells from obese mice (Fig. 1D). Interestingly, we also observed a significant percent weight gain (122.0% ± 6.92%) in regular-diet-fed miR-140 KO mice compared to that of WT-RD mice, similar to our observations for WT-HFD mice (Fig. 1A). These data show that miR-140 is downregulated in stromal cells from obese mice and that stromal cells from both obese and miR-140 KO mice have increased expression of αSMA, suggesting that obesity and downregulation of miR-140 may promote myofibroblast differentiation.
FIG 1.
High-fat diet downregulates miR-140 in mammary stromal cells. Female wild-type C57BL/6 mice were fed a regular chow diet (WT-RD) or a high-fat diet (WT-HFD) ad libitum for 16 weeks. (A) Percent weight gained after 16 weeks of a regular or high-fat diet. (B) Left panel, WT-RD, WT-HFD, and miR-140 KO mice after the respective diets for 16 weeks (left panel). Right panels, hematoxylin-and-eosin (H&E) staining of stromal areas of the mammary fat pads isolated from WT-RD, WT-HFD, and miR-140 KO mice. Bottom panel, quantification of adipocyte size. (C) miR-140 is downregulated in mammary stromal cells from high-fat-diet mice. Top panel, miR-140 expression was measured using qRT-PCR of RNA isolated from mammary fat pad SVF cells from WT-RD and obese mice. Bottom panels, RNA in situ staining for miR-140 expression in the mammary fat pads of WT-RD and WT-HFD mice verified downregulation in stromal cells. (D) Immunofluorescence detection of myofibroblast marker αSMA showed upregulation in SVF cells isolated from the mammary fat pads of obese and miR-140 KO mice 10 days after adipogenesis was induced in vitro. *, P < 0.05; **, P < 0.005; ***, P < 0.0001.
High-fat-diet-induced miR-140 downregulation is associated with increased ECM deposition.
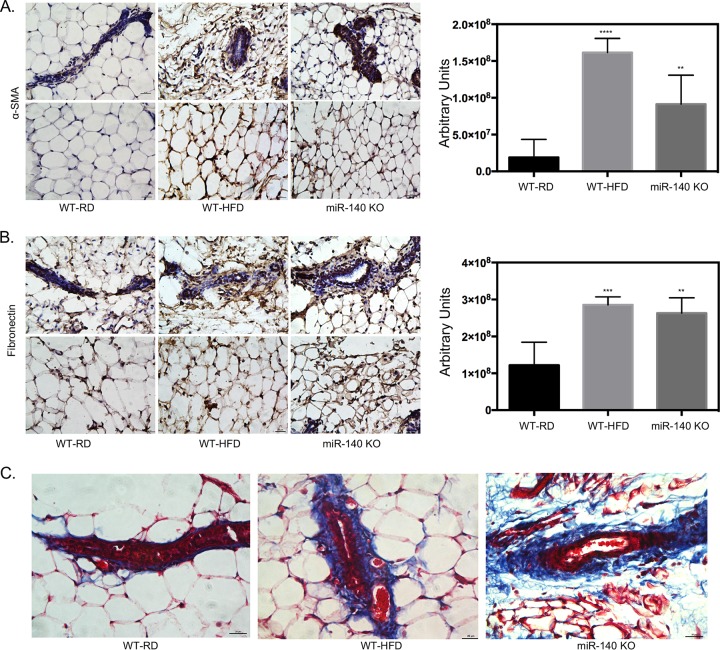
To assess the impact of a high-fat diet on mammary adipose tissue myofibroblast differentiation and ECM deposition, we examined mammary adipose tissue for myofibroblast accumulation and ECM changes related to fibrosis. Using immunohistochemistry analysis, we showed that mammary adipose tissue in obese mice is enriched for αSMA (Fig. 1A). One of the key pathologies of fibrosis is the increased production of ECM proteins such as collagen and fibronectin, resulting in excessive scar tissue accumulation (28). Therefore, we tested the expression of these two proteins in the mammary fat pad, focusing both on the stromal areas surrounding mammary ducts and on non-duct-associated stromal cells. We observed that fibronectin expression was increased in WT-HFD mouse mammary adipose tissue, especially in the stromal areas directly surrounding the mammary ducts (Fig. 2B). Finally, using a modified Masson's trichrome stain, we found that there was increased collagen deposition in the ductal microenvironment of WT-HFD mice compared to WT-RD mice, consistent with the excessive ECM production of the fibrotic phenotype (Fig. 2C). Mammary adipose tissue isolated from miR-140 KO mice similarly exhibited enhanced myofibroblast differentiation and fibrotic ECM deposition (Fig. 2A to C). These results show that obesity causes fibrosis of the mammary gland and suggest that the loss of miR-140 alone is sufficient to induce myofibroblast differentiation fibrosis.
FIG 2.
High-fat diet-induced miR-140 downregulation is associated with increased ECM deposition. Staining for markers of fibrosis in mammary fat pad tissue sections isolated from WT-RD, WT-HFD, and miR-140 KO mice is shown. (A) Immunohistochemistry staining for αSMA shows increased αSMA expression in the stromal cells and surrounding the mammary ducts of WT-HFD and miR-140 KO mouse mammary fat pads. (B) Immunohistochemistry staining for fibronectin shows increased fibronectin expression in the stromal cells and surrounding the mammary ducts of WT-HFD and miR-140 KO mouse mammary adipose tissue. (C) Modified Masson's trichrome stain shows increased collagen deposition (blue) in WT-HFD and miR-140 KO mouse mammary adipose tissue. **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001.
miR-140 deficiency contributes to myofibroblast differentiation.
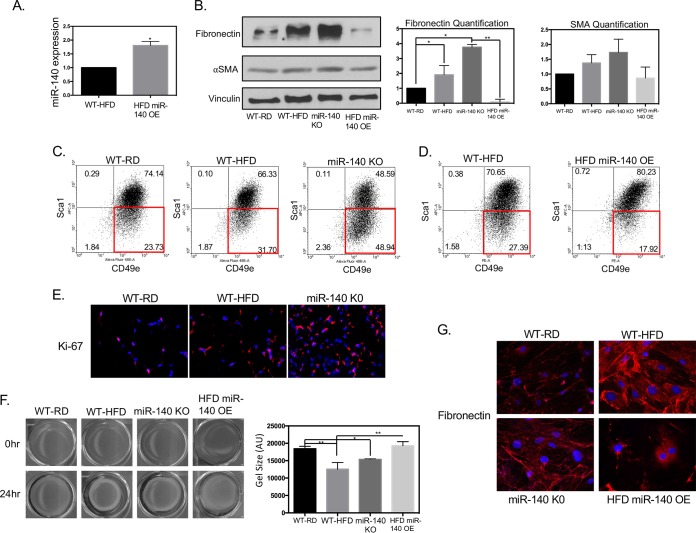
As we observed increased fibrosis in the WT-HFD and miR-140 KO mouse mammary glands, we next investigated the cellular and molecular effects of high-fat-diet-induced miR-140 downregulation. We isolated stromal vascular cells from WT-RD, WT-HFD, and miR-140 KO mouse mammary adipose tissue and through transient transfection rescued miR-140 in WT-HFD mouse SVF cells (Fig. 3A). Western blotting demonstrated that the myofibroblast marker αSMA was slightly increased in WT-HFD and miR-140 KO mouse SVF cells and that ECM protein fibronectin was significantly upregulated in both obese and miR-140 KO mouse SVF cells (Fig. 3B). miR-140 rescue resulted in the almost complete downregulation of fibronectin, consistent with our previous findings that fibronectin is a miR-140 target (unpublished data). miR-140 overexpression also reduced the protein level of αSMA, indicating that miR-140 overexpression causes an abrogation of the myofibroblast phenotype. While we observed a positive trend in αSMA expression in WT-HFD and miR-140 KO mouse SVF cells, primary SVF expression of αSMA was not a conclusive indicator of myofibroblast differentiation. Studies from our laboratory and others have established that murine myofibroblasts can be identified using fluorescence-activated cell sorting (FACS) for the markers SCA1low CD49ehigh (29). Using these markers, we found that 23.73% of WT-RD mouse SVF cells were myofibroblasts, with an increase to 31.7% in the obese mouse SVF cell population and 48.94% in the miR-140 KO mouse SVF cells (Fig. 3C). When miR-140 was transiently overexpressed in WT-HFD cells, there was a 33% decrease in the myofibroblast population compared to that of the WT-HFD population (Fig. 3D). These data indicate an increase in the myofibroblast population due to a long-term high-fat diet and miR-140 deficiency and that miR-140 downregulation is necessary for the myofibroblast phenotype.
FIG 3.
miR-140 deficiency contributes to myofibroblast differentiation. Protein, cellular, and functional markers of myofibroblast differentiation were examined in SVF cells isolated from the mammary fat pads of WT-RD, WT-HFD, and miR-140 KO mice. (A) Transient transfection was performed to rescue miR-140 expression in WT-HFD mouse SVF cells. (B) Expression levels of proteins associated with fibrosis and myofibroblast differentiation were analyzed using Western blotting. SVF cells from obese and miR-140 KO mice had higher expression levels of fibronectin and αSMA compared to WT-RD. Transient overexpression (OE) of miR-140 in SVF cells from obese mice resulted in decreased expression of all markers examined. (C) Myofibroblast formation was assessed using Sca-1 and CD49e markers using FACS analysis. SVF cells from obese and miR-140 KO mice demonstrated increased myofibroblast populations (SCA-1low CD49ehigh). (D) FACS sorting for myofibroblasts in WT-HFD mouse SVF and WT-HFD mouse SVF overexpressing miR-140. (E) Ki-67 expression was examined in mammary fat pad tissue sections. Ki-67 was found to be upregulated in WT-HFD and miR-140 KO mouse mammary adipose tissue. (F) Evaluation of myofibroblast differentiation using a collagen gel contraction assay. Cells from obese and miR-140 KO mice exhibited higher contractile ability than those from WT-RD mice. Overexpression of miR-140 almost completely ablated contractile ability. The contraction capacity was calculated by measuring the gel area with ImageJ 24 h after the stress was lifted. (G) The amount of fibronectin secreted by stromal vascular fraction cells was examined by immunofluorescence. Data are means ± standard deviations (SD) (n = 3). *, P < 0.05; **, P < 0.005.
We next examined the key functional abilities of myofibroblasts: proliferation, contraction, and ECM deposition. We first examined the proliferative ability of mammary fat pad stromal cells through Ki-67 staining. Using tissue slides isolated from our animal models, we performed immunofluorescence staining for Ki-67. We found that the mammary adipose tissue from obese mice had increased expression of Ki-67, with the mammary adipose tissue from miR-140 KO mice demonstrating even higher staining (Fig. 3D). To determine the contractile ability of the primary SVF cells, we conducted a 24-h collagen gel contraction assay. SVF cells isolated from mammary adipose tissue from both obese and miR-140 KO mice exhibited significantly higher contractile ability than SVF cells from regular-diet control mice (Fig. 3C), consistent with their increased myofibroblast populations. As observed in our FACS data, knockdown of miR-140 in WT-HFD cells almost completely abrogated the contractile ability of the WT-HFD cells. Finally, to examine ECM deposition, we plated SVF cells on coverslips and allowed them to grow in culture for 72 h. We then performed immunofluorescence staining for fibronectin. SVF cells from both obese and miR-140 KO mice produced an ECM enriched in fibronectin, and dense fibronectin deposition is seen especially around cells from obese mice (Fig. 3E). Similar to what we observed through Western blotting, when miR-140 was overexpressed, fibronectin expression was dramatically decreased.
Our results have shown that miR-140 is downregulated in SVF cells from obese mice and that miR-140 knockout alone increases myofibroblast differentiation and fibrosis without external factors such as a high-fat diet. To investigate whether miR-140 prevents myofibroblast differentiation and fibrosis development, we transiently overexpressed miR-140 in SVF cells from obese mice. The results suggest that miR-140 loss is critical for myofibroblast differentiation in SVF cells from obese mice and that miR-140 expression inhibits myofibroblast differentiation and protects against the development of fibrosis (Fig. 3A).
TGF-β1 signaling and miR-140 form a negative-feedback loop.
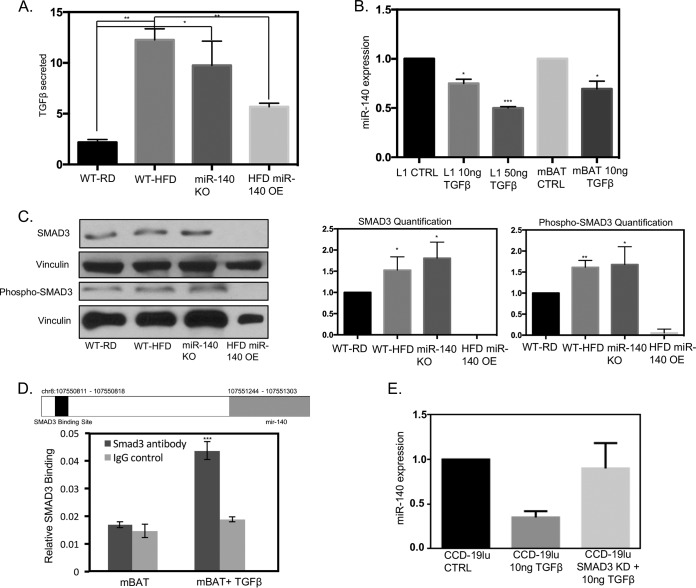
Having identified a functional relationship between miR-140 and myofibroblast differentiation, we next sought to understand how miR-140 is downregulated in mammary adipose tissue from obese mice. To this end, we first examined the activation of the TGF-β1 pathway. We investigated the expression of TGF-β1, a primary driver of myofibroblast differentiation, in 48-h-conditioned medium from SVF cells from WT-RD, obese, and miR-140 KO mice using a TGF-β1 ELISA. Myofibroblasts have been shown to produce and secrete higher levels of TGF-β1 than normal fibroblasts, activating a positive-feedback loop for myofibroblast differentiation (4). Therefore, as WT-HFD and miR-140 KO mouse mammary fat pads had increased myofibroblast differentiation, we expected that the primary SVF cells isolated from them would secrete higher levels of TGF-β1. As predicted, conditioned medium from SVF cells from WT-HFD and miR-140 KO mice had high levels of TGF-β1 compared to those from WT-RD mice. When miR-140 was transiently overexpressed (Fig. 3A), we observed a decrease of TGF-β1 in the conditioned medium (Fig. 4A). These data further confirm that miR-140 downregulation promotes myofibroblast differentiation and demonstrate that miR-140 regulates TGF-β1 signaling in stromal cells. We next investigated whether TGF-β1 mediates miR-140 downregulation. We treated the mouse preadipocyte cell lines 3T3-L1 and mBAT with TGF-β1 for 24 and 48 h and found that miR-140 was downregulated by TGF-β1 treatment in a dose-dependent manner (Fig. 4B). To identify the mechanism of miR-140 downregulation, we first examined epigenetic means. TGF-β1 has been shown to induce DNA methylation (30), and we have previously found that miR-140 is epigenetically regulated in breast cancer (21). However, when we examined the CpG islands in the miR-140 locus that we previously identified (21), we found no evidence that TGF-β1 regulates miR-140 expression through aberrant methylation (data not shown).
FIG 4.
TGF-β1 signaling and miR-140 form a negative-feedback loop. TGF-β1 downregulates miR-140 through SMAD3 binding to the miR-140 upstream sequence. (A) High levels of TGF-β1 are secreted by SVF cells from obese and miR-140 KO mice. (B) TGF-β1 decreases miR-140 expression in a dose-dependent manner in 3T3-L1 and mBAT mouse preadipocytes. (C) SMAD3 and phospho-SMAD3 protein expression is upregulated in SVF cells from obese and miR-140 KO mice, indicating increased pathway activity. miR-140 overexpression (OE) in WT-HFD mouse cells completely downregulated SMAD3/pSMAD3 expression. (D) Putative SMAD3 binding site in the upstream sequence of miR-140. ChIP demonstrates increased SMAD3 binding to miR-140 when mBAT cells are treated with 10 ng/ml of TGF-β1. (E) SMAD3 knockdown prevented TGF-β1-mediated miR-140 downregulation in CCD-19lu human lung fibroblasts. *, P < 0.05; **, P < 0.01.
As TGF-β1 did not inhibit miR-140 through epigenetic mechanisms, we next wondered if TGF-β1-activated transcription factors directly inhibited miR-140 expression. We examined the expression of SMAD3, a common TGF-β1-activated transcription factor that was shown to bind to the miR-140 locus in human osteoarthritis (31). Western blotting demonstrated that the SMAD3 protein level was increased in SVF cells from both obese and miR-140 KO mice compared to that in WT-RD mice (Fig. 4C). When the TGF-β1 pathway is active, SMAD3 is phosphorylated before being translocated to the nucleus, where it acts as a transcription factor. Therefore, we investigated whether the activated form of SMAD3, phospho-SMAD3, was also increased in SVF cells from WT-HFD and miR-140 KO mice. We observed an increase in pSMAD3 expression through Western blotting, indicating pathway activation. SMAD3 has previously been shown to bind and inhibit miR-140 in embryonic cell lines (31); however, researchers were unable to replicate these findings in mature chondrocytes (32). We were unable to identify the previously reported binding site (31), potentially due to the different tissues and model systems examined. miR-140 is located in an intronic region of WWP2, an E3 ubiquitin ligase gene (32). Therefore, to determine whether SMAD3 binds the WWP2/miR-140 locus, we performed sequence alignment analysis for the SMAD3 binding site GTCTAAGC and found a previously unidentified SMAD3 binding site (chromosome 8: 107550811 to 107550818) in the upstream region of miR-140 (Fig. 4D, upper panel). To interrogate the potential binding site, we treated mBAT mouse preadipocyte cells with 10 ng/ml TGF-β1 for 48 h to induce SMAD3 pathway activation. We then conducted chromatin immunoprecipitation using a SMAD3 antibody, followed by qRT-PCR, and found that TGF-β1 treatment resulted in a 2-fold increase in SMAD3 binding to the putative binding site (Fig. 4D, lower panel). As we found that SMAD3 binds to miR-140, we next verified that SMAD3 is necessary for TGF-β1-mediated miR-140 downregulation. We transiently knocked down SMAD3 in fibroblasts before treating them with 10 ng TGF-β1 for 48 h, as shown in Fig. 4B. SMAD3 knockdown prevented TGF-β1-mediated miR-140 downregulation, confirming that SMAD3 mediates TGF-β1 inhibition of miR-140.
These data demonstrate that TGF-β1 signaling downregulates miR-140 in a SMAD3-dependent manner. As SVF cells from obese and miR-140 KO mice had increased SMAD3 protein expression compared to WT-RD mice, we next wanted to determine if miR-140 targets SMAD3 for degradation, as previously shown in lung tissue (25). When we overexpressed miR-140 in SVF cells from obese mice, we observed the complete downregulation of SMAD3 and phospho-SMAD3 (Fig. 4C). Taken together, these data demonstrate the existence of a novel TGF-β1/SMAD3/miR-140 negative-feedback loop in the obese-mouse mammary microenvironment. Increased TGF-β1 production by SVF cells from obese mice activates SMAD3 downregulation of miR-140, which removes the inhibition of SMAD3 by miR-140, resulting in higher protein expression of SMAD3.
Effect of a long-term high-fat diet on mammary epithelial cells.
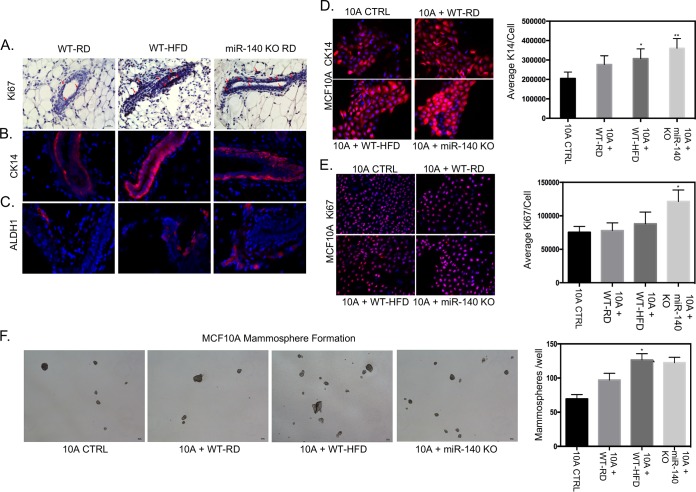
To determine whether the increased myofibroblast differentiation and fibrotic ECM seen in WT-HFD and miR-140 KO mouse mammary adipose tissue impact mammary ductal epithelial cells, we assayed the mammary ducts for molecular markers of premalignancy. We first examined the proliferation of ductal cells using immunohistochemistry staining for Ki-67 and found that ductal epithelial cells in both obese and miR-140 KO mouse mammary adipose tissue exhibited high staining for Ki-67 (Fig. 5A). We next stained for cytokeratin 14 (CK14), a marker for the basal epithelial lineage that is required for breast cancer invasion (33). Using immunofluorescence, we found that ductal expression of CK14 was upregulated in the ducts of both WT-HFD and miR-140 KO mice (Fig. 5B). Finally, we examined expression of ADLH1, a marker for both normal and malignant breast stem cells (34). ALDH1 expression was very low in WT-RD mouse mammary adipose tissue, with increases seen in both WT-HFD and miR-140 KO mouse mammary ducts (Fig. 5C).
FIG 5.
Effects of a high-fat diet on mammary epithelial cells. To investigate the effect of mammary fat pad fibrosis and high-fat diet on mammary epithelial cells, markers of proliferation, basal cell lineage, and stemness were examined in ductal epithelial tissue sections isolated from WT-RD, WT-HFD, and miR-140 KO mice. The immortalized epithelial cell line MCF10A was examined for the same markers after being treated with control SVF medium (10A CTRL) or conditioned medium from SVF cell lines. (A) Immunohistochemistry staining for Ki-67 demonstrated an increase in ductal epithelial cells from obese and miR-140 KO mice. (B) Immunofluorescence staining for basal cell lineage marker CK14 found increased expression in ductal epithelial cells from obese and miR-140 KO mice. (C) Immunofluorescence staining showed that stem cell marker ALDH1 was upregulated in ductal epithelial cells from obese and miR-140 KO mice. (D and E) MCF10A cells were treated with conditioned medium from SVF cells for 48 h before immunofluorescence staining. (D) MCF10A cells treated with conditioned medium from obese and miR-140 KO mouse SVF cells had significantly increased CK14 expression. (E) MCF10A cells treated with conditioned medium from obese-mouse SVF cells had a small increase in Ki-67 expression. MCF10A cells treated with conditioned medium from SVF cells from miR-140 KO mice had significantly increased Ki-67 expression. (F) MCF10A cells treated with conditioned medium from SVF cells from obese and miR-140 KO mice demonstrated significantly increased mammosphere formation. Data are means ± SD (n = 3). *, P < 0.05; **, P < 0.005.
Because we observed the changes to epithelial cells in a whole-animal knockout or high-fat-diet model, the impact seen may be due to the direct effect of the high-fat diet or miR-140 knockout on mammary epithelial cells. To precisely examine the effect of SVF cells from obese and miR-140 KO mice on mammary epithelial cells, we treated the nonmalignant immortalized mammary epithelial cell line MCF10A with conditioned medium from WT-RD, obese, and miR-140 KO mouse SVF cells for 48 h, after which we repeated the staining done in tissue (Fig. 5A to C) in vitro. CK14 was significantly increased in MCF10A cells cocultured with SVF cells from both obese and miR-140 KO mice, consistent with our in vivo results (Fig. 5D). While Ki-67 was highly increased in MFC10A cells treated with conditioned medium from miR-140 KO mouse SVF cells, it was only slightly increased in MCF10A treated with conditioned medium from obese-mouse SVF cells (Fig. 5E). Finally, ALDH1 staining was negative in MCF10A cells under all conditions (data not shown). To determine the functional self-renewal ability of these cells, we performed a mammosphere assay and found that MCF10A treated with conditioned medium from both obese and miR-140 KO mouse SVF cells formed significantly more mammospheres than MCF10A treated with conditioned medium from WT-RD mouse SVF cells or by themselves, indicating increased self-renewal and stemness (Fig. 5F).
Mouse mammary stromal cells isolated from mice on a high-fat diet and miR-140 KO mice promote breast cancer.
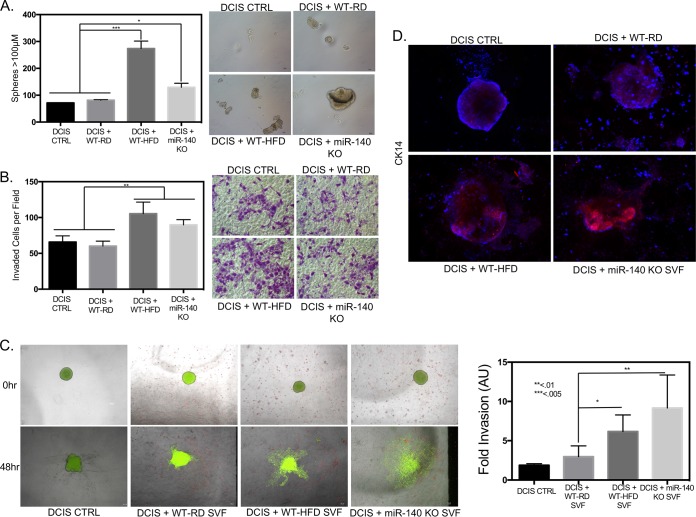
The progression of noninvasive ductal carcinoma in situ (DCIS) tumors to invasive ductal carcinoma (IDC) is thought to be driven by a population of self-renewing cancer stem cells (21). To examine the impact of high-fat diet downregulation of miR-140 on DCIS stemness and progression, we cocultured SVF cells with the early-stage noninvasive breast cancer cell line MCF10DCIS (DCIS cells) and performed mammosphere and transwell invasion assays. After 48 h of coculture, DCIS cells that had been cocultured with WT-HFD stromal cells demonstrated the highest level of mammosphere formation out of all groups (Fig. 6A). The hallmark of DCIS progression is invasive ability. To determine the effect of obese and miR-140 KO mouse SVF cells on DCIS cell invasion, we performed a transwell Matrigel invasion assay. Similar to the results seen in the mammosphere formation assay, DCIS cells that had been cocultured with SVF cells from obese mice showed the highest invasion, followed by those cocultured with miR-140 KO mouse SVF cells (Fig. 6B). These data show that the SVF cells from obese mice promote both self-renewal and the invasive ability of DCIS cells. Increased self-renewal ability of the DCIS cell population indicates an increase in the proportion of cancer stem cells in the DCIS whole-cell population and matches the increased self-renewal seen both in vitro and in MCF10A epithelial cells (Fig. 5C and F). While these experiments suggest that SVF cell-secreted factors from obese and miR-140 KO mice promote DCIS stemness and invasion, the ability of myofibroblasts to contract and remodel the ECM likely also plays a role. We next investigated the impact of ECM remodeling and contraction by SVF cells from WT-HFD and miR-140 KO mice on promoting DCIS invasion. We implanted DCIS cell organoids in Matrigel, a gel-like substrate that contains proteins and growth factors present in the basement membrane, in the presence of SVF cells from WT-RD, obese, or miR-140 KO mice. The addition of SVF cells makes the three-dimensional (3D) Matrigel culture a close mimic of the in vivo stromal microenvironment. The organoid size was measured at the time of implantation and after 48 h, and the invasive spread of the organoid was quantified to determine the invasive ability. When the DCIS organoid was in Matrigel without the presence of SVF cells, we observed almost a complete absence of spheroid growth and invasion. The WT-RD mouse SVF cells slightly increased the invasive ability of the DCIS organoid, and when the organoid was plated in the presence of SVF cells from obese mice, there was a significant increase in organoid invasion. When the DCIS organoid was plated alongside miR-140 KO mouse SVF cells, organoid invasion was increased, with almost complete disaggregation of the organoid (Fig. 6C). Finally, organoids were stained for CK14, which has been demonstrated to be highly enriched in the invasive fronts of DCIS cell organoid invasion (33). CK14 expression matched the invasive ability of organoids, with the DCIS organoid alone and in the presence of WT-RD mouse SVF cells expressing very low levels of CK14. When the organoid was plated in the Matrigel culture with SVF cells from obese mice, CK14 was enriched, and when miR-140 KO mouse stromal cells were present, CK14 was highly expressed in the DCIS organoid (Fig. 6D).
FIG 6.
Mammary stromal cells from high-fat-diet and miR-140 KO mice promote breast cancer. DCIS cells were cocultured with SVF cells and then replated for functional assays to examine the effect of fibrotic SVF cells on DCIS invasiveness and aggression. (A) DCIS cells were cocultured with SVF cells for 48 h and then replated for mammosphere formation. DCIS cells cocultured with SVF cells from obese and miR-140 KO mice had significantly increased mammosphere formation. (B) After coculture, DCIS cells were plated for a transwell Matrigel invasion assay. (C) DCIS organoids were plated in a 3D Matrigel culture with SVF cells. After 48 h, organoid invasion was measured using ImageJ. DCIS organoids in 3D culture with SVF cells from obese and miR-140 KO mice had much higher levels of 3D invasion than DCIS cells cultured with SVF cells from WT-RD mice or alone. (D) At 48 h after organoid implantation, DCIS organoids in the 3D Matrigel culture with SVF cells were stained for a marker of basal lineage and tumor invasion/aggression, CK14. DCIS organoids cultured with SVF cells from obese and miR-140 KO mice had increased CK14 expression. Data are means ± SD (n = 3). DCIS CTRL, DCIS cells without coculture. *, P < 0.01; **, P < 0.005; ***, P < 0.001.
DISCUSSION
miR-140 was first identified as a regulator of cartilage development (18, 19) and has been shown to target TGF-β1 signaling through the degradation of SMAD3 in embryonic cell lines (25). The ability of mesenchymal stem cells within the SVF to differentiate among a variety of lineages, including adipogenic, chondrogenic, and osteogenic, has been extensively studied; however, the mechanisms regulating myofibroblast differentiation from stem cells and transdifferentiation of mature SVF cells are largely unknown. Obesity results in a high basal level of inflammation and has recently been shown to promote myofibroblast differentiation within the mammary fat pad (12). Our results demonstrate that obesity causes the downregulation of miR-140 in the mature mammary adipose tissue. While it has previously been shown that miR-140 is a tumor suppressor miRNA that is dysregulated in breast cancer (21), these are the first data showing pathogenic dysregulation of miR-140 in the mammary microenvironment.
In breast cancer, we have identified two different mechanisms of miR-140 downregulation. In basal-like breast cancers, it is silenced via methylation (21), whereas we have identified estrogen-mediated transcriptional inhibition in estrogen receptor-positive breast cancers (22). We found that in mammary adipose tissue, miR-140 is downregulated through aberrant TGF-β1 signaling and SMAD3 binding to the miR-140 locus. TGF-β1 is known to be a potent stimulator of myofibroblast differentiation, but its role in mammary adipose tissue is unknown. Here we identified a novel TGF-β1/SMAD3/miR-140 negative-feedback loop, wherein obesity-mediated increases in TGF-β1 signaling activate SMAD3, which binds to and inhibits miR-140. As miR-140 targets SMAD3 for degradation, the downregulation of miR-140 results in dysregulated expression of SMAD3, which promotes myofibroblast differentiation.
The ECM remodeling and microenvironmental changes enacted by miR-140-deficient stromal vascular fraction cells from obese mice impact both normal mammary ductal epithelial cells and early-stage breast cancer cells. We observed that adipose tissue from obese and miR-140 KO mice induces high expression of premalignancy markers in nontumorigenic epithelial cells. Importantly, we also observed that coculture with SVF cells from obese or miR-140 KO mice increased the stemness and invasive abilities of early-stage breast cancer cells, suggesting that SVF cells from obese individuals may promote breast cancer progression.
miR-140 may be a novel target for prevention of fibrosis in the high-fat mammary microenvironment. Adipocytes from obese individuals actively shed miRNA-containing exosomes (35), which may affect signaling in nearby cells. We have previously shown that exosomes secreted by preadipocytes contain high levels of miR-140 and that treatment with shikonin can induce high levels of miR-140 in preadipocyte exosomes (23). The use of miR-140-promoting drugs such as shikonin or paclitaxel may be a novel therapeutic microenvironmental targeting strategy for the prevention of fibrosis and its associated diseases. In future studies, we will use high-fat-diet-induced obesity in athymic nude mice to perform mammary gland xenografts and use these models to characterize the impact of miR-140-upregulating drugs and therapies on the inhibition of myofibroblast differentiation to prevent fibrosis and fibrosis-related diseases.
MATERIALS AND METHODS
Cell culture, reagents, transfection.
MCF10A and MCF10DCIS (DCIS) (Asterand, Detroit, MI) cells were grown in Dulbecco's modified Eagle medium (DMEM)–F-12 supplemented with 10 μg/ml insulin, 100 ng/ml cholera toxin, 0.5 μg/ml hydrocortisone (Sigma, St. Louis, MO), 20 ng/ml epidermal growth factor (EGF), and 5% horse serum (Invitrogen, Carlsbad, CA). 3T3-L1 (L1) and mBAT mouse preadipocytes were grown in DMEM supplemented with 5% fetal bovine serum (FBS) (HyClone, Rockford, IL) and 1% l-glutamine (Invitrogen, Carlsbad, CA). mBAT cells were obtained from the laboratory of Da-Wei Gong (Department of Medicine, University of Maryland School of Medicine, Baltimore, MD). Cells were incubated in 5% CO2 at 37°C. Human normal lung fibroblast cells (CCD-19Lu) were purchased from the American Type Culture Collection (ATCC). They were cultured in Eagle's minimum essential medium (EMEM) (ATCC, Manassas, VA) supplemented with 10% fetal bovine serum (FBS) (HyClone, Rockford, IL). The SMAD3 overexpression plasmid pQCXIH-Flag Smad3 WT was a gift from Joan Massague (Addgene plasmid 27025) (36). All transfection assays were performed as described previously by Li et al. (20).
Animal models and isolation of adipose tissue SVF.
Wild-type C57BL/6 mice were obtained from the University of Maryland Baltimore Veterinary Resources. miR-140 KO mice on a C57BL/6 background were a generous gift from Hiroshi Asahara, who developed the knockout model (27). Four-week-old female C57BL/6 mice were randomized and fed a regular chow diet (WT-RD mice) or a high-fat diet (WT-HFD mice) (Research Diets; D12450Bi, 60 kcal% from fat) ad libitum for 16 weeks. Body weight was reported as a percent change relative to baseline. The adipose tissue stromal vascular fraction (SVF) was isolated from the mammary fat pads of 20-week-old, wild-type, regular-diet mice, wild-type, high-fat-diet mice, and miR-140 KO regular-diet mice as previously described (24). Mammary fat pads were formalin fixed for immunohistochemistry evaluation or were processed for cell isolation. Tissues for cell isolation were cut up and digested with collagenase at 37°C for 1 to 2 h with gentle vortexing every 15 min. After digestion, the solution was filtered through 250-μm tissue strainers (Thermo Scientific, Rockford, IL) and collected in a sterile tube. After centrifugation at 200 × g for 10 min, the filtrate was comprised of mature adipocytes (top layer) and SVF cells (bottom layer). The pellet (SVF cells) was washed twice with phosphate-buffered saline (PBS) and resuspended in culture medium (DMEM–10% FBS–1% penicillin-streptomycin). The cell solution was filtered through a 40-μm cell strainer and transferred to a 10-cm tissue culture dish. All studies were done in accordance with federal guidelines and institutional policies of the University of Maryland Animal Care and Use Committee.
Cellular immunofluorescence.
Cells were fixed with 4% paraformaldehyde for 10 min, washed twice with PBS, and blocked with 10% goat serum in PBS at room temperature for 1 h, followed by primary antibody incubation overnight at 4°C. After washes in PBS, cells were incubated with secondary antibody for 1 h, followed by washes in PBS. Cells were then counterstained with Hoechst 33342 in PBS for 10 min at room temperature, mounted using mounting medium (KPL, Gaithersburg, MD), and visualized with an Olympus IX81 spinning-disk confocal microscope. Quantification was performed using ImageJ (NIH) analysis (37).
Tissue immunofluorescence.
Paraffin-embedded tissue sections were deparaffinized in xylene, rehydrated in ethanol, and washed twice with distilled water (dH2O). Antigen retrieval was performed using 10 mM sodium citrate, pH 6.0, at 95°C for 10 min, after which slides were allowed to cool for 30 min at room temperature. Samples were quenched using 3% hydrogen peroxide, followed by washing and blocking with 10% goat serum in PBS for 1 h. Primary antibody was applied, and slides were incubated overnight at 4°C. Samples were washed in PBS and then incubated with secondary antibody for 1 h, followed by washes in PBS and counterstaining with Hoechst 33342 in PBS for 10 min at room temperature. Samples were mounted using mounting medium (KPL, Gaithersburg, MD) and visualized with an Olympus IX81 spinning-disk confocal microscope. Quantification was performed using ImageJ (NIH) analysis.
Immunohistochemistry.
Paraffin-embedded tissue sections were deparaffinized in xylene and rehydrated in ethanol and washed twice with dH2O. Antigen retrieval was performed using 10 mM sodium citrate, pH 6.0, at 95°C for 10 min and allowed to cool for 30 min at room temperature. Samples were quenched using 3% hydrogen peroxide, followed by washing and blocking with 10% goat serum in PBS. Samples were incubated with the primary antibody overnight at 4°C in antibody diluent, washed, and incubated with secondary antibody for 1 h at room temperature. The substrate reaction was performed using the 3,3-diaminobenzidine–horseradish peroxidase (DAB-HRP) substrate (Vector Laboratories, CA) for peroxidase. Sections were mounted using DPX (Sigma, USA) and visualized using a Nikon Eclipse Ti-U microscope. Quantification was performed using ImageJ (NIH) thresholding analysis (38).
RNA in situ hybridization.
In situ hybridization for miR-140 was performed as previously described using a 5′-digoxigenin-tagged probe (21, 39). Paraffin-embedded tissue sections were deparaffinized in xylene, rehydrated in ethanol, and washed twice with dH2O. A colorimetric detection reaction was performed using NBT-BCIP (nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate) (Roche, Indianapolis, IN) for 48 h. The slides were stained with nuclear fast red (Sigma, St. Louis, MO), and images were captured using a Nikon Eclipse Ti microscope (Nikon Instruments Inc., Melville, NY).
Masson's trichrome stain.
To determine the collagen deposition in mammary adipose tissue, we performed a modified Masson's staining according to the manufacturer's instructions (ScyTek Laboratories, West Logan, UT; catalog no. TRM-1). Briefly, after tissue rehydration, sections were fixed in Bouin's fluid, stained with Weigert's hematoxylin, and incubated in Biebrich scarlet-acid fuschin. They were then washed in a phosphomolybdic-phophotungstic acid solution and counterstained in aniline blue and acetic acid. Sections were then dehydrated and visualized using a Nikon Eclipse Ti microscope (Nikon Instruments Inc., Melville, NY).
Contraction assay.
A collagen gel contraction assay was set up according to the manufacturer's instructions (CBA-201; Cell Biolabs, San Diego, CA) and as previously described (39). In brief, a cell suspension of 2.0 × 106 cells/ml was obtained. One hundred microliters of the cell suspension was mixed with 400 μl of neutralized collagen solution, added to one well of a 24-well cell culture plate, and allowed to solidify for 1 h at 37°C. After polymerization, 1 ml of complete medium was added to each well, and the cells were incubated for 48 h. The stress was released by running a sterile pipette tip along the sides of the well. The culture dish was then scanned immediately after the stress was released (time zero) and at the other time points that were determined. The area of the collagen gel was then measured using ImageJ software (NIH).
TGF-β1 ELISA.
The protein levels of TGF-β1 in 48-h-conditioned medium from SVF cells from WT-RD, WT-HFD and miR-140 KO mice were assayed using an enzyme-linked immunosorbent assay (ELISA) kit (eBioscience, San Diego, CA; catalog no. EBS-88-8350-22) in accordance with the manufacturer' protocol.
Fluorescence-activated cell sorting (FACS).
Cells were blocked with anti-mouse FcR antibody (CD16/CD32) (BD Biosciences, San Jose, CA; catalog no. 553131) for 15 min at 4°C in PBS with 2% FBS and 1 mM EDTA. Cells were then stained with SCA1-Alexa Fluor 647 (Biolegend, San Diego, CA; catalog no. 108118) and CD49e-Alexa Fluor 488 (Biolegend catalog no. 103810) for 20 min on ice. After antibody incubation, the cells were washed twice in PBS with 2% FBS and 1 mM EDTA, resuspended, and analyzed using a FACSAria II cell sorter (Beckman Coulter, Fullerton, CA).
Western blotting.
Total cell lysates were separated by SDS-PAGE and blotted onto a polyvinylidene difluoride membrane. The membrane was incubated with specific primary antibody overnight at 4°C, followed by the HRP-conjugated secondary antibody, and visualized by a ECL Western blotting detection system (Thermo Scientific, Rockford, IL). Vinculin (Sigma, St. Louis, MO) was used as loading control at a 1:15,000 dilution. Fibronectin (Millipore, Billerica, MA), αSMA (Santa Cruz Biotechnology, Dallas, TX), and SMAD3 (Santa Cruz Biotechnology, Dallas, TX) were used at 1:500, 1:250, and 1:100 dilutions, respectively.
qRT-PCR.
qRT-PCR analysis of miRNA expression was performed as described previously with normalization to U6 small nuclear RNA (40).
Coculture, conditioned-medium treatment, invasion, and migration assays.
Coculture assays were performed using Transwell chambers with a 0.4-μm pore size (Costar, Cambridge MA). A total of 0.5 × 105 SVF cells/ml were seeded in the top well in SVF medium, and 0.5 × 105 cells/ml were plated in the bottom well. Cells were incubated for 48 h before DCIS cells were resuspended for replating in functional assays.
Medium conditioned for 48 h was removed from cell plates and used to treat MCF10A cells plated in chamber well slides. MCF10A cells were treated with conditioned medium for 48 h, after which immunofluorescence staining was performed.
Transwell invasion assays were performed using Transwell migration chambers with an 8-μm pore size (Costar, Cambridge, MA). The top well was coated with 1 mg/ml Matrigel (BD Biosciences) in cell culture medium and incubated for 30 min at 37°C. MCF10DCIS cells (0.5 × 105 cells/ml) were seeded in the upper chamber (1.5 ml). To facilitate invasion, the lower chamber contained 1.5 ml DMEM with 10% FBS. The cells were allowed to migrate toward the 10% FBS gradient overnight. The migrated cells were stained with 1% crystal violet in methanol-PBS and counted using light microscopy.
Transwell migration assays were performed using Transwell migration chambers with an 8-μm pore size (Costar, Cambridge, MA). MCF10DCIS cells (0.5 × 105 cells/ml) were seeded in the upper chamber (1.5 ml). To facilitate migration, the lower chamber contained 1.5 ml DMEM with 10% FBS. The cells were allowed to migrate toward the 10% FBS gradient overnight. The migrated cells were stained with 1% crystal violet in methanol-PBS and counted using light microscopy.
Mammosphere formation.
MCF10A or MCF10DCIS single cells were obtained using 40-μm cell strainers (Fisher Scientific, Pittsburgh, PA) and counted. A total of 10,000 cells/ml were seeded in six-well plates coated with 2% poly(2-hydroxyethyl methacrylate) (poly-HEMA; Sigma St. Louis, MO) in DMEM–F-12 containing 2% B27, 20 ng/ml EGF, 4 μg/ml insulin, and 0.4% BSA. After 7 days of culture, spheres larger than 100 μm were quantified by light microscopy.
Chromatin immunoprecipitation.
The transcription factor binding site was identified through sequence alignment. Chromatin immunoprecipitation (ChIP) was performed as described previously (24). To induce SMAD3 pathway activation, cells were treated for 48 h with 10 ng/ml TGF-β1. Cells were cross-linked with 1% formaldehyde and sonicated. Chromatin was incubated with antibodies against SMAD3 (H300; Santa Cruz Biotechnology, Santa Cruz, CA) at 4°C for immunoprecipitation. Rabbit IgG was used as the negative control. Immunoprecipitated chromatin was analyzed by real-time qPCR. The following primers were used: 5′-CCCCTGGCTTTCCTTCTATG-3′ and 5′-ACAGGACATGCAGCAGGAGT-3′.
DCIS organoid formation and 3D invasion.
DCIS organoid 3D invasion was performed as previously described (41). Ninety-six-well plates were coated with 0.75% agarose and incubated for 30 min at 37°C. DCIS cells were stained with green fluorescent linker PKH67 (catalog no. MINI67-1KT; Sigma, St. Louis, MO) according to the manufacturer's instructions. A total of 10,000 DCIS cells/well were plated in a total volume of 100 μl culture medium. The cells were allowed to aggregate overnight in cell incubators at 5% CO2 and 37°C. SVF cells were stained with red fluorescent linker PKH26 (catalog no. MINI26-1KT; Sigma, St. Louis, MO) according to the manufacturer's instructions. ECM solution was prepared by mixing 100 μl Matrigel (4.5 to 6 mg/ml) with 20,000 SVF cells resuspended in 100 μl plain cell culture medium. The 3D invasion assay was performed in an 8-well chamber slide (Lab-Tek II catalog no. 154534; Thermo Fisher, Waltham, MA). After 100 μl ECM-cell mixture was pipetted into the well, 1 DCIS spheroid was gently embedded in the ECM mixture, followed by the addition of 100 μl ECM-cell mixture. After 30 min of incubation, 100 μl prewarmed medium was added to the well. The plate was returned to the incubator, and invasion was monitored at the desired time points. For fluorescent staining, spheroids were fixed in 4% paraformaldehyde for 20 min at room temperature. After two PBS washes, cell spheroids were permeabilized with 1× PBS–0.5% Tween 20 (PBST) for 15 min at room temperature, followed by two washes with PBST. Spheroids were then blocked in 1× PBS with 10% goat serum for 1 h at room temperature. Primary antibody was diluted in 1× PBS–1% goat serum overnight at 4°C. Spheroids were washed twice, and fluorochrome-conjugated secondary antibody (Life Technologies) was added in 1× PBS–1% goat serum for 1 h at room temperature. After spheroids were washed in PBST three times, Hoechst 33342 stain was added for 10 min. Slides were mounted and visualized with an Olympus IX81 spinning-disk confocal microscope.
Statistical analysis.
Statistical analysis was performed by Student's t test. P values of <0.05 were considered significant. Data are presented as means ± standard errors (SE). Data were analyzed using GraphPad Prism software (version 6.0).
ACKNOWLEDGMENTS
This work was supported by grants from the National Cancer Institute (R01) (CA163820A1 and CA157779A1) (Q.Z.) and the American Cancer Society (RSG-12-006-01-CNE) (Q.Z.).
REFERENCES
- 1.Gimble JM, Guilak F, Nuttall ME, Sathishkumar S, Vidal M, Bunnell BA. 2008. In vitro differentiation potential of mesenchymal stem cells. Transfus Med Hemother 35:228–238. doi: 10.1159/000124281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalluri R, Zeisberg M. 2006. Fibroblasts in cancer. Nat Rev Cancer 6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 3.Chandler EM, Seo BR, Califano JP, Andresen Eguiluz RC, Lee JS, Yoon CJ, Tims DT, Wang JX, Cheng L, Mohanan S, Buckley MR, Cohen I, Nikitin AY, Williams RM, Gourdon D, Reinhart-King CA, Fischbach C. 2012. Implanted adipose progenitor cells as physicochemical regulators of breast cancer. Proc Natl Acad Sci U S A 109:9786–9791. doi: 10.1073/pnas.1121160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kojima Y, Acar A, Eaton EN, Mellody KT, Scheel C, Ben-Porath I, Onder TT, Wang ZC, Richardson AL, Weinberg RA, Orimo A. 2010. Autocrine TGF-beta and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor-promoting mammary stromal myofibroblasts. Proc Natl Acad Sci U S A 107:20009–20014. doi: 10.1073/pnas.1013805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolters PJ, Collard HR, Jones KD. 2014. Pathogenesis of idiopathic pulmonary fibrosis. Annu Rev Pathol 9:157–179. doi: 10.1146/annurev-pathol-012513-104706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yarnold J, Vozenin Brotons M-C. 2010. Pathogenetic mechanisms in radiation fibrosis. Radiother Oncol 97:149–161. doi: 10.1016/j.radonc.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Weigel C, Schmezer P, Plass C, Popanda O. 2015. Epigenetics in radiation-induced fibrosis. Oncogene 34:2145–2155. doi: 10.1038/onc.2014.145. [DOI] [PubMed] [Google Scholar]
- 8.Cutting GR. 2015. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet 16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lijnen PJ, Petrov VV, Fagard RH. 2000. Induction of cardiac fibrosis by transforming growth factor-β1. Mol Genet Metab 71:418–435. doi: 10.1006/mgme.2000.3032. [DOI] [PubMed] [Google Scholar]
- 10.Porter KE, Turner NA. 2009. Cardiac fibroblasts: at the heart of myocardial remodeling. Pharmacol Ther 123:255–278. doi: 10.1016/j.pharmthera.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Sun K-H, Chang Y, Reed NI, Sheppard D. 4 March 2016. αSMA is an inconsistent marker of fibroblasts responsible for force dependent TGFβ activation or collagen production across multiple models of organ fibrosis. Am J Physiol Lung Cell Mol Physiol doi: 10.1152/ajplung.00350.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seo BR, Bhardwaj P, Choi S, Gonzalez J, Andresen Eguiluz RC, Wang K, Mohanan S, Morris PG, Du B, Zhou XK, Vahdat LT, Verma A, Elemento O, Hudis CA, Williams RM, Gourdon D, Dannenberg AJ, Fischbach C. 2015. Obesity-dependent changes in interstitial ECM mechanics promote breast tumorigenesis. Sci Transl Med 7:301ra130. doi: 10.1126/scitranslmed.3010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pessin JE, Kwon H. 2012. How does high-fat diet induce adipose tissue fibrosis? J Investig Med 60:1147–1150. doi: 10.2310/JIM.0b013e318271fdb9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subbaramaiah K, Howe LR, Bhardwaj P, Du B, Gravaghi C, Yantiss RK, Zhou XK, Blaho VA, Hla T, Yang P, Kopelovich L, Hudis CA, Dannenberg AJ. 2011. Obesity is associated with inflammation and elevated aromatase expression in the mouse mammary gland. Cancer Prev Res (Phila) 4:329–346. doi: 10.1158/1940-6207.CAPR-10-0381. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Park J, Scherer PE. 2012. Adipocyte-derived endotrophin promotes malignant tumor progression. J Clin Invest 122:4243–4256. doi: 10.1172/JCI63930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harvey KF, Zhang X, Thomas DM. 2013. The Hippo pathway and human cancer. Nat Rev Cancer 13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 17.Kamikawa A, Ichii O, Yamaji D, Imao T, Suzuki C, Okamatsu-Ogura Y, Terao A, Kon Y, Kimura K. 2009. Diet-induced obesity disrupts ductal development in the mammary glands of nonpregnant mice. Dev Dyn 238:1092–1099. doi: 10.1002/dvdy.21947. [DOI] [PubMed] [Google Scholar]
- 18.Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, Dalmay T. 2006. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett 580:4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 19.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RHA. 2005. MicroRNA expression in zebrafish embryonic development. Science 309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 20.Li Q, Eades G, Yao Y, Zhang Y, Zhou Q. 2014. Characterization of a stem-like subpopulation in basal-like ductal carcinoma in situ (DCIS) lesions. J Biol Chem 289:1303–1312. doi: 10.1074/jbc.M113.502278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q, Yao Y, Eades G, Liu Z, Zhang Y, Zhou Q. 2014. Downregulation of miR-140 promotes cancer stem cell formation in basal-like early stage breast cancer. Oncogene 33:2589–2600. doi: 10.1038/onc.2013.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Eades G, Yao Y, Li Q, Zhou Q. 2012. Estrogen receptor α signaling regulates breast tumor-initiating cells by down-regulating miR-140 which targets the transcription factor SOX2. J Biol Chem 287:41514–41522. doi: 10.1074/jbc.M112.404871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gernapudi R, Yao Y, Zhang Y, Wolfson B, Roy S, Duru N, Eades G, Yang P, Zhou Q. 2015. Targeting exosomes from preadipocytes inhibits preadipocyte to cancer stem cell signaling in early-stage breast cancer. Breast Cancer Res Treat 150:685–695. doi: 10.1007/s10549-015-3326-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gernapudi R, Wolfson B, Zhang Y, Yao Y, Yang P, Asahara H, Zhou Q. 12 October 2015. miR-140 promotes expression of long non-coding RNA NEAT1 in adipogenesis. Mol Cell Biol doi: 10.1128/MCB.00702-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C, Song X, Li Y, Han F, Gao S, Wang X, Xie S, Lv C. 2013. Low-dose paclitaxel ameliorates pulmonary fibrosis by suppressing TGF-β1/Smad3 pathway via miR-140 upregulation. PLoS One 8:e70725-11. doi: 10.1371/journal.pone.0070725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muir LA, Neeley CK, Meyer KA, Baker NA, Brosius AM, Washabaugh AR, Varban OA, Finks JF, Zamarron BF, Flesher CG, Chang JS, DelProposto JB, Geletka L, Martinez-Santibañez G, Kaciroti N, Lumeng CN, O'Rourke RW. 2016. Adipose tissue fibrosis, hypertrophy, and hyperplasia: correlations with diabetes in human obesity. Obesity 24:597–605. doi: 10.1002/oby.21377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyaki S, Sato T, Inoue A, Otsuki S, Ito Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, Takada S, Lotz MK, Ueno-Kudo H, Asahara H. 2010. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev 24:1173–1185. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wynn TA, Ramalingam TR. 2012. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med 18:1028–1040. doi: 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akamatsu T, Arai Y, Kosugi I, Kawasaki H, Meguro S, Sakao M, Shibata K, Suda T, Chida K, Iwashita T. 2013. Direct isolation of myofibroblasts and fibroblasts from bleomycin-injured lungs reveals their functional similarities and differences. Fibrogenesis Tissue Repair 6:15. doi: 10.1186/1755-1536-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cardenas H, Vieth E, Lee J, Segar M, Liu Y, Nephew KP, Matei D. 2014. TGF-β induces global changes in DNA methylation during the epithelial-to-mesenchymal transition in ovarian cancer cells. Epigenetics 9:1461–1472. doi: 10.4161/15592294.2014.971608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tardif G, Pelletier J-P, Fahmi H, Hum D, Zhang Y, Kapoor M, Martel-Pelletier J. 2013. NFAT3 and TGF-β/SMAD3 regulate the expression of miR-140 in osteoarthritis. Arthritis Res Ther 15:R197. doi: 10.1186/ar4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Qin S, Yi C, Ma G, Zhu H, Zhou W, Xiong Y, Zhu X, Wang Y, He L, Guo X. 2011. MiR-140 is co-expressed with Wwp2-C transcript and activated by Sox9 to target Sp1 in maintaining the chondrocyte proliferation. FEBS Lett 585:2992–2997. doi: 10.1016/j.febslet.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 33.Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. 2013. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell 155:1639–1651. doi: 10.1016/j.cell.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. 2007. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrante SC, Nadler EP, Pillai DK, Hubal MJ, Wang Z, Wang JM, Gordish-Dressman H, Koeck E, Sevilla S, Wiles AA, Freishtat RJ. 2015. Adipocyte-derived exosomal miRNAs: a novel mechanism for obesity-related disease. Pediatr Res 77:447–454. doi: 10.1038/pr.2014.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao S, Alarcón C, Sapkota G, Rahman S, Chen P-Y, Goerner N, Macias MJ, Erdjument-Bromage H, Tempst P, Massagué J. 2009. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol Cell 36:457–468. doi: 10.1016/j.molcel.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A. 2014. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle 13:1400–1412. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen EC. 2013. Quantitative analysis of histological staining and fluorescence using ImageJ. Anat Rec (Hoboken) 296:378–381. doi: 10.1002/ar.22641. [DOI] [PubMed] [Google Scholar]
- 39.Duru N, Gernapudi R, Zhang Y, Yao Y, Lo P-K, Wolfson B, Zhou Q. 2015. NRF2/miR-140 signaling confers radioprotection to human lung fibroblasts. Cancer Lett 369:184–191. doi: 10.1016/j.canlet.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eades G, Yao Y, Yang M, Zhang Y, Chumsri S, Zhou Q. 2011. miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J Biol Chem 286:25992–26002. doi: 10.1074/jbc.M111.229401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duru N, Gernapudi R, Lo P-K, Yao Y, Wolfson B, Zhang Y, Zhou Q. 2016. Characterization of the CD49f+/CD44+/CD24− single-cell derived stem cell population in basal-like DCIS cells. Oncotarget 7:47511–47525. [DOI] [PMC free article] [PubMed] [Google Scholar]